Ectopic Expression of Perilla frutescens WRI1 Enhanced Storage Oil Accumulation in Nicotiana benthamiana Leaves
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Isolation and Subcellular Localization of PfWRI1A and PfWRI1B
2.3. Protein Sequence Alignment and Phylogenetic Tree Analysis of WRI1 Orthologs from Various Plant Species
2.4. RT-PCR and RT-qPCR Analyses
2.5. Transient Expression of PfWRI1A and PfWRI1B in N. benthamiana Leaves and Nile Red Staining
2.6. TLC and GC Analyses
3. Results
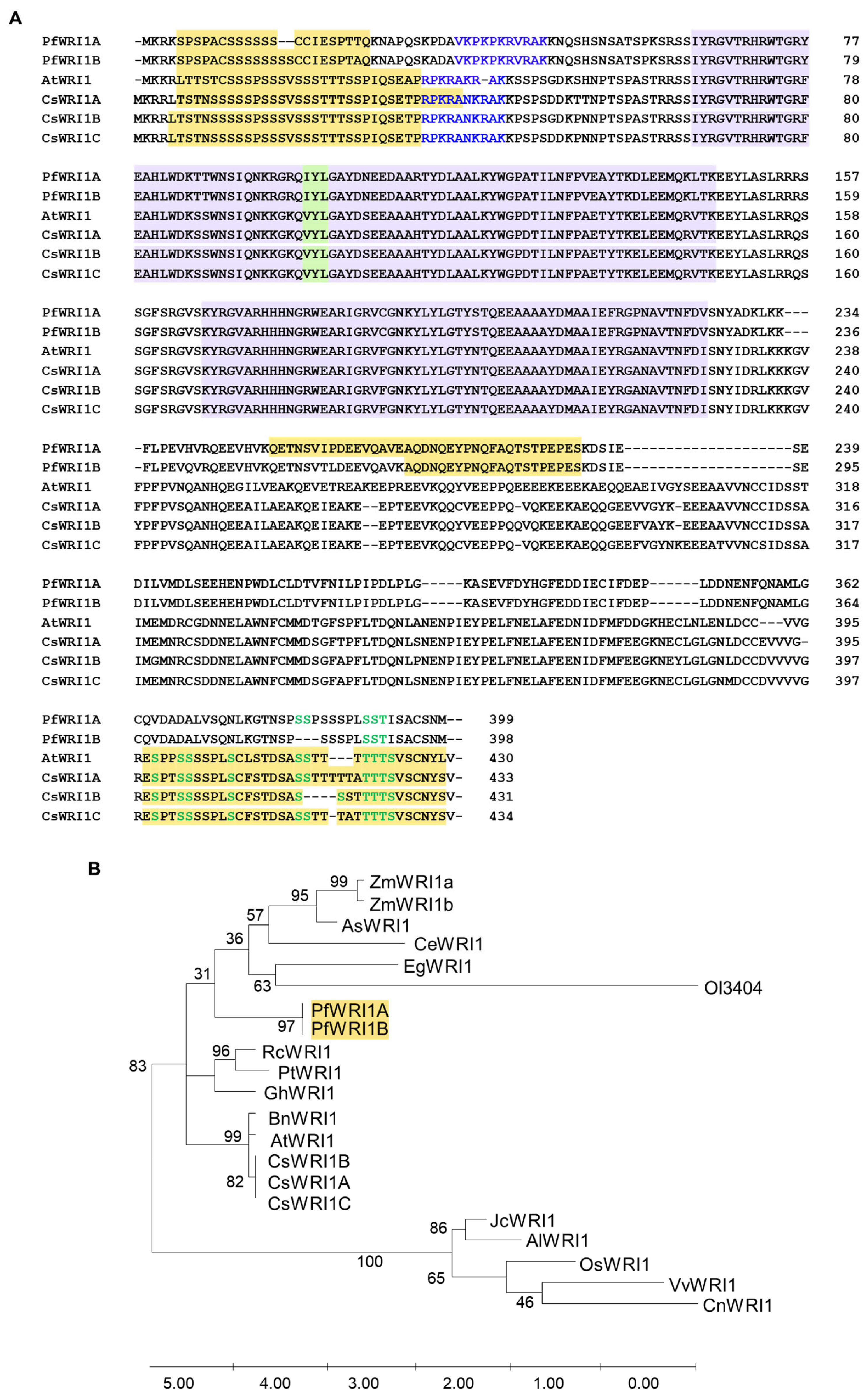
3.1. Nucleotide and Amino Acid Sequences of P. frutescens WRI1 Isoforms and Phylogenetic Tree in WRI1 Orthologs from Various Plant Resources
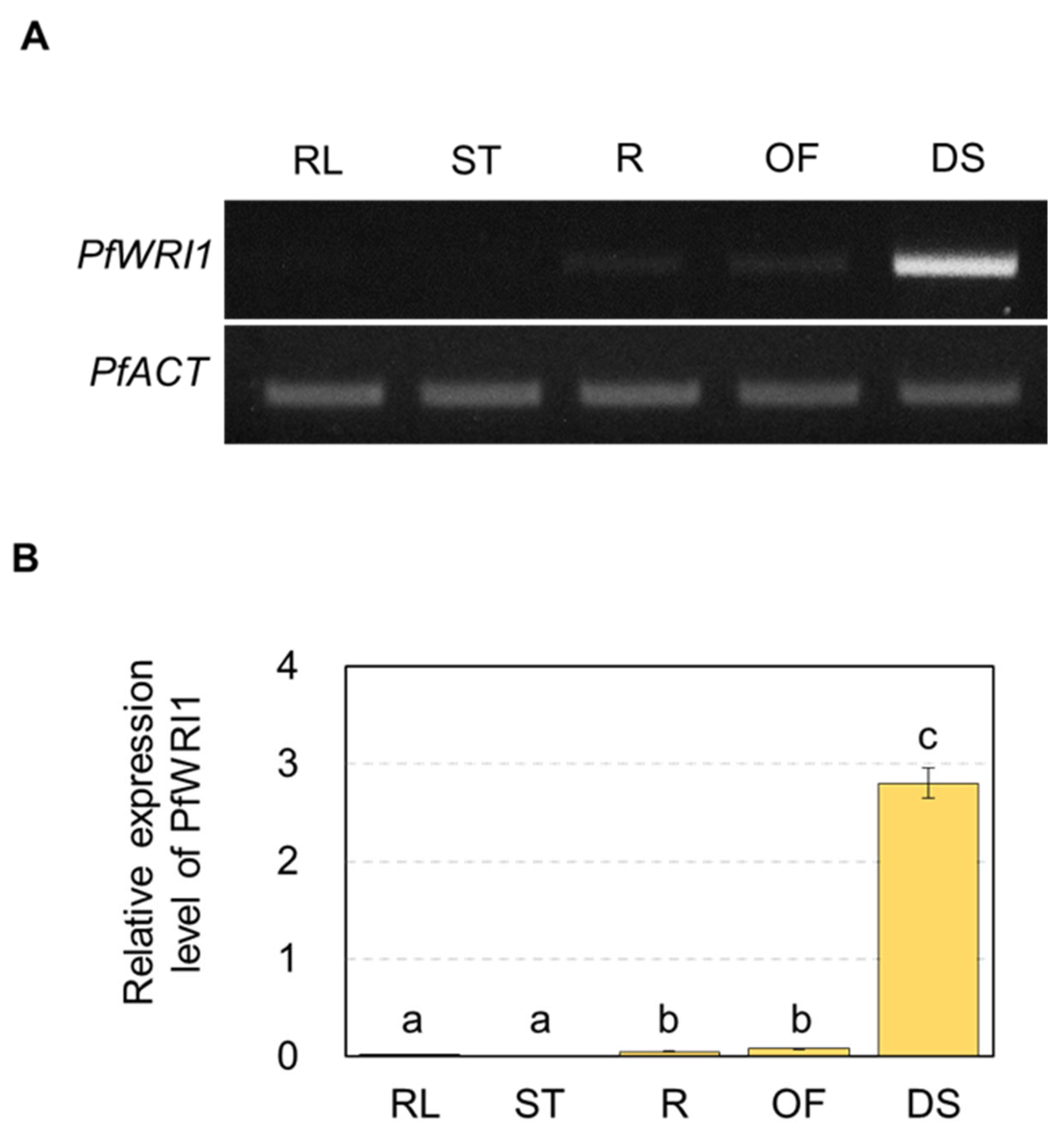
3.2. Expression of PfWRI1A and PfWRI1B in Various Perilla Organs
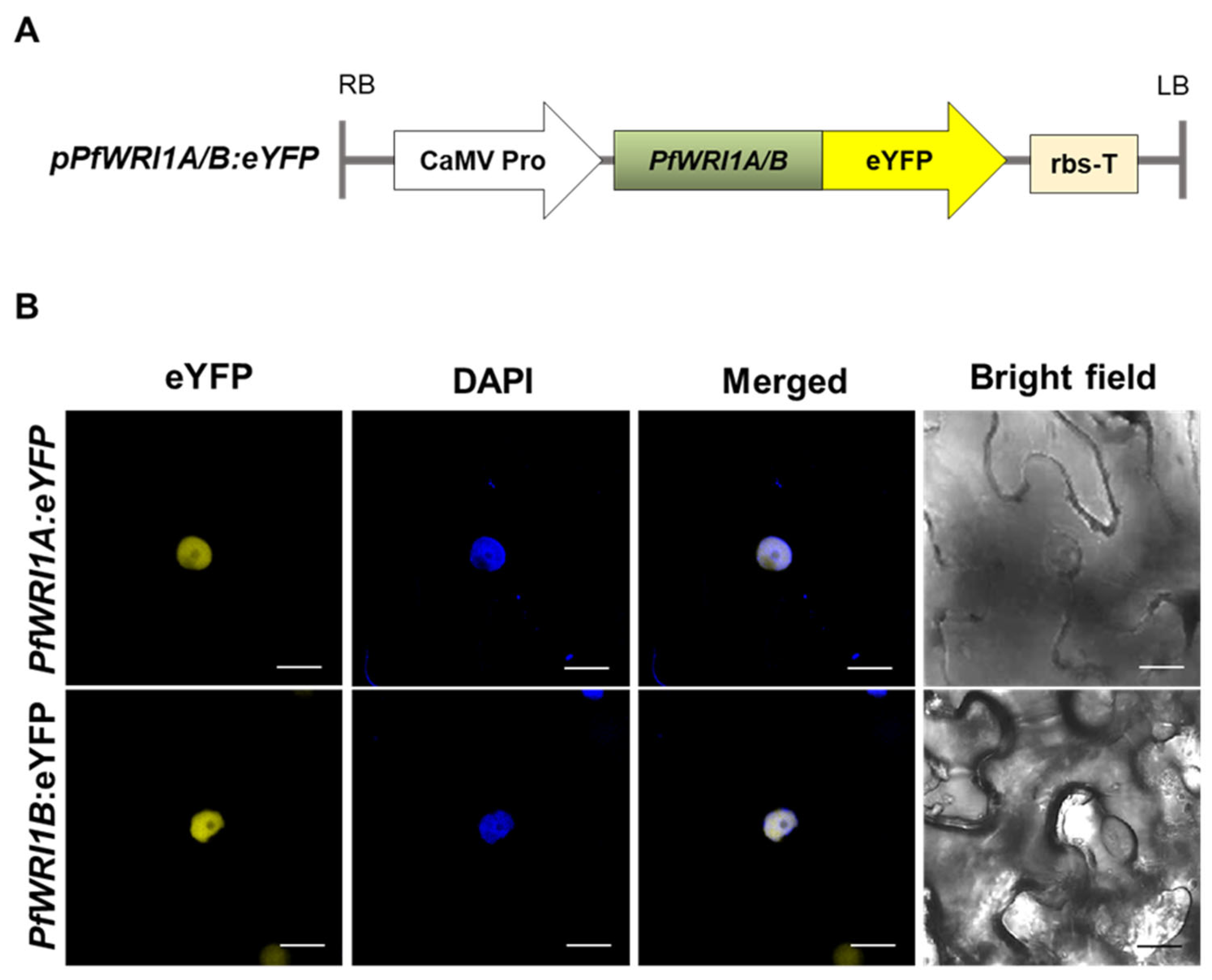
3.3. Subcellular Localization of PfWRI1A:eYFP and PfWRI1B:eYFP in N. benthamiana Leaves
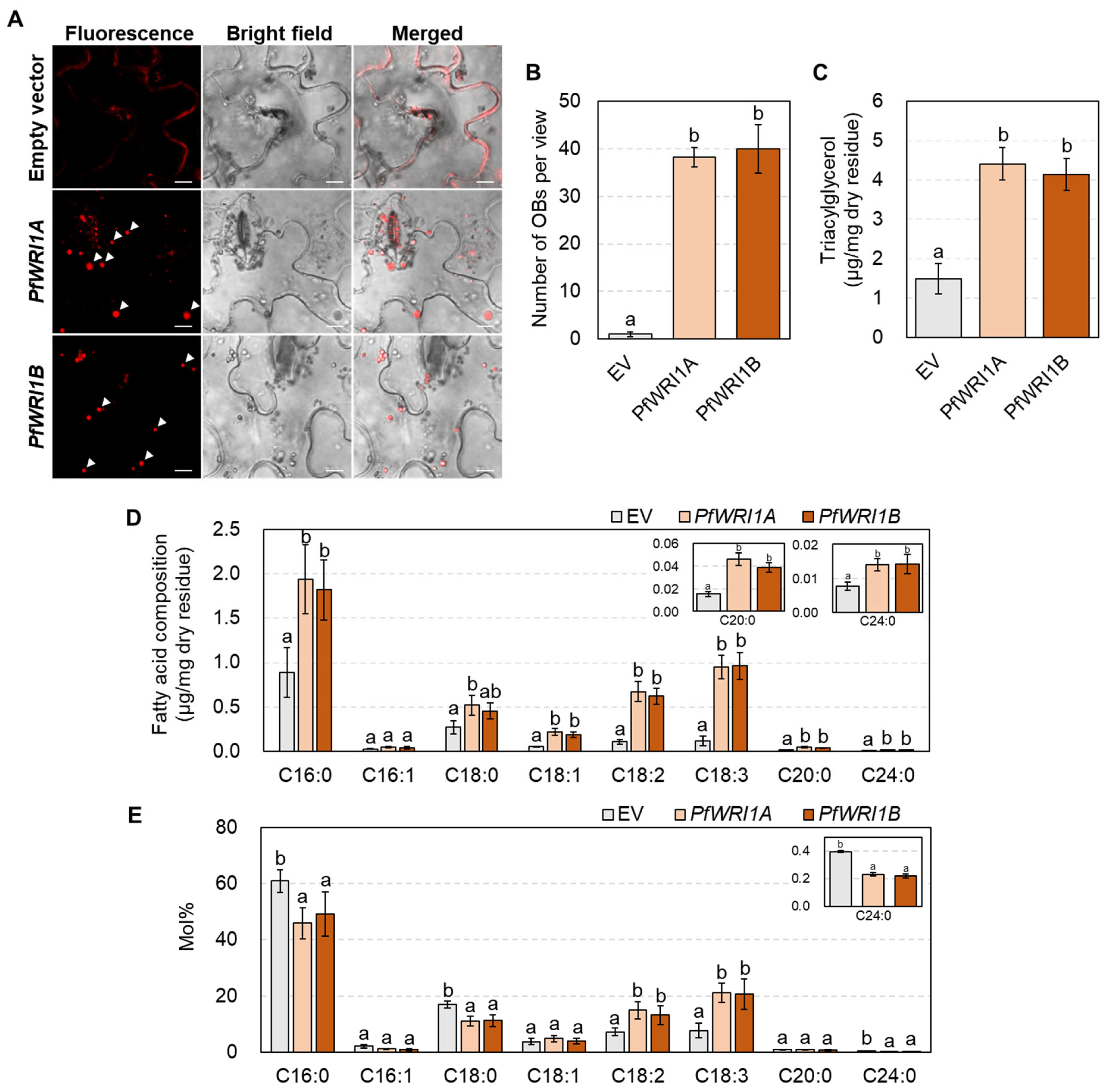
3.4. Oil Body Formation and TAG Measurement in N. benthamiana Leaves Expressing PfWRI1A or PfWRI1B
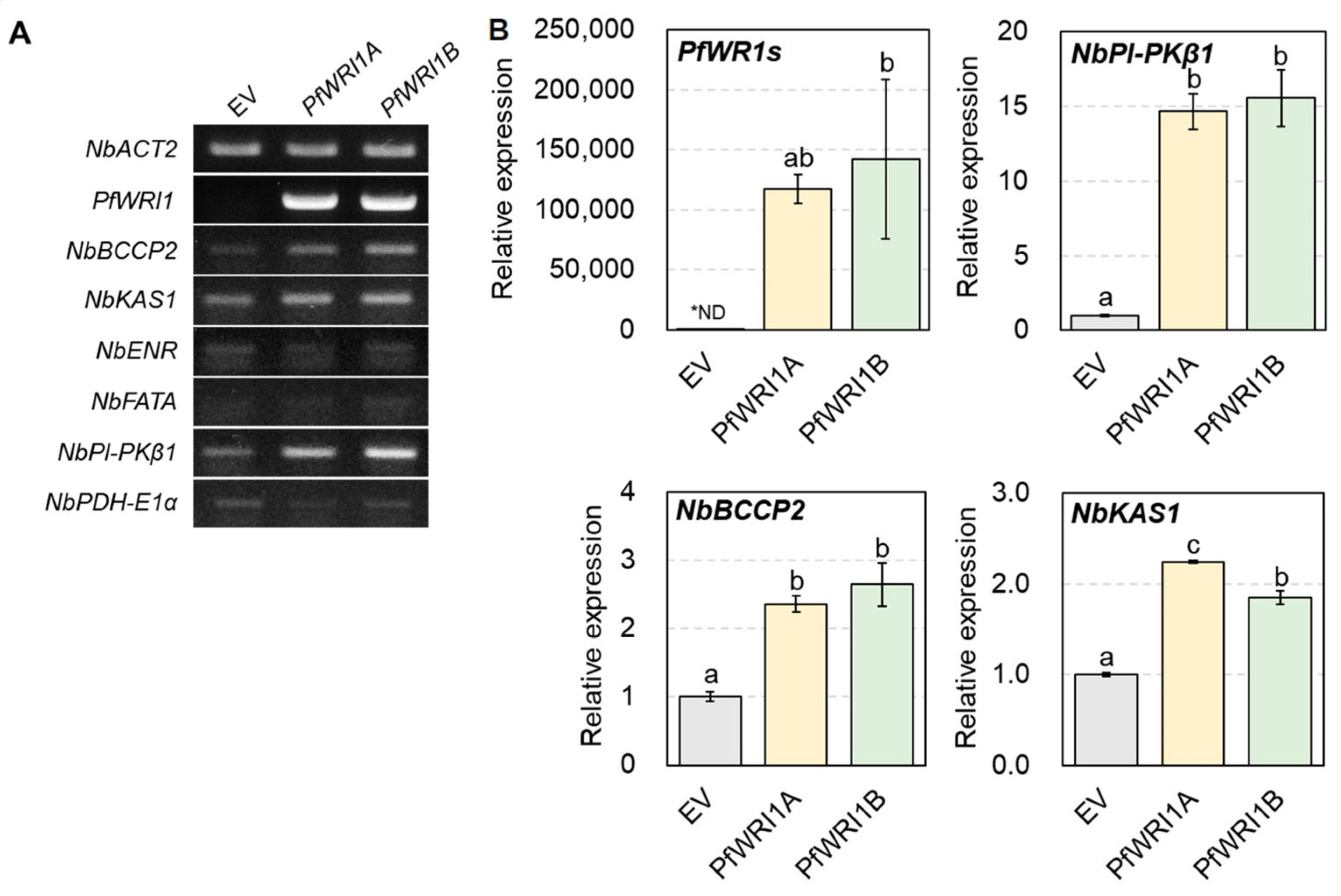
3.5. Induced Expzression of WRI1′s Target Genes in N. benthamiana Leaves Expressing PfWRI1A or PfWRI1B
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graham, I.A. Seed storage oil mobilization. Annu. Rev. Plant Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef] [PubMed]
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008, 54, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Dyer, J.M.; Stymne, S.; Green, A.G.; Carlsson, A.S. High-value oils from plants. Plant J. 2008, 54, 640–655. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.A. Can we feed the world & sustain the planet? Sci. Am. 2011, 305, 60–65. [Google Scholar] [PubMed]
- Goncalves, E.C.; Wilkie, A.C.; Kirst, M.; Rathinasabapathi, B. Metabolic regulation of triacylglycerol accumulation in the green algae: Identification of potential targets for engineering to improve oil yield. Plant Biotechnol. J. 2016, 14, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Cahoon, E.B.; Shockey, J.M.; Dietrich, C.R.; Gidda, S.K.; Mullen, R.T.; Dyer, J.M. Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: Solving bottlenecks in fatty acid flux. Curr. Opin. Plant Biol. 2007, 10, 236–244. [Google Scholar] [CrossRef]
- Vigeolas, H.; Waldeck, P.; Zank, T.; Geigenberger, P. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol. J. 2007, 5, 431–441. [Google Scholar] [CrossRef]
- Kim, H.; Park, J.; Kim, D.; Kim, A.; Suh, M. Functional analysis of diacylglycerol acyltransferase1 genes from Camelina sativa and effects of CsDGAT1B overexpression on seed mass and storage oil content in C. sativa. Plant Biotechnol. Rep. 2016, 10, 141–153. [Google Scholar] [CrossRef]
- Chhikara, S.; Abdullah, H.M.; Akbari, P.; Schnell, D.; Dhankher, O.P. Engineering Camelina sativa (L.) Crantz for enhanced oil and seed yields by combining diacylglycerol acyltransferase1 and glycerol-3-phosphate dehydrogenase expression. Plant Biotechnol. J. 2018, 16, 1034–1045. [Google Scholar] [CrossRef]
- Jing, G.; Tang, D.; Yao, Y.; Su, Y.; Shen, Y.; Bai, Y.; Jing, W.; Zhang, Q.; Lin, F.; Guo, D.; et al. Seed specifically over-expressing DGAT2A enhances oil and linoleic acid contents in soybean seeds. Biochem. Biophys. Res. Comm. 2021, 568, 143–150. [Google Scholar] [CrossRef]
- Browse, J.; Slack, C.R. Fatty-acid synthesis in plastids from maturing safflower and linseed cotyledons. Planta 1985, 166, 74–80. [Google Scholar] [CrossRef]
- Rawsthorne, S. Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 2002, 41, 182–196. [Google Scholar] [CrossRef]
- DeBrosse, S.D.; Kerr, D.S. Chapter 12—Pyruvate dehydrogenase complex deficiency. In Mitochondrial Case Studies; Saneto, R.P., Parikh, S., Cohen, B.H., Eds.; Academic Press: Boston, MA, USA, 2016; pp. 93–101. [Google Scholar]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef]
- Jung, S.H.; Kim, R.J.; Kim, K.J.; Lee, D.H.; Suh, M.C. Plastidial and mitochondrial malonyl CoA-ACP malonyltransferase is essential for cell division and its overexpression increases storage oil content. Plant Cell Physiol. 2019, 60, 1239–1249. [Google Scholar] [CrossRef]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940. [Google Scholar]
- Clough, R.C.; Matthis, A.L.; Barnum, S.R.; Jaworski, J.G. Purification and characterization of 3-ketoacyl-acyl carrier protein synthase III from spinach. A condensing enzyme utilizing acetyl-coenzyme A to initiate fatty acid synthesis. J. Biol. Chem. 1992, 267, 20992–20998. [Google Scholar] [CrossRef]
- Bonaventure, G.; Salas, J.J.; Pollard, M.R.; Ohlrogge, J.B. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 2003, 15, 1020–1033. [Google Scholar] [CrossRef]
- Shockey, J.M.; Fulda, M.S.; Browse, J.A. Arabidopsis contains nine long-chain acyl-Coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 2002, 129, 1710–1722. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. In The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2013; Volume 11, p. e0161. [Google Scholar]
- Dahlqvist, A.; Stahl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 2009, 21, 3885–3901. [Google Scholar] [CrossRef]
- Tumaney, A.W.; Shekar, S.; Rajasekharan, R. Identification, purification, and characterization of monoacylglycerol acyltransferase from developing peanut cotyledons. J. Biol. Chem. 2001, 276, 10847–10852. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Enugutti, B.; Rajakumari, S.; Rajasekharan, R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 2006, 141, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.L.; Whitehead, L.; He, Z.; Gazda, V.; Gilday, A.; Kozhevnikova, E.; Vaistij, F.E.; Larson, T.R.; Graham, I.A. A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol. 2012, 160, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Shockey, J.M.; Klasson, K.T.; Chapital, D.C.; Mason, C.B.; Scheffler, B.E. Developmental regulation of diacylglycerol acyltransferase family gene expression in tung tree tissues. PLoS ONE 2013, 8, e76946. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.R.; Kim, Y.M.; Yeo, Y.; Kim, S.; Suh, M.C. Camelina cytosol-localized diacylglycerol acyltransferase 3 contributes to the accumulation of seed storage oils. Ind. Crop. Prod. 2022, 189, 115808. [Google Scholar] [CrossRef]
- Focks, N.; Benning, C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoët, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef]
- Baud, S.; Wuillème, S.; To, A.; Rochat, C. Lepiniec, Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 2009, 60, 933–947. [Google Scholar] [CrossRef]
- Maeo, K.; Tokuda, T.; Ayame, A.; Mitsui, N.; Kawai, T.; Tsukagoshi, H.; Ishiguro, S.; Nakamura, K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009, 60, 476–487. [Google Scholar] [CrossRef]
- Fukuda, N.; Ikawa, Y.; Aoyagi, T.; Kozaki, A. Expression of the genes coding for plastidic acetyl-CoA carboxylase subunits is regulated by a location-sensitive transcription factor binding site. Plant Mol. Biol. 2013, 82, 473–483. [Google Scholar] [CrossRef]
- Marchive, C.; Nikovics, K.; To, A.; Lepiniec, L.; Baud, S. Transcriptional regulation of fatty acid production in higher plants: Molecular bases and biotechnological outcomes. Eur. J. Lipid Sci. Technol. 2014, 116, 1332–1343. [Google Scholar] [CrossRef]
- Cernac, A.; Benning, C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.; Py, C.; Gendrot, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Arondel, V.; Kilaru, A.; Bates, P.D.; Thrower, N.A.; Benning, C.; Ohlrogge, J.B. WRINKLED1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 2013, 8, e68887. [Google Scholar] [CrossRef]
- An, D.; Kim, H.; Ju, S.; Go, Y.S.; Kim, H.U.; Suh, M.C. Expression of camelina WRINKLED1 isoforms rescue the seed phenotype of the Arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front. Plant Sci. 2017, 8, 34. [Google Scholar] [CrossRef]
- Ye, J.; Wang, C.; Sun, Y.; Qu, J.; Mao, H.; Chua, N.-H. Overexpression of a transcription factor increases lipid content in a woody perennial Jatropha curcas. Front. Plant Sci. 2018, 9, 1479. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, G.; Li, P.; Yang, J.; Guo, L. Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol. J. 2020, 18, 155–171. [Google Scholar] [CrossRef]
- Wu, X.L.; Liu, Z.H.; Hu, Z.H.; Huang, R.Z. BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. J. Integr. Plant Biol. 2014, 56, 582–593. [Google Scholar] [CrossRef]
- An, D.; Suh, M.C. Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 2015, 9, 137–148. [Google Scholar] [CrossRef]
- Grimberg, Å.; Carlsson, A.S.; Marttila, S.; Bhalerao, R.; Hofvander, P. Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 2015, 15, 192. [Google Scholar] [CrossRef]
- Asif, M. Health effects of omega-3,6,9 fatty acids: Perilla frutescens is a good example of plant oils. Orient. Pharm. Exp. Med. 2011, 11, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Honda, G.; Yuba, A.; Kojima, T.; Tabata, M. Chemotaxonomic and cytogenetic studies on Perilla frutescens var. citriodora (‘Lemon Egoma’). Nat. Med. 1994, 48, 185–190. [Google Scholar]
- Nitta, M.; Lee, J.K.; Kang, C.W.; Katsuta, M.; Yasumoto, S.; Liu, D.; Nagamine, T.; Ohnishi, O. The distribution of Perilla species. Genet. Resour. Crop Evol. 2005, 52, 797–804. [Google Scholar] [CrossRef]
- Lee, K.R.; Lee, Y.; Kim, E.H.; Lee, S.B.; Roh, K.H.; Kim, J.B.; Kang, H.C.; Kim, H.U. Functional identification of oleate 12-desaturase and ω-3 fatty acid desaturase genes from Perilla frutescens var. frutescens. Plant Cell Rep. 2016, 35, 2523–2537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, Q.; Leng LZhang, D.; Chen, S.; Shi, Y.; Ning, Z.; Chen, S. Incipient diploidization of the medicinal plant Perilla within 10,000 years. Nat. Comm. 2021, 12, 5508. [Google Scholar] [CrossRef]
- Oñate-Sánchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes. 2008, 1, 93. [Google Scholar] [CrossRef]
- Holsters, M.; De Waele, D.; Depicker, A.; Messens, E.; Van Montagu, M.; Schell, J. Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 1978, 163, 181–187. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The sol genomics network (SGN)--from genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, 1036–1041. [Google Scholar] [CrossRef]
- Ji, X.J.; Mao, X.; Hao, Q.T.; Liu, B.L.; Xue, J.A.; Li, R.Z. Splice variants of the castor WRI1 gene upregulate fatty acid and oil biosynthesis when expressed in tobacco leaves. Int. J. Mol. Sci. 2018, 19, 146. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Grix, M.; Mantyla, J.J.; Yang, Y.; Benning, C.; Ohlrogge, J.B. Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015, 83, 864–874. [Google Scholar] [CrossRef]
- Vanhercke, T.; Tahchy, A.E.; Shrestha, P.; Zhou, X.R.; Singh, S.P.; Petrie, J.R. Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 2013, 587, 364–369. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef]
- Mano, F.; Aoyanagi, T.; Kozaki, A. Atypical splicing accompanied by skipping conserved micro-exons produces unique WRINKLED1, An AP2 domain transcription factor in rice plants. Plants 2019, 8, 207. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, L.; Ma, W. WRINKLED1, a “master regulator” in transcriptional control of plant oil biosynthesis. Plants 2019, 8, 238. [Google Scholar] [CrossRef]
- Chen, L.; Lee, J.H.; Weber, H.; Tohge, T.; Witt, S.; Roje, S.; Hellmann, H. Arabidopsis BPM proteins function as substrate adaptors to a CULLIN3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 2013, 25, 2253–2264. [Google Scholar] [CrossRef]
- Li, Q.; Shao, J.; Tang, S.; Shen, Q.; Wang, T.; Chen, W.; Hong, Y. Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front Plant Sci. 2015, 6, 1015. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lee, K.-R.; Suh, M.C. Ectopic Expression of Perilla frutescens WRI1 Enhanced Storage Oil Accumulation in Nicotiana benthamiana Leaves. Plants 2023, 12, 1081. https://doi.org/10.3390/plants12051081
Kim S, Lee K-R, Suh MC. Ectopic Expression of Perilla frutescens WRI1 Enhanced Storage Oil Accumulation in Nicotiana benthamiana Leaves. Plants. 2023; 12(5):1081. https://doi.org/10.3390/plants12051081
Chicago/Turabian StyleKim, Semi, Kyeong-Ryeol Lee, and Mi Chung Suh. 2023. "Ectopic Expression of Perilla frutescens WRI1 Enhanced Storage Oil Accumulation in Nicotiana benthamiana Leaves" Plants 12, no. 5: 1081. https://doi.org/10.3390/plants12051081
APA StyleKim, S., Lee, K.-R., & Suh, M. C. (2023). Ectopic Expression of Perilla frutescens WRI1 Enhanced Storage Oil Accumulation in Nicotiana benthamiana Leaves. Plants, 12(5), 1081. https://doi.org/10.3390/plants12051081






