Improvements for Tissue-Chopping-Based Immunofluorescence Staining Method of Chloroplast Proteins
Abstract
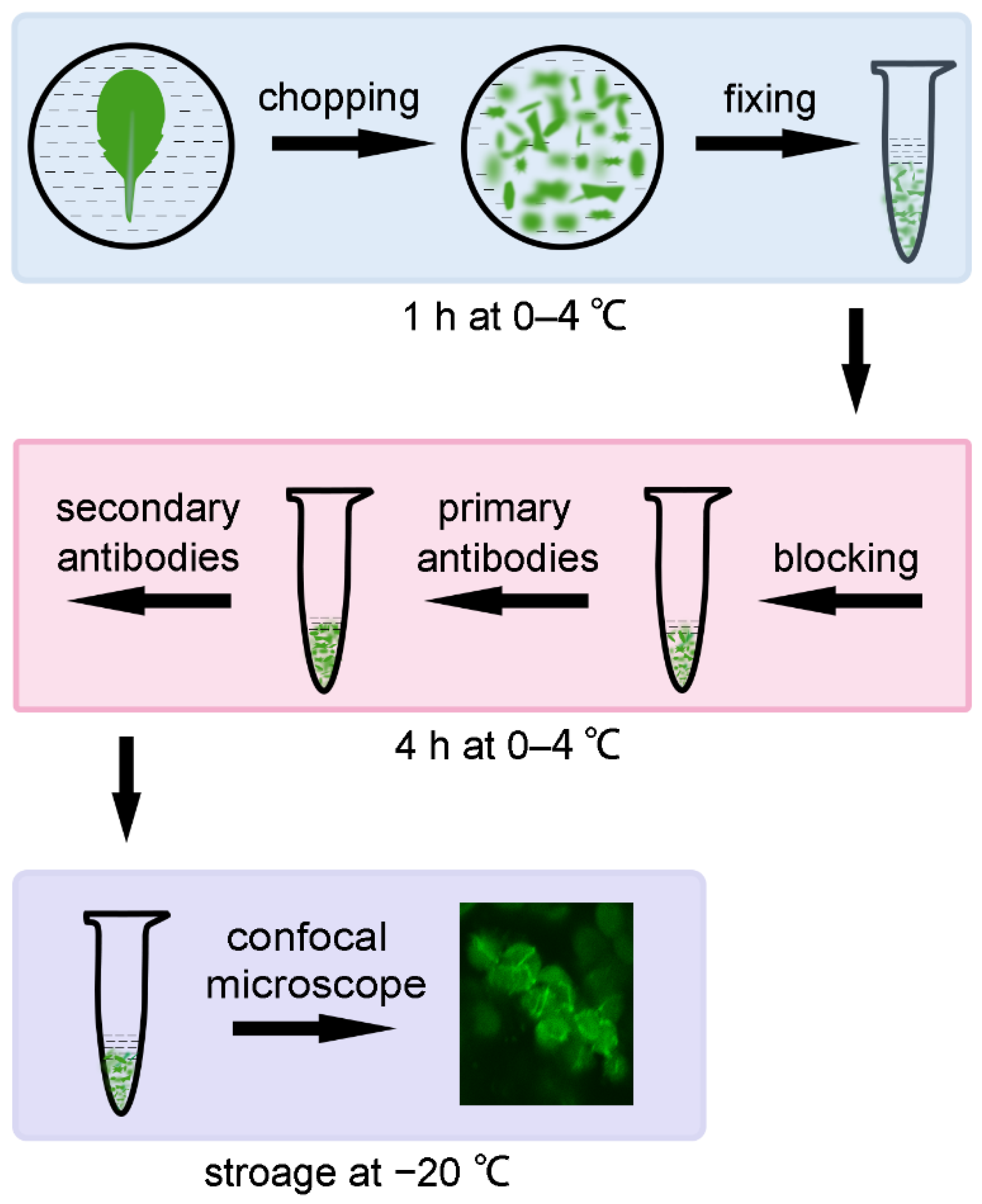
:1. Introduction
2. Results
2.1. Direct Observation with a Confocal Microscope Can Shorten the Experimental Time
2.2. Low Temperature Can Reduce Chlorophyll Autofluorescence and the Background Noise Signal
2.3. Storage at −20 °C Can Effectively Prolong the Lifetime of the Sample
2.4. The Improved Immunofluorescence Staining Method Can Be Widely Used in Other Species
3. Discussion
4. Material and Methods
4.1. Plant Materials and Growth Conditions
4.2. Tissue Breaking and Fixing of Leaves
4.3. Immunofluorescence Staining
4.4. Storage of Samples
4.5. Confocal Microscopy and Image Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Osteryoung, K.W.; Nunnari, J. The division of endosymbiotic organelles. Science 2003, 302, 1698–1704. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; MacCready, J.S.; Ducat, D.C.; Osteryoung, K.W. The Molecular Machinery of Chloroplast Division. Plant Physiol. 2018, 176, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Pyke, K.A. Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 2014, 65, 443–472. [Google Scholar] [CrossRef] [PubMed]
- Stokes, K.D.; Osteryoung, K.W. Early divergence of the FtsZ1 and FtsZ2 plastid division gene families in photosynthetic eukaryotes. Gene 2003, 320, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; McAndrew, R.S. The Plastid Division Machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 315–333. [Google Scholar] [CrossRef] [PubMed]
- Vitha, S.; Froehlich, J.E.; Koksharova, O.; Pyke, K.A.; van Erp, H.; Osteryoung, K.W. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 2003, 15, 1918–1933. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Froehlich, J.E.; Osteryoung, K.W. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 2006, 18, 2517–2530. [Google Scholar] [CrossRef] [PubMed]
- Vitha, S.; McAndrew, R.S.; Osteryoung, K.W. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 2001, 153, 111–120. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Q.; Feng, Y.; Liu, X.; Gao, H. An improved immunofluorescence staining method for chloroplast proteins. Plant Cell Rep. 2016, 35, 2285–2293. [Google Scholar] [CrossRef]
- Wang, W.; Li, J.; Sun, Q.; Yu, X.; Zhang, W.; Jia, N.; An, C.; Li, Y.; Dong, Y.; Han, F.; et al. Structural insights into the coordination of plastid division by the ARC6-PDV2 complex. Nat. Plants 2017, 3, 17011. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; An, J.; Wang, L.; Sun, Q.; An, C.; Wu, B.; Hong, C.; Wang, X.; Dong, S.; Guo, J.; et al. A novel amphiphilic motif at the C-terminus of FtsZ1 facilitates chloroplast division. Plant Cell 2022, 34, 419–432. [Google Scholar] [CrossRef]
- Wang, L.; Tang, M.; Huang, W.; An, J.; Liu, X.; Gao, H. A Tissue-Chopping Based Immunofluorescence Staining Method for Chloroplast Proteins. Front. Plant Sci. 2022, 13, 910569. [Google Scholar] [CrossRef] [PubMed]
- Hanson, D.A.; Ziegler, S.F. Fusion of green fluorescent protein to the C-terminus of granulysin alters its intracellular localization in comparison to the native molecule. J. Negat. Results Biomed. 2004, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Ovečka, M.; Samajová, O.; Baluška, F.; Samaj, J. Immunofluorescent localization of MAPKs in Steedman’s wax sections. Methods Mol. Biol. 2014, 1171, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Schmitz, A.J.; Kadirjan-Kalbach, D.K.; Terbush, A.D.; Osteryoung, K.W. Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in arabidopsis through interaction with FtsZ2. Plant Cell 2013, 25, 1787–1802. [Google Scholar] [CrossRef] [PubMed]
- Pyke, K.A.; Leech, R.M. A Genetic Analysis of Chloroplast Division and Expansion in Arabidopsis thaliana. Plant Physiol. 1994, 104, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Kadirjan-Kalbach, D.; Froehlich, J.E.; Osteryoung, K.W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 2003, 100, 4328–4333. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| A List of the Plant Species with Signals |
|---|
| Sophora japonica Sophora japonica ‘Oligophylla’ Styphnolobium japonicum ‘Pendula’ Yulania denudata Lonicera maackii (Rupr.) Maxim. Viburnum opulus subsp. calvescens (Rehder) Sugimoto Kolkwitzia amabilis Graebn. Jasminum nudiflorum Lindl. Syringa reticulata subsp. amurensis (Ruprecht) P. S. Green and M. C. Chang Aesculus chinensis Bunge Koelreuteria paniculata Laxm. Kerria japonica (L.) DC. Amygdalus persica ‘Atropurpurea’ Prunus triloba ‘Multiplex’ Diospyros lotus Sambucus williamsii Hance Hibiscus syriacus L. Campsis grandiflora (Thunb.) Schum. Parthenocissus quinquefolia Solanum lycopersicum L. Scadoxus pole-evansii (Oberm.) Friis and Nordal Chenopodium album L. Taraxacum mongolicum Hand.-Mazz. Erigeron canadensis L. Inula japonica Thunb. Lactuca indica L. Chrysanthemum × morifolium Lythrum salicaria L. Viola philippica Cav. Orychophragmus violaceus (Linnaeus) O. E. Schulz Hosta plantaginea Sagittaria trifolia subsp. leucopetala (Miquel) Q. F. Wang ‘Florepleno’ Alisma plantago-aquatica L. Epipremnum aureum (Linden et Andre) Bunting Lemna minor L. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Chen, Y.; Niu, D.; Tang, M.; An, J.; Xue, S.; Liu, X.; Gao, H. Improvements for Tissue-Chopping-Based Immunofluorescence Staining Method of Chloroplast Proteins. Plants 2023, 12, 841. https://doi.org/10.3390/plants12040841
Wang L, Chen Y, Niu D, Tang M, An J, Xue S, Liu X, Gao H. Improvements for Tissue-Chopping-Based Immunofluorescence Staining Method of Chloroplast Proteins. Plants. 2023; 12(4):841. https://doi.org/10.3390/plants12040841
Chicago/Turabian StyleWang, Lulu, Yajuan Chen, Di Niu, Mingdong Tang, Jinjie An, Shanshan Xue, Xiaomin Liu, and Hongbo Gao. 2023. "Improvements for Tissue-Chopping-Based Immunofluorescence Staining Method of Chloroplast Proteins" Plants 12, no. 4: 841. https://doi.org/10.3390/plants12040841







