Floral Homeotic Factors: A Question of Specificity
Abstract
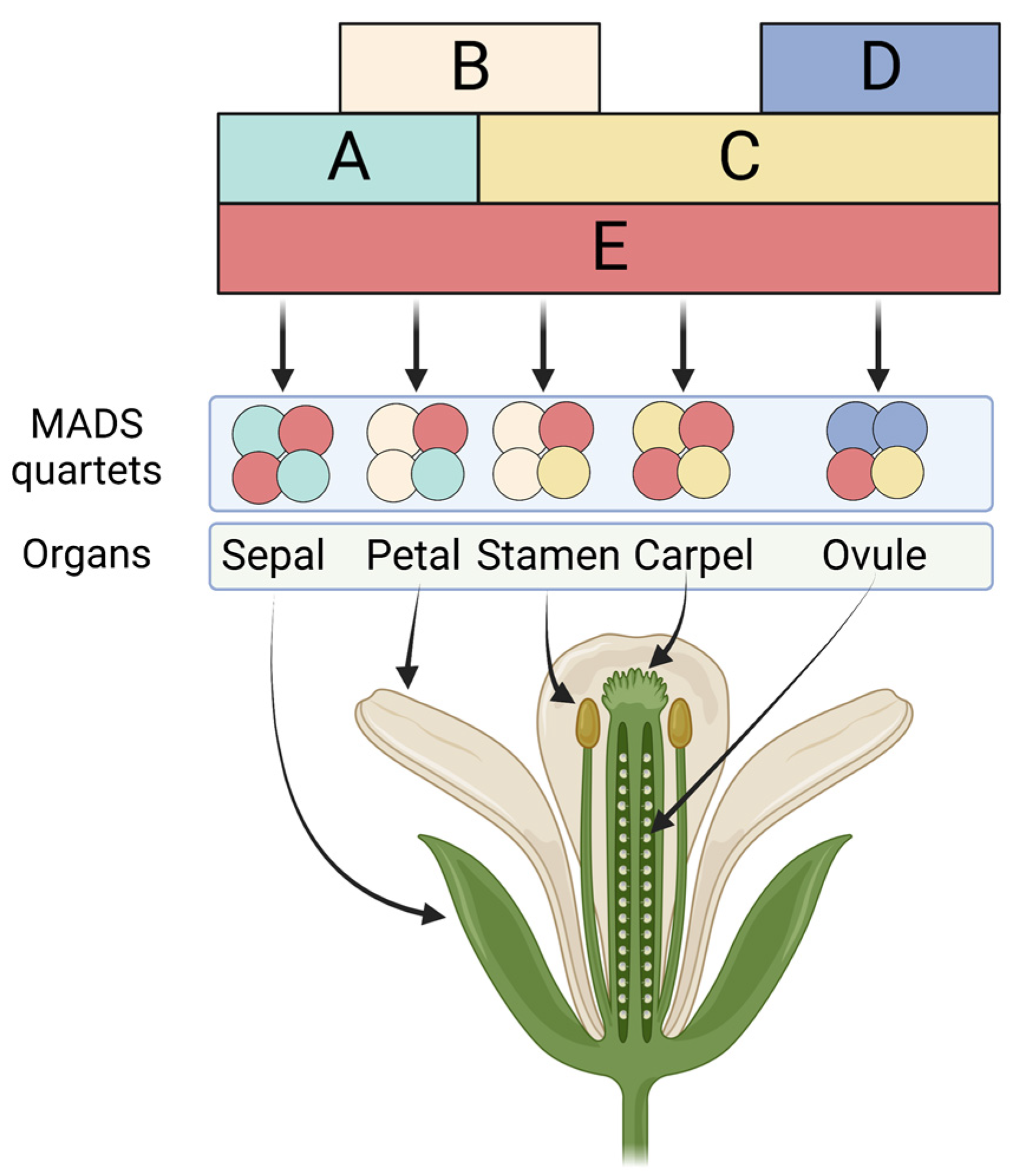
:1. Introduction
2. DNA-Binding Preferences as a Determinant of Specificity
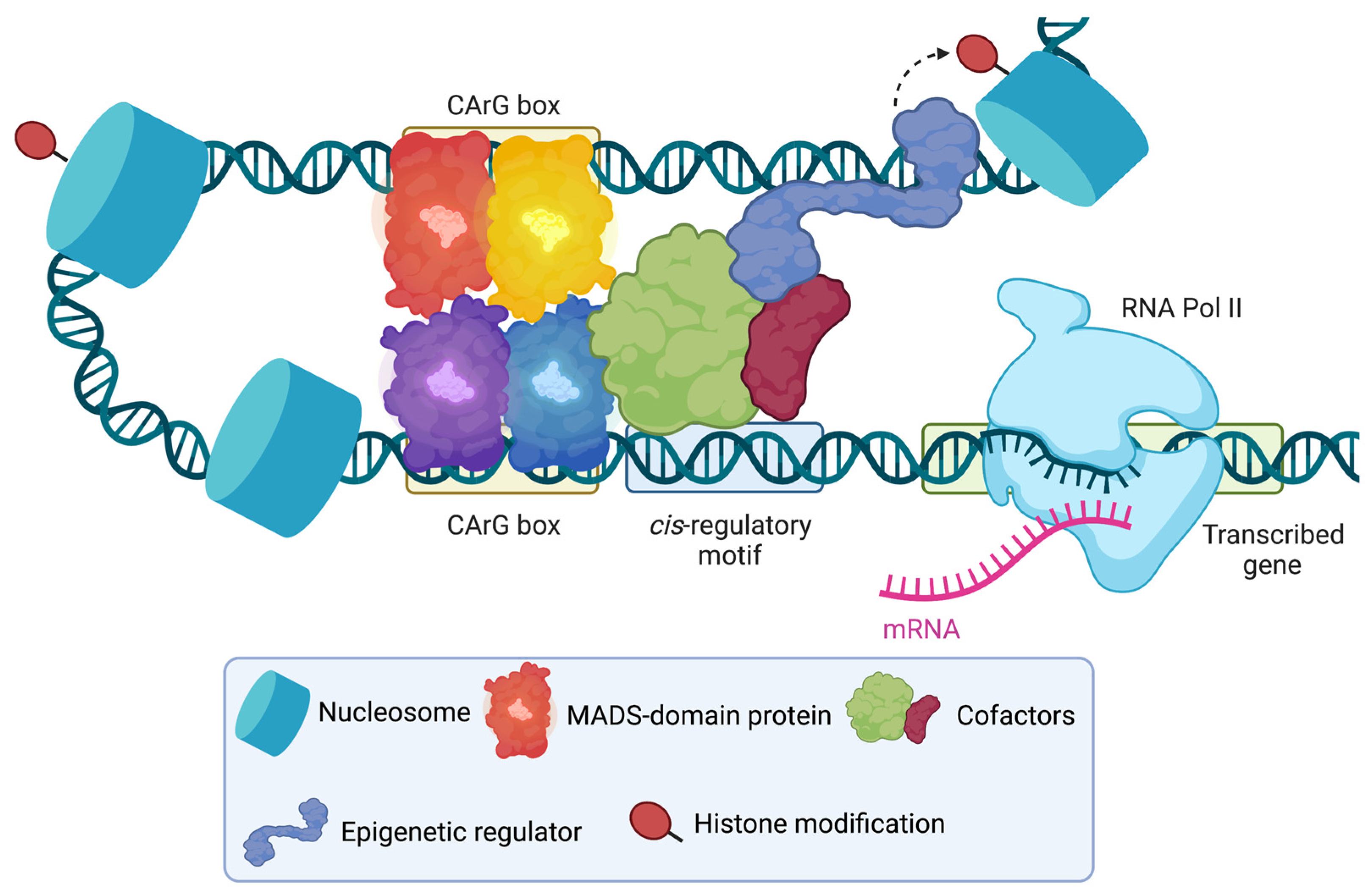
3. Cofactors as Determinants for Specificity
4. Other Possible Determinants for Specificity
5. Lessons from Hox genes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. The ABC model of flower development: Then and now. Development 2012, 139, 4095–4098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Causier, B.; Schwarz-Sommer, Z.; Davies, B. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 2010, 21, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellmer, F.; Graciet, E.; Riechmann, J.L. Specification of floral organs in Arabidopsis. J. Exp. Bot. 2014, 65, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Chen, D.; Kaufmann, K. Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 2016, 29, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ng, K.H.; Lim, T.S.; Yu, H.; Meyerowitz, E.M. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 2007, 19, 3516–3529. [Google Scholar] [CrossRef] [Green Version]
- Wuest, S.E.; O’Maoileidigh, D.S.; Rae, L.; Kwasniewska, K.; Raganelli, A.; Hanczaryk, K.; Lohan, A.J.; Loftus, B.; Graciet, E.; Wellmer, F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 2012, 109, 13452–13457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Maoileidigh, D.S.; Wuest, S.E.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwasniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Mena, C.; de Folter, S.; Costa, M.M.; Angenent, G.C.; Sablowski, R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 2005, 132, 429–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellmer, F.; Riechmann, J.L.; Alves-Ferreira, M.; Meyerowitz, E.M. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 2004, 16, 1314–1326. [Google Scholar] [CrossRef] [Green Version]
- Alves-Ferreira, M.; Wellmer, F.; Banhara, A.; Kumar, V.; Riechmann, J.L.; Meyerowitz, E.M. Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 2007, 145, 747–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Maoileidigh, D.S.; Graciet, E.; Wellmer, F. Genetic Control of Arabidopsis Flower Development. In Advances in Botanical Research; Fornara, F., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 72, pp. 159–190. [Google Scholar]
- de Folter, S.; Immink, R.G.; Kieffer, M.; Parenicova, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive Interaction Map of the Arabidopsis MADS Box Transcription Factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef] [Green Version]
- Theissen, G.; Melzer, R.; Rumpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.; Muino, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [Green Version]
- Hugouvieux, V.; Silva, C.S.; Jourdain, A.; Stigliani, A.; Charras, Q.; Conn, V.; Conn, S.J.; Carles, C.C.; Parcy, F.; Zubieta, C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res. 2018, 46, 4966–4977. [Google Scholar] [CrossRef] [Green Version]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef] [Green Version]
- Aerts, N.; de Bruijn, S.; van Mourik, H.; Angenent, G.C.; van Dijk, A.D.J. Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 131. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Muino, J.M.; Chen, D.; Angenent, G.C.; Kaufmann, K. Differences in DNA Binding Specificity of Floral Homeotic Protein Complexes Predict Organ-Specific Target Genes. Plant Cell 2017, 29, 1822–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kappel, S.; Eggeling, R.; Rumpler, F.; Groth, M.; Melzer, R.; Theissen, G. DNA-binding properties of the MADS-domain transcription factor SEPALLATA3 and mutant variants characterized by SELEX-seq. Plant Mol. Biol. 2021, 105, 543–557. [Google Scholar] [CrossRef]
- Kaufmann, K.; Wellmer, F.; Muino, J.M.; Ferrier, T.; Wuest, S.E.; Kumar, V.; Serrano-Mislata, A.; Madueno, F.; Krajewski, P.; Meyerowitz, E.M.; et al. Orchestration of floral initiation by APETALA1. Science 2010, 328, 85–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajoro, A.; Madrigal, P.; Muino, J.M.; Matus, J.T.; Jin, J.; Mecchia, M.A.; Debernardi, J.M.; Palatnik, J.F.; Balazadeh, S.; Arif, M.; et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014, 15, R41. [Google Scholar] [CrossRef]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 2017, 6, e21856. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Muino, J.M.; Jauregui, R.; Airoldi, C.A.; Smaczniak, C.; Krajewski, P.; Angenent, G.C. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009, 7, e1000090. [Google Scholar] [CrossRef] [Green Version]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Regulation of the arabidopsis floral homeotic gene APETALA1. Cell 1994, 76, 131–143. [Google Scholar] [CrossRef]
- Mandel, M.A.; Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell 1997, 8, 1243–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, X.; Vega-Leon, R.; Hugouvieux, V.; Blanc-Mathieu, R.; van der Wal, F.; Lucas, J.; Silva, C.S.; Jourdain, A.; Muino, J.M.; Nanao, M.H.; et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021, 12, 4760. [Google Scholar] [CrossRef] [PubMed]
- O’Maoileidigh, D.S.; Thomson, B.; Raganelli, A.; Wuest, S.E.; Ryan, P.T.; Kwasniewska, K.; Carles, C.C.; Graciet, E.; Wellmer, F. Gene network analysis of Arabidopsis thaliana flower development through dynamic gene perturbations. Plant J. 2015, 83, 344–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Smith, N.C.; Matthews, J.M. Mechanisms of DNA-binding specificity and functional gene regulation by transcription factors. Curr. Opin. Struct. Biol. 2016, 38, 68–74. [Google Scholar] [CrossRef]
- Junion, G.; Spivakov, M.; Girardot, C.; Braun, M.; Gustafson, E.H.; Birney, E.; Furlong, E.E. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 2012, 148, 473–486. [Google Scholar] [CrossRef] [Green Version]
- Lloret-Fernandez, C.; Maicas, M.; Mora-Martinez, C.; Artacho, A.; Jimeno-Martin, A.; Chirivella, L.; Weinberg, P.; Flames, N. A transcription factor collective defines the HSN serotonergic neuron regulatory landscape. Elife 2018, 7, e32785. [Google Scholar] [CrossRef]
- Posern, G.; Treisman, R. Actin’ together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006, 16, 588–596. [Google Scholar] [CrossRef]
- Onuh, J.O.; Qiu, H. Serum response factor-cofactor interactions and their implications in disease. FEBS J. 2021, 288, 3120–3134. [Google Scholar] [CrossRef]
- Nagy, G.; Nagy, L. Motif grammar: The basis of the language of gene expression. Comput. Struct. Biotechnol. J. 2020, 18, 2026–2032. [Google Scholar] [CrossRef]
- Trigg, S.A.; Garza, R.M.; MacWilliams, A.; Nery, J.R.; Bartlett, A.; Castanon, R.; Goubil, A.; Feeney, J.; O’Malley, R.; Huang, S.C.; et al. CrY2H-seq: A massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 2017, 14, 819–825. [Google Scholar] [CrossRef] [Green Version]
- Simonini, S.; Roig-Villanova, I.; Gregis, V.; Colombo, B.; Colombo, L.; Kater, M.M. Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell 2012, 24, 4163–4172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera-Ubaldo, H.; Campos, S.E.; Lopez-Gomez, P.; Luna-Garcia, V.; Zuniga-Mayo, V.M.; Armas-Caballero, G.E.; Gonzalez-Aguilera, K.L.; DeLuna, A.; Marsch-Martinez, N.; Espinosa-Soto, C.; et al. The protein-protein interaction landscape of transcription factors during gynoecium development in Arabidopsis. Mol. Plant 2023, 16, 260–278. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Interactome Mapping, C. Evidence for network evolution in an Arabidopsis interactome map. Science 2011, 333, 601–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, R.; Jez, J.M.; Basra, A.; Zhang, B.; Schachtman, D.P. 14-3-3 proteins fine-tune plant nutrient metabolism. FEBS Lett. 2011, 585, 143–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brambilla, V.; Battaglia, R.; Colombo, M.; Masiero, S.; Bencivenga, S.; Kater, M.M.; Colombo, L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 2007, 19, 2544–2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altmann, M.; Altmann, S.; Rodriguez, P.A.; Weller, B.; Elorduy Vergara, L.; Palme, J.; Marin-de la Rosa, N.; Sauer, M.; Wenig, M.; Villaecija-Aguilar, J.A.; et al. Extensive signal integration by the phytohormone protein network. Nature 2020, 583, 271–276. [Google Scholar] [CrossRef]
- Liu, C.; Xi, W.; Shen, L.; Tan, C.; Yu, H. Regulation of floral patterning by flowering time genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef] [Green Version]
- Sang, Q.; Pajoro, A.; Sun, H.; Song, B.; Yang, X.; Stolze, S.C.; Andres, F.; Schneeberger, K.; Nakagami, H.; Coupland, G. Mutagenesis of a Quintuple Mutant Impaired in Environmental Responses Reveals Roles for CHROMATIN REMODELING4 in the Arabidopsis Floral Transition. Plant Cell 2020, 32, 1479–1500. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, V.V.; Surendrarao, A.; Liu, Z. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 2006, 133, 3159–3166. [Google Scholar] [CrossRef] [Green Version]
- Mendes, M.A.; Guerra, R.F.; Berns, M.C.; Manzo, C.; Masiero, S.; Finzi, L.; Kater, M.M.; Colombo, L. MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell 2013, 25, 2560–2572. [Google Scholar] [CrossRef] [Green Version]
- Filtz, T.M.; Vogel, W.K.; Leid, M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol. Sci. 2014, 35, 76–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popescu, S.C.; Popescu, G.V.; Bachan, S.; Zhang, Z.; Gerstein, M.; Snyder, M.; Dinesh-Kumar, S.P. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009, 23, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Molkentin, J.D.; Li, L.; Olson, E.N. Phosphorylation of the MADS-Box transcription factor MEF2C enhances its DNA binding activity. J. Biol. Chem. 1996, 271, 17199–17204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, D.; Chang, D.; Marx, J.; Wei, L.; Olson, E.N.; Parmacek, M.S.; Balasubramanyam, A.; Schwartz, R.J. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc. Natl. Acad. Sci. USA 2006, 103, 4516–4521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patharkar, O.R.; Macken, T.A.; Walker, J.C. Serine 231 and 257 of Agamous-like 15 are phosphorylated in floral receptacles. Plant Signal. Behav. 2016, 11, e1199314. [Google Scholar] [CrossRef]
- Yalovsky, S.; Kulukian, A.; Rodriguez-Concepcion, M.; Young, C.A.; Gruissem, W. Functional requirement of plant farnesyltransferase during development in Arabidopsis. Plant Cell 2000, 12, 1267–1278. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, E.W.; Mohamed, A.; Litt, A. Functional Divergence of APETALA1 and FRUITFULL is due to Changes in both Regulation and Coding Sequence. Front. Plant Sci. 2015, 6, 1076. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.; Marquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 35. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Rao, X.; Wang, Y.; Feng, W.; Liang, H.; Liu, Y. Roles of alternative splicing in modulating transcriptional regulation. BMC Syst. Biol. 2017, 11, 89. [Google Scholar] [CrossRef] [Green Version]
- Severing, E.I.; van Dijk, A.D.; Morabito, G.; Busscher-Lange, J.; Immink, R.G.; van Ham, R.C. Predicting the impact of alternative splicing on plant MADS domain protein function. PLoS ONE 2012, 7, e30524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pose, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ryu, H.S.; Chung, K.S.; Pose, D.; Kim, S.; Schmid, M.; Ahn, J.H. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 2013, 342, 628–632. [Google Scholar] [CrossRef]
- Mann, R.S.; Lelli, K.M.; Joshi, R. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009, 88, 63–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Merabet, S.; Mann, R.S. To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins. Trends Genet. 2016, 32, 334–347. [Google Scholar] [CrossRef] [Green Version]
- De Kumar, B.; Darland, D.C. The Hox protein conundrum: The “specifics” of DNA binding for Hox proteins and their partners. Dev. Biol. 2021, 477, 284–292. [Google Scholar] [CrossRef]
- Meyerowitz, E.M. Plants compared to animals: The broadest comparative study of development. Science 2002, 295, 1482–1485. [Google Scholar] [CrossRef] [Green Version]
- Porcelli, D.; Fischer, B.; Russell, S.; White, R. Chromatin accessibility plays a key role in selective targeting of Hox proteins. Genome Biol. 2019, 20, 115. [Google Scholar] [CrossRef] [Green Version]
- Cain, B.; Gebelein, B. Mechanisms Underlying Hox-Mediated Transcriptional Outcomes. Front. Cell Dev. Biol. 2021, 9, 787339. [Google Scholar] [CrossRef]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Higueras, C.; Rastogi, C.; Voutev, R.; Bussemaker, H.J.; Mann, R.S.; Hombria, J.C. In vivo Hox binding specificity revealed by systematic changes to a single cis regulatory module. Nat. Commun. 2019, 10, 3597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Protein Alias | Family | Interacting with | Evidence | Reference | |
|---|---|---|---|---|---|
| AT1G02065 | SPL8 | SQUAMOSA-promoter binding protein | AG, AP1 | IP-MS | [19] |
| AT1G08970 | NF-YC9 | NF-Y | AG | Two-hybrid | [42] |
| AT1G13400 | NUB | Zinc finger | AG | Two-hybrid | [44] |
| AT1G19220 | ARF19 | Auxin Response Factor | SEP2 | Two-hybrid | [42] |
| AT1G63910 | MYB103 | MYB | AG | Two-hybrid | [42] |
| AT1G68240 | - | Basic helix-loop-helix | SEP2 | Two-hybrid | [42] |
| AT1G69690 | TCP15 | TCP | AG, AP1, PI, SEP4 | Two-hybrid | [42,44,45] |
| AT1G77450 | NAC032 | NAC domain containing protein | SEP2 | Two-hybrid | [42] |
| AT2G01930 | BPC1 | Basic pentacysteine | AP1 | Two-hybrid | [43] |
| AT2G17950 | WUS | Homeodomain | SEP3 | Two-hybrid | [44] |
| AT2G35940 | BLH1 | Homeodomain | AP1 | IP-MS | [19] |
| AT2G37630 | AS1 | MYB | AG | Two-hybrid | [44] |
| AT3G11100 | VFP3 | SANT and trihelix | SEP4 | Two-hybrid | [42] |
| AT3G15030 | TCP4 | TCP | AP1, SEP4 | Two-hybrid | [42] |
| AT3G19070 | - | Homeodomain-like | SEP1 | Two-hybrid | [42] |
| AT3G24490 | - | MYB/SANT-like | SEP1 | Two-hybrid | [42] |
| AT3G46600 | - | GRAS | AG, SEP1 | Two-hybrid | [42] |
| AT3G47620 | TCP14 | TCP | AP1, SEP4 | Two-hybrid | [42] |
| AT3G51080 | GATA6 | GATA | PI | Two-hybrid | [42] |
| AT4G01580 | - | AP2/B3-like | PI | Two-hybrid | [42] |
| AT4G03250 | - | Homeodomain-like | SEP1 | Two-hybrid | [42] |
| AT4G14225 | - | Zinc finger | PI | Two-hybrid | [42] |
| AT4G15250 | BBX9 | Zinc finger | AG | Two-hybrid | [42] |
| AT4G18390 | TCP2 | TCP | AG | Two-hybrid | [42] |
| AT4G37740 | GRF2 | Growth regulating factor | PI | Two-hybrid | [42] |
| AT5G02030 | RPL | Homeodomain | AP1 | IP-MS | [19] |
| AT5G05120 | - | Zinc finger | PI | Two-hybrid | [42] |
| AT5G09750 | HEC3 | Basic helix-loop-helix | PI | Two-hybrid | [42] |
| AT5G11270 | OCP3 | Homeodomain | SEP1 | Two-hybrid | [45] |
| AT5G24470 | PRR5 | Pseudo-response regulator | PI | Two-hybrid | [42] |
| AT5G38480 | GRF3 | Growth regulating factor | PI | IP-MS | [46] |
| AT5G41410 | BEL1 | Homeodomain | AG, SEP3 | Reconstituted Complex | [47] |
| AT5G41920 | SCL23 | GRAS | SEP4 | Two-hybrid | [48] |
| AT5G61850 | LFY | FLO/LFY | SEP3 | Reconstituted Complex | [49] |
| Gene ID | Protein | Family | Interacting with | Evidence | Reference |
|---|---|---|---|---|---|
| AT5G44800 | CHR4 | CHD3 | AG, AP1, PI, SEP3 | IP-MS | [19,50] |
| AT3G06400 | CHR11 | ISWI | AG, AP1, AP3, PI, SEP3 | IP-MS | [19] |
| AT4G39100 | SHL1 | PHD finger, BAH-domain | AP3 | Two-hybrid | [42] |
| AT1G43850 | SEU | adn1/SEU | AP1, AP3, SEP3 | Two-hybrid; Reconstituted Complex, IP-MS | [19,51] |
| AT5G18620 | CHR17 | ISWI | AG, AP1, PI, SEP3 | IP-MS | [19] |
| AT3G48430 | REF6 | JHDM3 histone demethylase | AG, AP1, SEP3 | IP-MS | [19] |
| AT2G25170 | PKL | CHD3 | SEP3 | IP-MS | [19] |
| AT2G32700 | LUH | WD40/ LUFS domain | AP1, SEP3 | IP-MS | [19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goslin, K.; Finocchio, A.; Wellmer, F. Floral Homeotic Factors: A Question of Specificity. Plants 2023, 12, 1128. https://doi.org/10.3390/plants12051128
Goslin K, Finocchio A, Wellmer F. Floral Homeotic Factors: A Question of Specificity. Plants. 2023; 12(5):1128. https://doi.org/10.3390/plants12051128
Chicago/Turabian StyleGoslin, Kevin, Andrea Finocchio, and Frank Wellmer. 2023. "Floral Homeotic Factors: A Question of Specificity" Plants 12, no. 5: 1128. https://doi.org/10.3390/plants12051128
APA StyleGoslin, K., Finocchio, A., & Wellmer, F. (2023). Floral Homeotic Factors: A Question of Specificity. Plants, 12(5), 1128. https://doi.org/10.3390/plants12051128





