Marker-Assisted Pyramiding of Blast-Resistance Genes in a japonica Elite Rice Cultivar through Forward and Background Selection
Abstract
:1. Introduction
2. Results
2.1. Development of Molecular Markers
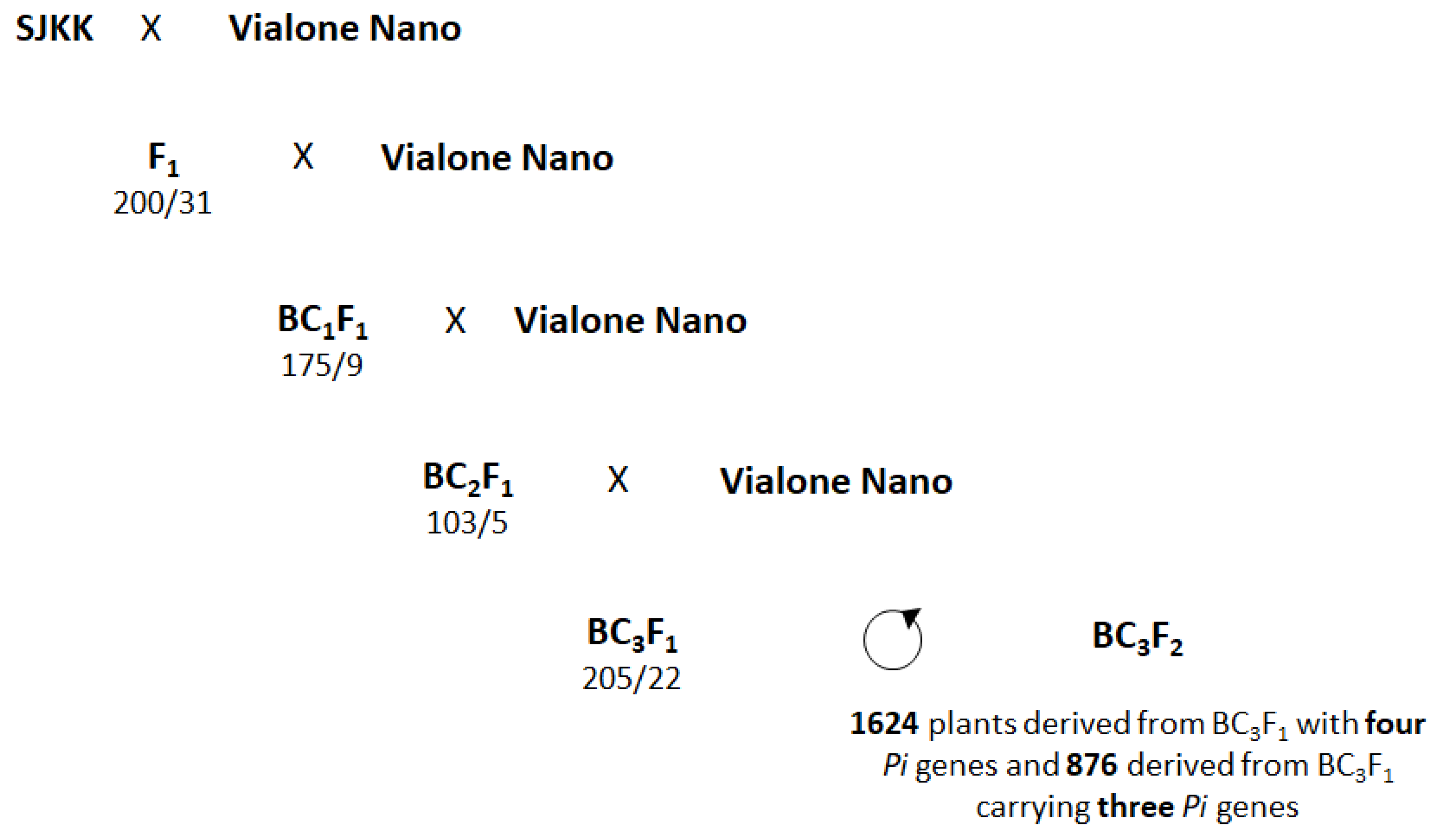
2.2. MABC
2.3. Foreground Selection
2.4. Background Selection
2.5. Phenotyping
2.6. Checking for the Presence of the Pita2/Ptr Gene in the Introgression Lines
3. Discussion
4. Materials and Methods
4.1. Plant Material and DNA Extraction
4.2. Molecular Marker Development and Analysis
4.3. Foreground Selection by KASP Marker Assays
4.4. Background Selection by KASP Marker Assays
4.5. Phenotyping
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
References
- Couch, B.C.; Kohn, L.M. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 2002, 94, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Luo, J.; Rossman, A.Y.; Aoki, T.; Chuma, I.; Crous, P.W.; Dean, R.; de Vries, R.; Donofrio, N.; Hyde, K.D.; et al. Generic names in Magnaporthales. IMA Fungus 2016, 7, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S.; Jena, K.K. Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). In Advances in Genetics, Genomics and Control of Rice Blast Disease; Wang, G.L., Valent, B., Eds.; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar] [CrossRef]
- Pennisi, E. Armed and Dangerous. Science 2010, 327, 5967. [Google Scholar] [CrossRef]
- Faivre-Rampant, O.; Bruschi, G.; Abbruscato, P.; Cavigiolo, S.; Picco, A.M.; Borgo, L.; Lupotto, E.; Piffanelli, P. Assessment of genetic diversity in Italian rice germplasm related to agronomic traits and blast resistance (Magnaporthe oryzae). Mol. Breed. 2011, 27, 233–246. [Google Scholar] [CrossRef]
- Tacconi, G.; Baldassarre, V.; Lanzanova, C.; Faivre-Rampant, O.; Cavigiolo, S.; Urso, S.; Lupotto, E.; Valè, G. Polymorphism analysis of genomic regions associated with broad-spectrum effective blast resistance genes for marker development in rice. Mol. Breed. 2011, 26, 595–617. [Google Scholar] [CrossRef]
- Volante, A.; Tondelli, A.; Desiderio, F.; Abbruscato, P.; Menin, B.; Biselli, C.; Casella, L.; Singh, N.; McCouch, S.R.; Tharreau, D.; et al. Genome wide association studies for japonica rice resistance to blast in field and controlled conditions. Rice 2020, 13, 71. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Miah, G.; Sahebi, M.; Azizi, P.; Tanweer, F.A.; Akhtar, M.S.; Nasehi, A. Molecular Breeding Strategy and Challenges Towards Improvement of Blast Disease Resistance in Rice Crop. Front. Plant Sci. 2015, 6, 886. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Mizukami, Y.; Koga, H.; Yamanouchi, U.; Yoshioka, Y.; Hayashi, N.; Ebana, K.; Mizobuchi, R.; Yano, M. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 2015, 5, 7773. [Google Scholar] [CrossRef] [Green Version]
- Xiao, N.; Wu, Y.; Pan, C.; Yu, L.; Chen, Y.; Liu, G.; Li, Y.; Zhang, X.; Wang, Z.; Dai, Z.; et al. Improving of Rice Blast Resistances in Japonica by Pyramiding Major R Genes. Front. Plant Sci. 2017, 7, 1918. [Google Scholar] [CrossRef]
- Zhu, D.; Kang, H.; Li, Z.; Liu, M.; Zhu, X.; Wang, Y.; Wang, D.; Wang, Z.; Liu, W.; Wang, G.-L. A Genome-Wide Association Study of Field Resistance to Magnaporthe oryzae in Rice. Rice 2016, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.-H.; Ebbole, D.J.; Wang, Z.-H. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integr. Agric. 2017, 16, 2746–2760. [Google Scholar] [CrossRef]
- Sahu, P.K.; Sao, R.; Choudhary, D.K.; Thada, A.; Kumar, V.; Mondal, S.; Das, B.K.; Jankuloski, L.; Sharma, D. Advancement in the Breeding, Biotechnological and Genomic Tools towards Development of Durable Genetic Resistance against the Rice Blast Disease. Plants 2022, 11, 2386. [Google Scholar] [CrossRef]
- Sreewongchai, T.; Toojinda, T.; Thanintorn, N.; Kosawang, C.; Vanavichit, A.; Tharreau, D.; Sirithunya, P. Development of elite indica rice lines with wide spectrum of resistance to Thai blast isolates by pyramiding multiple resistance QTLs. Plant Breed. 2010, 129, 176–180. [Google Scholar] [CrossRef]
- Ashkani, S.; Yusop, M.R.; Shabanimofrad, M.; Azadi, A.; Ghasemzadeh, A.; Azizi, P.; Latif, M.A. Allele Mining Strategies: Principles and Utilisation for Blast Resistance Genes in Rice (Oryza sativa L.). Curr. Issues Mol. Biol. 2015, 17, 57–74. [Google Scholar] [CrossRef]
- Orasen, G.; Greco, R.; Puja, E.; Pozzi, C.; Stile, M.R. Blast resistance R genes pyramiding in temperate japonica rice. Euphytica 2020, 216, 40. [Google Scholar] [CrossRef]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- Werner, K.; Friedt, W.; Ordon, F. Strategies for Pyramiding Resistance Genes Against the Barley Yellow Mosaic Virus Complex (BaMMV, BaYMV, BaYMV-2). Mol. Breed. 2005, 16, 45–55. [Google Scholar] [CrossRef]
- Srivastava, D.; Shamim; Kumar, M.; Mishra, A.; Pandey, P.; Kumar, D.; Yadav, P.; Siddiqui, M.H.; Singh, K.N. Current Status of Conventional and Molecular Interventions for Blast Resistance in Rice. Rice Sci. 2017, 24, 299–321. [Google Scholar] [CrossRef]
- Allard, R.W. Principles of Plant Breeding; John Wiley & Sons Inc.: New York, NY, USA, 1960. [Google Scholar]
- Hasan, M.M.; Rafii, M.Y.; Ismail, M.R.; Mahmood, M.; Rahim, H.A.; Alam, M.A.; Ashkani, S.; Malek, M.A.; Latif, M.A. Marker-assisted backcrossing: A useful method for rice improvement. Biotechnol. Biotechnol. Equip. 2015, 29, 237–254. [Google Scholar] [CrossRef]
- Francia, E.; Tacconi, G.; Crosatti, C.; Barabaschi, D.; Bulgarelli, D.; Dall’Aglio, E.; Valè, G. Marker assisted selection in crop plants. Plant Cell Tissue Organ Cult. (PCTOC) 2005, 82, 317–342. [Google Scholar] [CrossRef]
- Reyes, V.P.; Angeles-Shim, R.B.; Mendioro, M.S.; Manuel, M.C.C.; Lapis, R.S.; Shim, J.; Sunohara, H.; Nishiuchi, S.; Kikuta, M.; Makihara, D.; et al. Marker-Assisted Introgression and Stacking of Major QTLs Controlling Grain Number (Gn1a) and Number of Primary Branching (WFP) to NERICA Cultivars. Plants 2021, 10, 844. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Shim, R.B.; Reyes, V.P.; del Valle, M.M.; Lapis, R.S.; Shim, J.; Sunohara, H.; Jena, K.K.; Ashikari, M.; Doi, K. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast. Rice Sci. 2020, 27, 113–123. [Google Scholar] [CrossRef]
- Reyes, V.P.; Angeles-Shim, R.B.; Lapis, R.S.; Shim, J.; Sunohara, H.; Jena, K.K.; Ashikari, M.; Doi, K. Improvement of Asian Rice Cultivars through Marker-Assisted Introgression of Yield QTLs Grain Number 1a (Gn1a) and Wealthy Farmer’s Panicle (WFP). Philipp. J. Biochem. Mol. Biol. 2021, 2, 29. [Google Scholar] [CrossRef]
- Hospital, F.; Charcosset, A. Marker-Assisted Introgression of Quantitative Trait Loci. Genetics 1997, 147, 1469–1485. [Google Scholar] [CrossRef]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.; Matsumoto, T.; Ono, K.; Yano, M. Two Adjacent Nucleotide-Binding Site–Leucine-Rich Repeat Class Genes Are Required to Confer Pikm-Specific Rice Blast Resistance. Genetics 2008, 180, 2267–2276. [Google Scholar] [CrossRef]
- De la Concepcion, J.C.; Franceschetti, M.; Maqbool, A.; Saitoh, H.; Terauchi, R.; Kamoun, S.; Banfield, M.J. Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat. Plants 2018, 4, 576–585. [Google Scholar] [CrossRef]
- Maqbool, A.; Saitoh, H.; Franceschetti, M.; Stevenson, C.; Uemura, A.; Kanzaki, H.; Kamoun, S.; Terauchi, R.; Banfield, M. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 2015, 4, e08709. [Google Scholar] [CrossRef]
- Costanzo, S.; Jia, Y. Sequence variation at the rice blast resistance gene Pikm locus: Implications for the development of allele specific markers. Plant Sci. 2010, 178, 523–530. [Google Scholar] [CrossRef]
- Wang, Z.-X.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999, 19, 55–64. [Google Scholar] [CrossRef]
- Hayashi, K.; Hashimoto, N.; Daigen, M.; Ashikawa, I. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor. Appl. Genet. 2004, 108, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.T.; Wu, K.-S.; Farrall, L.; Jia, Y.; Hershey, H.P.; McAdams, S.A.; Faulk, K.N.; Donaldson, G.K.; Tarchini, R.; Valent, B. A Single Amino Acid Difference Distinguishes Resistant and Susceptible Alleles of the Rice Blast Resistance Gene Pi-ta. Plant Cell 2000, 12, 2033–2045. [Google Scholar] [CrossRef]
- Hayashi, K.; Yoshida, H.; Ashikawa, I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 2006, 113, 251–260. [Google Scholar] [CrossRef]
- Zhou, B.; Qu, S.; Liu, G.; Dolan, M.; Sakai, H.; Lu, G.; Bellizzi, M.; Wang, G.-L. The Eight Amino-Acid Differences Within Three Leucine-Rich Repeats Between Pi2 and Piz-t Resistance Proteins Determine the Resistance Specificity to Magnaporthe grisea. Mol. Plant-Microbe Interact. 2006, 19, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Xiao, G.; Telebanco-Yanoria, M.J.; Siazon, P.M.; Padilla, J.; Opulencia, R.; Bigirimana, J.; Habarugira, G.; Wu, J.; Li, M.; et al. The broad-spectrum rice blast resistance (R) gene Pita2 encodes a novel R protein unique from Pita. Rice 2020, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, X.; Jia, Y.; Minkenberg, B.; Wheatley, M.; Fan, J.; Jia, M.H.; Famoso, A.; Edwards, J.D.; Wamishe, Y.; et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Tanweer, F.A.; Rafii, M.Y.; Sijam, K.; Rahim, H.A.; Ahmed, F.; Latif, M.A. Current advance methods for the identification of blast resistance genes in rice. Comptes Rendus Biol. 2015, 338, 321–334. [Google Scholar] [CrossRef]
- Herzog, E.; Frisch, M. Selection strategies for marker-assisted backcrossing with high-throughput marker systems. Theor. Appl. Genet. 2011, 123, 251–260. [Google Scholar] [CrossRef]
- Rafalski, A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002, 5, 94–100. [Google Scholar] [CrossRef]
- Schlötterer, C. The evolution of molecular markers—Just a matter of fashion? Nat. Rev. Genet. 2004, 5, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2013, 33, 1–14. [Google Scholar] [CrossRef]
- Biscarini, F.; Cozzi, P.; Casella, L.; Riccardi, P.; Vattari, A.; Orasen, G.; Perrini, R.; Tacconi, G.; Tondelli, A.; Biselli, C.; et al. Genome-Wide Association Study for Traits Related to Plant and Grain Morphology, and Root Architecture in Temperate Rice Accessions. PLoS ONE 2016, 11, e0155425. [Google Scholar] [CrossRef]
- Volante, A.; Desiderio, F.; Tondelli, A.; Perrini, R.; Orasen, G.; Biselli, C.; Riccardi, P.; Vattari, A.; Cavalluzzo, D.; Urso, S.; et al. Genome-Wide Analysis of japonica Rice Performance under Limited Water and Permanent Flooding Conditions. Front. Plant Sci. 2017, 8, 1862. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- A Mather, K.; Caicedo, A.L.; Polato, N.R.; Olsen, K.M.; McCouch, S.; Purugganan, M.D. The Extent of Linkage Disequilibrium in Rice (Oryza sativa L.). Genetics 2007, 177, 2223–2232. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, A.; Mithra, S.V.A.; Krishnamurthy, S.L.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S.K.; et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, X.; Ge, S.; Jensen, J.D.; Hu, F.; Dong, Y.; Gutenkunst, R.N.; Fang, L.; Huang, L.; Li, J.; et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2011, 30, 105–111. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Islam, K.; Latif, M.A. A Review of Microsatellite Markers and Their Applications in Rice Breeding Programs to Improve Blast Disease Resistance. Int. J. Mol. Sci. 2013, 14, 22499–22528. [Google Scholar] [CrossRef] [Green Version]
- Cruz, C.M.V.; Bai, J.; Oña, I.; Leung, H.; Nelson, R.J.; Mew, T.-W.; Leach, J.E. Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. USA 2000, 97, 13500–13505. [Google Scholar] [CrossRef]
- Hittalmani, S.; Parco, A.; Mew, T.V.; Zeigler, R.S.; Huang, N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000, 100, 1121–1128. [Google Scholar] [CrossRef]
- Chen, Z.; Guan, H.; Wang, X.; Dong, R.; Zhuo, C.; Mao, D.; Pan, R.; Zhou, Y.; Wu, W. Pyramiding of 3-resistant-gene to improve rice blast resistance of a restorer line, Fuhui 673. Sheng Wu Gong Cheng Xue Bao 2019, 35, 837–846. [Google Scholar] [PubMed]
- Chen, Y.-C.; Hu, C.-C.; Chang, F.-Y.; Chen, C.-Y.; Chen, W.-L.; Tung, C.-W.; Shen, W.-C.; Wu, C.-W.; Cheng, A.-S.; Liao, D.-J.; et al. Marker-Assisted Development and Evaluation of Monogenic Lines of Rice cv. Kaohsiung 145 Carrying Blast Resistance Genes. Plant Dis. 2021, 105, 3858–3868. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Li, Z.; Liu, J.; Shen, Z.; Gao, G.; Zhang, Q.; He, Y. Development and evaluation of improved lines with broad-spectrum resistance to rice blast using nine resistance genes. Rice 2019, 12, 29. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, N.; Chen, Y.; Yu, L.; Pan, C.; Li, Y.; Zhang, X.; Huang, N.; Ji, H.; Dai, Z.; et al. Comprehensive evaluation of resistance effects of pyramiding lines with different broad-spectrum resistance genes against Magnaporthe oryzae in rice (Oryza sativa L.). Rice 2019, 12, 11. [Google Scholar] [CrossRef]
- McClung, A.M.; Marchetti, M.A.; Webb, B.D.; Bollich, C.N. Registration of ‘Jefferson’ rice. Crop Sci. 1997, 37, 629–630. [Google Scholar]
- Urso, S.; Desiderio, F.; Biselli, C.; Bagnaresi, P.; Crispino, L.; Piffanelli, P.; Abbruscato, P.; Assenza, F.; Guarnieri, G.; Cattivelli, L.; et al. Genetic analysis of durable resistance to Magnaporthe oryzae in the rice accession Gigante Vercelli identified two blast resistance loci. Mol. Genet. Genom. 2015, 291, 17–32. [Google Scholar] [CrossRef]
- Devanna, B.N.; Jain, P.; Solanke, A.U.; Das, A.; Thakur, S.; Singh, P.K.; Kumari, M.; Dubey, H.; Jaswal, R.; Pawar, D.; et al. Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions. J. Fungi 2022, 8, 584. [Google Scholar] [CrossRef]
- Risterucci, A.M.; Grivet, L.; N’Goran, J.A.K.; Pieretti, I.; Flament, M.H.; Lanaud, C. A high-density linkage map of Theobroma cacao L. Theor. Appl. Genet. 2000, 101, 948–955. [Google Scholar] [CrossRef]
- Gallet, R.; Fontaine, C.; Bonnot, F.; Milazzo, J.; Tertois, C.; Adreit, H.; Ravigné, V.; Fournier, E.; Tharreau, D. Evolution of Compatibility Range in the Rice−Magnaporthe oryzae System: An Uneven Distribution of R Genes Between Rice Subspecies. Phytopathology 2016, 106, 348–354. [Google Scholar] [CrossRef] [Green Version]
- Kiyosawa, S. The inheritance of resistance of the zenith type varieties of rice to the blast fungus. Jpn. J. Breed. 1967, 17, 99–107. [Google Scholar] [CrossRef]
- Biselli, C.; Cavalluzzo, D.; Perrini, R.; Gianinetti, A.; Bagnaresi, P.; Urso, S.; Orasen, G.; Desiderio, F.; Lupotto, E.; Cattivelli, L.; et al. Improvement of marker-based predictability of Apparent Amylose Content in japonica rice through GBSSI allele mining. Rice 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Primer Name | Sequence | Ta (°C) | Polymorphism Type | RE | Gene | Reference | Application in: | |
|---|---|---|---|---|---|---|---|---|
| MABC | KASP Marker /Sequencing | |||||||
| Pib5 f Pib5 r | CCTACTGCTCTCGCTCCGAATTCC CAGAATTTTGTCAGGAACCTGCC | 58 | InDel and SNPs | - | Pib | [7] | x | |
| Pib3 f Pib3 r | AGTAGTATCTCCCTACTCACACGACAC GGATGACCTGAACTGAAACTCACAGT | 58 | dCAPS | DdeI | Pib | [7] | x | |
| Z6050 f Z6050 r | CCCGAGCACTTGCAGATCTTGGGCGAAGCA ATCTCTCCGGCACGACCGAAGC | 60 | dCAPS | SphI/PaeI | Piz | [7,33] | x | |
| Z56592 f Z56592 r | GGACCCGCGTTTTCCACGTGTAA AGGAATCTATTGCTAAGCATGAC | 60 | SNPs | - | Piz | [36] | x | |
| Pik2-2AE f Pik2-2AE r | TCCTTAGCCCTGTCAACTGA TGCTGGATCTTGAGGACTGG | 53 | SNPs | - | Pik2 | This work | x | |
| Pik1-10 f Pik1-10 r | ACTGTAGTGCATACCATTGG GAGTGCTCCCCACATACAA | 50 | Pres/abs | - | Pik1 | This work | x | |
| Pita-10 f Pita-10 r | TATCTTGCAAATGCGTCCG GCCAAGAAGATGATCAGCA | 60 | CAPS | HincII | Pita | This work | x | x |
| Pita2/Ptr -5 f Pita2/Ptr-5 r | TTGCTTGTTCGGATCAGTGC TGTTGCTGATCGTCTTGCTG | 57 | SNPs | Pita2/Ptr | This work | x | ||
| Amplicon | Donor Parent Sequence SJKK | Recurrent Parent Sequence Vialone Nano |
|---|---|---|
| Pib5 | TCAAGAGAATTTGAGAAGCAAGAGAGGC TCTGATACCAGATTGTCAGGATCTCAAGAA ATCARCAANGARMAACAAGAACACACAA GGATTCAGGCAACTAGTTTGGATTGATCTG CTCCAACCCAACAGG | TCAAGAGAATTTGAGAAGCAAGAGAGGC TCTGATACCAGATTGTCAGGATCTCAAGAA ATTARCAANGARMAACAAGAACACACAA GGATTCAGGCAACTAGTTTGGATTGATCTG CTCCAACCCAACAGG |
| Z56592 | CGATGTTCGAGAGCCCATGGATGTTTAGTT GTTTAGACATGGTGTTGGACGGTCGAATGG TGGGCCTGTTGTAGGTATGGTGGCATCTGG CAACCAGTCAT | CGATGTTCGAGAGCCCATGGATGTTTAGTT GTTTAGACATGGTGTTGGACAGTCGAATGG TGGGCCTGTTGTAGGTATGGTGGCATCTGG CAACCAGTCAT |
| Pik2-2AE | CCTTGAGGYGACGGGTTTTTMATTGSCTTMTT WCTTTTTTCTTGAGGCAACCTCAGCCCCTTCC GTGTGTTTTTRTTCCCYCCAAGTATGCTAMTG ATCCGTTTTAGCTGGMCTAYWGACTTAGGCA GSTCCYYGACATATGTCTCCCTTATGT | CCTTGAGGYGACGGGTTTTTMATTGSCTTMTT WCTTTTTTCTTGAGGCAACCTCAGCCCCTTCC GTGTATTTTTRTTCCCYCCAAGTATGCTAMTG ATCCGTTTTAGCTGGMCTAYWGACTTAGGCA GSTCCYYGACATATGTCTCCCTTATGT |
| Pita-10 | GTCAGCGACAGAAACCGGCGGCGTTCGTTG CCGGCGGAGTCCTCGCGATCGTCGTCGTCGT CTTCTTCTCTCGGCCTCGAGCTCGAGGTGCG CCTGCCAAGATGGTAGCTC | GTCAGCGACAGAAACCGGCGGCGTTCGTTG CCGGCGGAGTCCTCGCGATCGTCGTCGTCGA CTTCTTCTCTCGGCCTCGAGCTCGAGGTGCG CCTGCCAAGATGGTAGCTC |
| Pita2/Ptr | TATACACAGTAGACAATATTGGATTGAGTTT CTGATGAACACAGTAGGAATTCTTCTCAATA CGGTGTGTGCGTGTAGAGAATAATCAACAAT GGTACGTCGTGCTGCTTGCCTCGGCGCACGG CAACA | TATACACAGTAGACAATATTGGATTGAGTTT CTGATGAACACAGTAGGAATTCTTCTCGATA CGGTGTGTGCGTGTAGAGAATAATCAACAAT GGTACGTCGTGCTGCTTGCCTCGGCGCACGG CAACA |
| Locus | Number of Plants | |||
|---|---|---|---|---|
| Piz | Pib | Pik | Pita | |
| + | + | + | + | 8 |
| + | + | + | - | 5 |
| + | + | - | + | 27 |
| - | + | + | + | 40 |
| + | - | + | + | 10 |
| - | + | - | + | 147 |
| + | - | - | + | 45 |
| - | + | + | - | 44 |
| - | - | + | + | 65 |
| + | - | + | - | 51 |
| + | + | - | - | 19 |
| - | - | - | + | 241 |
| - | + | - | - | 139 |
| + | - | - | - | 176 |
| - | - | + | - | 283 |
| Variety | Pi Gene(s) | Phenotypic Scores | |||
|---|---|---|---|---|---|
| BN0013 | BN0040 | NG0190 | TG0015 | ||
| Saber | Pib | R | R | R | R |
| BL1 | Pib | S | S | R | S |
| Katy | Pita and Pita2/Ptr | R | R | R | R |
| K1 | Pita | S | S | S | R |
| Kusabue | Pik | R | R | R | R |
| Kanto 51 | Pik | R | S | S | S |
| Jefferson | Piz | R | R | R | R |
| Zenith | Piz | S | R | S | S |
| Vialone Nano | - | S | S | S | S |
| Maratelli | - | S | S | S | S |
| 154/05/3/154/A | Piz + Pib + Pik + Pita + Pita2 | R | R | R | R |
| 154/06/04/127 | Piz + Pib + Pik + Pita+ Pita2 | R | R | R | R |
| 154/05/01/219/C | Piz + Pib + Pik | R | R | R | R |
| 154/05/01/185 | Piz + Pib + Pita+ Pita2 | R | R | R | ND |
| 154/05/11/135 | Piz + Pik + Pita+ Pita2 | R | R | R | R |
| 154/05/11/65 | Piz + Pib + Pik | ND | R | R | ND |
| 154/16/70/310 | Pib + Pik + Pita+ Pita2 | R | R | R | ND |
| Parameters | Line 154/06/04/127 | Vialone Nano |
|---|---|---|
| Amylose (% d.m.) | 25.90 | 26.80 |
| Grain length, L—milled (mm) | 5.57 | 5.51 |
| Grain width, W—milled (mm) | 3.13 | 3.21 |
| L/W (milled) | 1.78 | 1.72 |
| Grain length, L—hulled (mm) | 6.09 | 6.12 |
| Grain width, W—hulled (mm) | 3.30 | 3.56 |
| L/W (hulled) | 1.85 | 1.72 |
| Thousand kernel weight (g) | 37 | 36 |
| Sowing–flowering (days) | 102 | 103 |
| Grain yield (t/ha) | 9.1 | 5.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zampieri, E.; Volante, A.; Marè, C.; Orasen, G.; Desiderio, F.; Biselli, C.; Canella, M.; Carmagnola, L.; Milazzo, J.; Adreit, H.; et al. Marker-Assisted Pyramiding of Blast-Resistance Genes in a japonica Elite Rice Cultivar through Forward and Background Selection. Plants 2023, 12, 757. https://doi.org/10.3390/plants12040757
Zampieri E, Volante A, Marè C, Orasen G, Desiderio F, Biselli C, Canella M, Carmagnola L, Milazzo J, Adreit H, et al. Marker-Assisted Pyramiding of Blast-Resistance Genes in a japonica Elite Rice Cultivar through Forward and Background Selection. Plants. 2023; 12(4):757. https://doi.org/10.3390/plants12040757
Chicago/Turabian StyleZampieri, Elisa, Andrea Volante, Caterina Marè, Gabriele Orasen, Francesca Desiderio, Chiara Biselli, Marco Canella, Lorena Carmagnola, Joëlle Milazzo, Henri Adreit, and et al. 2023. "Marker-Assisted Pyramiding of Blast-Resistance Genes in a japonica Elite Rice Cultivar through Forward and Background Selection" Plants 12, no. 4: 757. https://doi.org/10.3390/plants12040757
APA StyleZampieri, E., Volante, A., Marè, C., Orasen, G., Desiderio, F., Biselli, C., Canella, M., Carmagnola, L., Milazzo, J., Adreit, H., Tharreau, D., Poncelet, N., Vaccino, P., & Valè, G. (2023). Marker-Assisted Pyramiding of Blast-Resistance Genes in a japonica Elite Rice Cultivar through Forward and Background Selection. Plants, 12(4), 757. https://doi.org/10.3390/plants12040757






