Abstract
The NAC (NAM, ATAF and CUC) gene family plays an important role in plant development and abiotic stress response. However, up to now, the identification and research of the NAC (PeNAC) family members of passion fruit are still lacking. In this study, 25 PeNACs were identified from the passion fruit genome, and their functions under abiotic stress and at different fruit-ripening stages were analyzed. Furthermore, we analyzed the transcriptome sequencing results of PeNACs under four various abiotic stresses (drought, salt, cold and high temperature) and three different fruit-ripening stages, and verified the expression results of some genes by qRT-PCR. Additionally, tissue-specific analysis showed that most PeNACs were mainly expressed in flowers. In particular, PeNAC-19 was induced by four various abiotic stresses. At present, low temperatures have seriously endangered the development of passion fruit cultivation. Therefore, PeNAC-19 was transformed into tobacco, yeast and Arabidopsis to study their function of resisting low temperature. The results show that PeNAC-19 responded to cold stress significantly in tobacco and Arabidopsis, and could improve the low temperature tolerance of yeast. This study not only improved the understanding of the PeNAC gene family characteristics and evolution, but also provided new insights into the regulation of the PeNAC gene at different stages of fruit maturation and abiotic stresses.
1. Introduction
Transcription factors (TFs) play an important role in regulating cell signaling, cell morphogenesis and plant resistance to external environmental stresses [1,2]. TFs regulate gene expression by binding to specific promoter cis-acting elements to activate or repress the transcriptional level of target genes [3,4]. In plants, the NAC (NAM, ATAF1/2 and CUC2) transcription factor family is named after three proteins: petunia apical meristem (NAM), Arabidopsis thaliana ATAF1/2 and cup cotyledon (CUC) [5,6], which is one of the largest and most plant-specific TF families. Typical NAC proteins include a highly conserved N-terminal region (NAC domain), while the C-terminal region contains a relatively distinct transcriptional activation/repression region (TAR or TRR) [5,7,8], which is a highly diverse transcriptional regulatory region [9], may be involved in protein–protein interactions and contribute to its regulatory specificity [10]. The N-terminus of the NAC protein contains a conserved domain of 150–160 amino acids involved in DNA binding, dimerization and localization [11], which is further divided into five subdomains (A-E), of which the A, C and D subdomains are highly conserved [12].
Since the first report of the NAC protein in 1996 [5], NAC protein families have been identified in several plant species, such as Arabidopsis thaliana [13], Oryza sativa [14], Musa acuminata [15], Medicago sativa [16], Dimocarpus longan [17], Actinidia chinensis [18], Populus trichocarpa [19], Vitis vinifera [20] and Pyrus pyrifolia [21]. Due to their powerful functions in plants, the NAC family has been extensively studied in recent years. The related reports indicate that NAC family members play an essential role in response to plant abiotic stress.
The overexpression of OsNAC10 in rice can improve the drought tolerance and grain yield of plants [22]. The overexpression of OsNAC6/SNAC2 can improve the drought tolerance, salt tolerance and cold tolerance of rice seedlings [23,24]. The transcription factor MbNAC25 of Siberian crab apple (Malus baccata Borkh) can improve cold tolerance in transgenic Arabidopsis [25]. Both SNAC2 of rice and PbeNAC1 of pear (Pyrus betulifolia Bunge) confer cold tolerance in rice and pears [26]. In tomatoes, SlNAM1 is induced by chilling stress and can improve the chilling resistance in transgenic plants [27].
In addition, there are also related studies showing that the NAC transcription factor is related to fruit development and ripening. Fruit development and ripening are complex processes regulated by various factors such as gene regulation, hormones, light and temperature The processes in the development and ripening of fruit are regulated by many factors such as light, temperature, hormone and gene regulation. In particular, they are regulated by a variety of transcription factors that affect the expression levels of downstream target genes [28]. Some studies have shown that NAC transcription factors can regulate the expression of the genes involved in hormone biosynthesis and signal transduction during fruit development and maturation [29]. In spruce, the overexpression of PaNAC03 affects plant embryonic development [30]. Furthermore, in Vitis vinifera, the development of seeds and fruits is affected by the interaction between VvNAC26 and VvMADS9 [31]. In tomatoes, SlNAC1 plays an important role in the softening process [32]. In strawberries (Fragaria chiloensis), the FcNAC1 protein is involved in pectin metabolism to soften fruit [33].
Passion fruit (Passiflora edulis Sim) is a perennial evergreen vine of the Passiflora genus of Passifloraceae, a tropical and rare fruit tree, which contains more than 100 kinds of aroma in its fruit pulp. It is native to central and northern South America, and is widely distributed throughout America, Australia and Africa [34]. According to the reports, Passiflora has about 520 varieties, most of which are used for ornamental purposes, and only a small number of 60 species can be eaten [35]. At present, the main countries for passion fruit are Brazil, Colombia, Ecuador, Australia, Vietnam, China, etc. Due to its unique flavor and short growth period (4–6 months), the planting area under cultivation gradually increased. Abiotic stresses seriously affect the normal development of the passion fruit industry. Therefore, it is important to excavate the function genes of stress resistance in passion fruit and analyze their mechanisms of action [36]. In this study, using the high-quality genomic data of passion fruit [37], 25 members of the passion fruit NAC (PeNAC) family were identified. In contrast to another previously published result [38], they used another genome [39]. The result of genome assemblies vary widely in the HR genome [37] and the MER genome [39]: contig N50 (3.1 Mb and 70 kb), complete BUSCOs (91.56% and 88.1%) and scaffold N50 (148,138.5 Mb and 126.4 Mb). More importantly, we also identified the expression patterns of this gene in different fruit-ripening stages and abiotic stresses, and validated them by qPCR. Additionally, one of the PeNAC genes exhibited resistance to cold stress. These results provide useful information for the genetic improvement of fruit quality and the improvement of the abiotic stress resistance of passion fruit, and lay a good foundation for the study of the regulation mechanism of fruit quality.
2. Results
2.1. Identification of the Passion Fruit NAC Family
In this research, 25 PeNACs have been identified. In addition, we analyzed the characteristics of the PeNACs (Table 1). The length of the PeNAC CDS sequence ranged from 174 bp (PeNAC-21) to 6126 bp (PeNAC-12). The identified PeNACs encoded proteins ranging from 57 amino acids of PeNAC-21 to 2041 amino acids of PeNAC-12. The MW ranged from 6.74Da (PeNAC-21) to 229.5Da (PeNAC-12). The isoelectric point ranged from 3.84 (PeNAC-9) to 10.44 (PeNAC-7). Subcellular localization predicted that all genes were located in the nucleus.

Table 1.
Basic information of NAC genes identified in passion fruit. MW: molecular weight. PI: isoelectric point.
2.2. Phylogenetic Analysis of PeNACs Protein
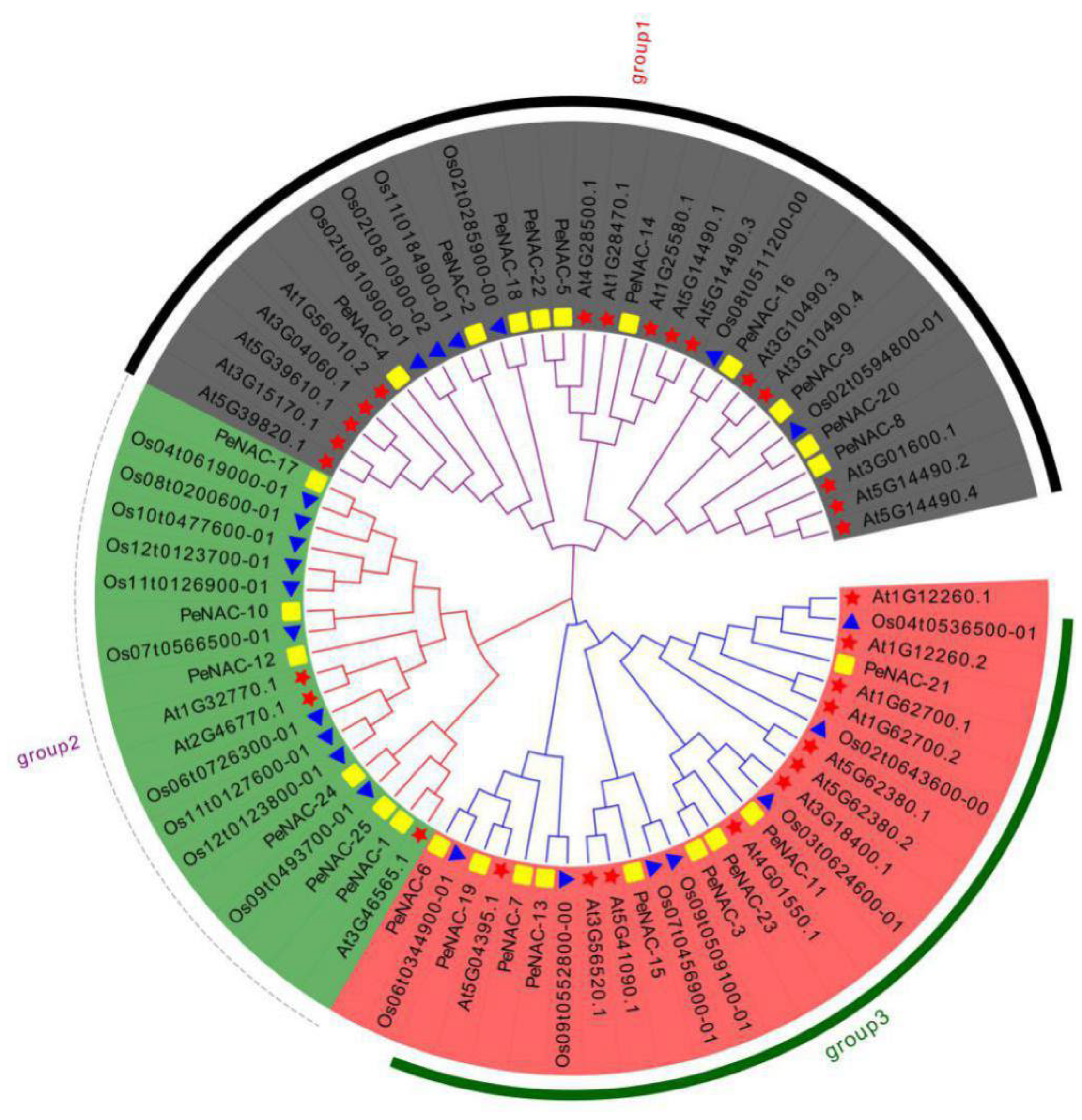
To study the classification and evolutionary relationships of NAC proteins in passion fruit, a phylogenetic tree was constructed by the protein sequences of 25 PeNACs (passion fruit), 29 AtNACs (Arabidopsis) and 23 OsNACs (rice) (Figure 1). According to the kinship of the members, PeNACs were divided into three subfamilies: group 1 (PeNAC-2/4/5/8/9/14/16/18/20/22), group 2 (PeNAC-1/10/12/17/24/25) and group 3 (PeNAC-3/6/7/11/13/15/19/21/23).
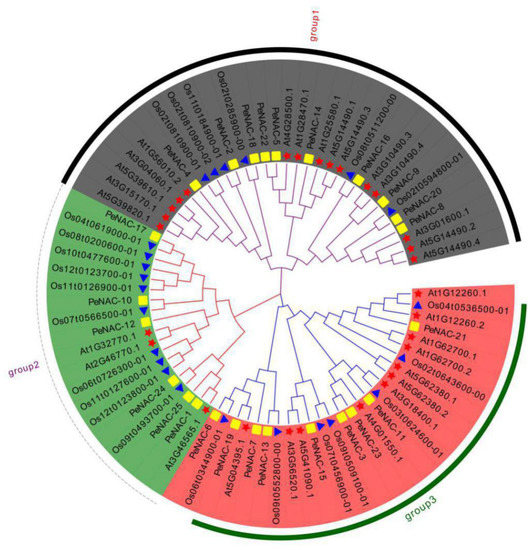
Figure 1.
Phylogenetic evolutionary tree of NACs among the passion fruit, Arabidopsis and rice was constructed by ClustalX 2.0 and MEGA 7.0. The square represents the NACs in passion fruit; the five-pointed star represents the NACs in Arabidopsis; the triangle represents the NACs in rice.
The homologous genes of NAC in passion fruit and Arabidopsis can be inferred due to the fact that passion fruit and Arabidopsis are both dicotyledones. For group 1, PeNAC-4 was the best orthology matches of At1G56010.2. PeNAC-8 was the most homogeneous genes of At3G01600.1. PeNAC-9 were phylogenetically closest to At3G10490.3 and At3G10490.4. PeNAC-14 exhibited the closest relationship with At1G25580.1. For group 2, PeNAC-1 was the best orthology match of At3G46565.1. PeNAC-12 was the most homogeneous gene of At1G32770.1 and At2G46770.1. For group 3, PeNAC-21 was the best orthology match of At1G12260.2. PeNAC-11 was phylogenetically closest to At3G18400.1. PeNAC-15 exhibited the closest relationship with At5G41090.1 and At3G56520.1.
2.3. Expression Pattern of NACs in Passion Fruit
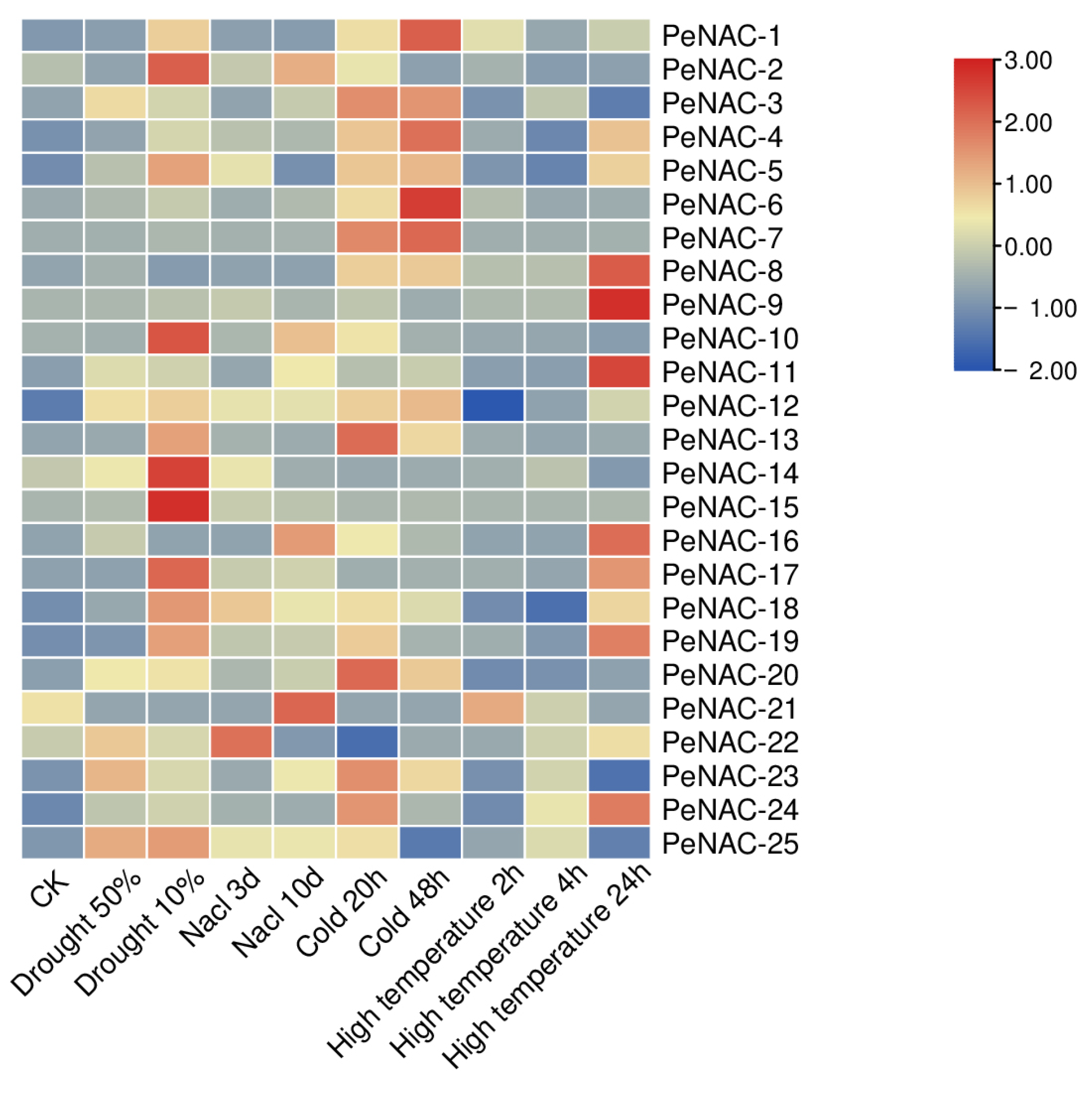
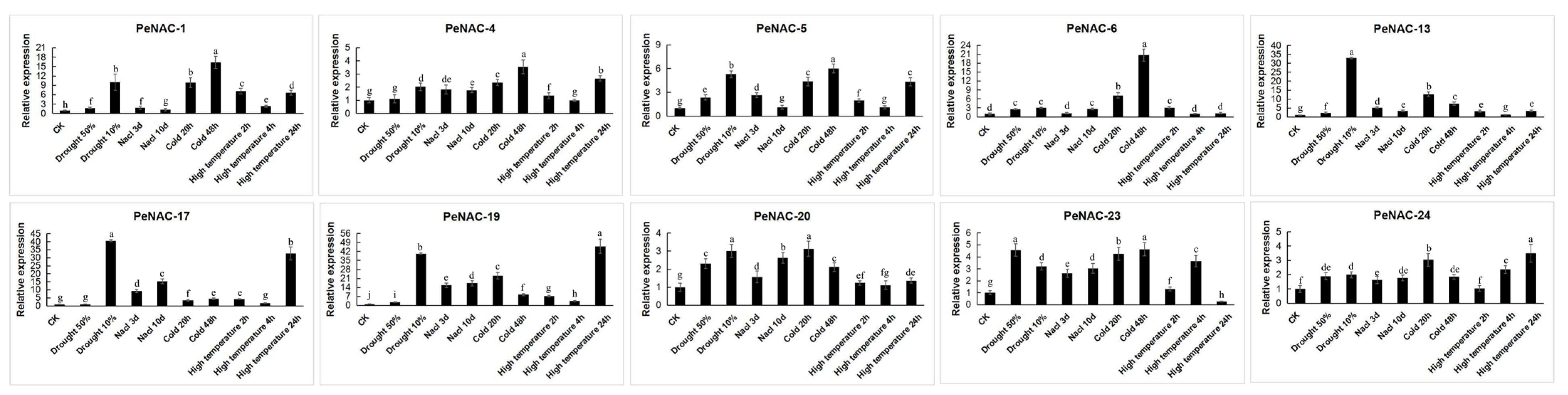
The result of the RNA-seq data showed that the NACs have different response degrees to various abiotic stresses (Figure 2). Nineteen genes were induced by drought stress. The expression of PeNAC-2/5/10/14/15/17/18/25 reached the highest levels when the soil water content was 10%. The expression level of some genes is induced to increase with the degree of salt stress, such as PeNAC-2/3/10/11/16/23/25. The transcript levels of PeNAC-1/6/7/8/13 did not change much with the increase in salt stress. Under cold stress, most genes were upregulated. Among them, the expression of PeNAC-1/3/4/6/7/13/20 displayed significant changes. Under high temperature stress, the expression of the PeNAC-1/4/5/8/9/11/16/17/18/19/24 increased with the degree of stress. The expression of some genes (PeNAC-6/7/10/13/14/15/20) displayed no visible change. The results show that most of the PeNACs can respond to various abiotic stresses (Figure 3).
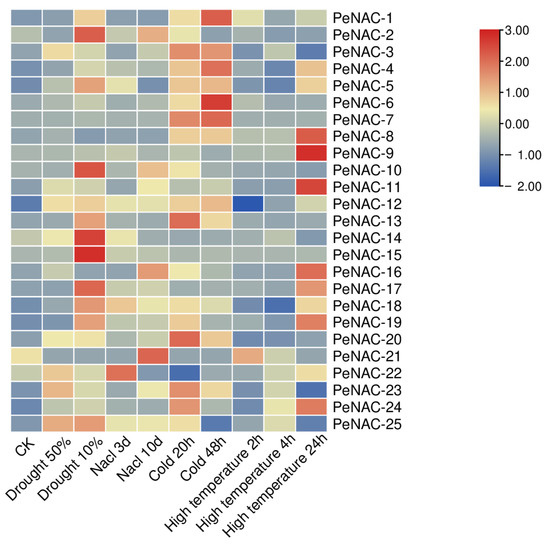
Figure 2.
Expression profiles of PeNACs genes responding to drought (soil moisture of 50% and 10%), salt (300 mM NaCl for 3 d and 10 d), cold (0 °C for 20 h and 48 h) and high temperature (45 °C for 2 h, 4 h and 24 h). The plants of about 1 m and with 8–10 functional leaves were used for abiotic stress treatments. The details are shown in Table S1. A low expression level is shown in blue and a high expression level is shown in red. CK means control check.
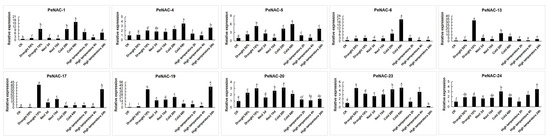
Figure 3.
Expression analysis of 10 PeNACs under different abiotic stresses in the passion fruit. The experimental materials are identical to those used for RNA-seq under the abiotic stresses. The details are shown in Table S2. Data are means ± SD of n = 3 biological replicates. Means denoted by the same letter are not significantly different at p < 0.05 as determined by Duncan’s multiple range test.
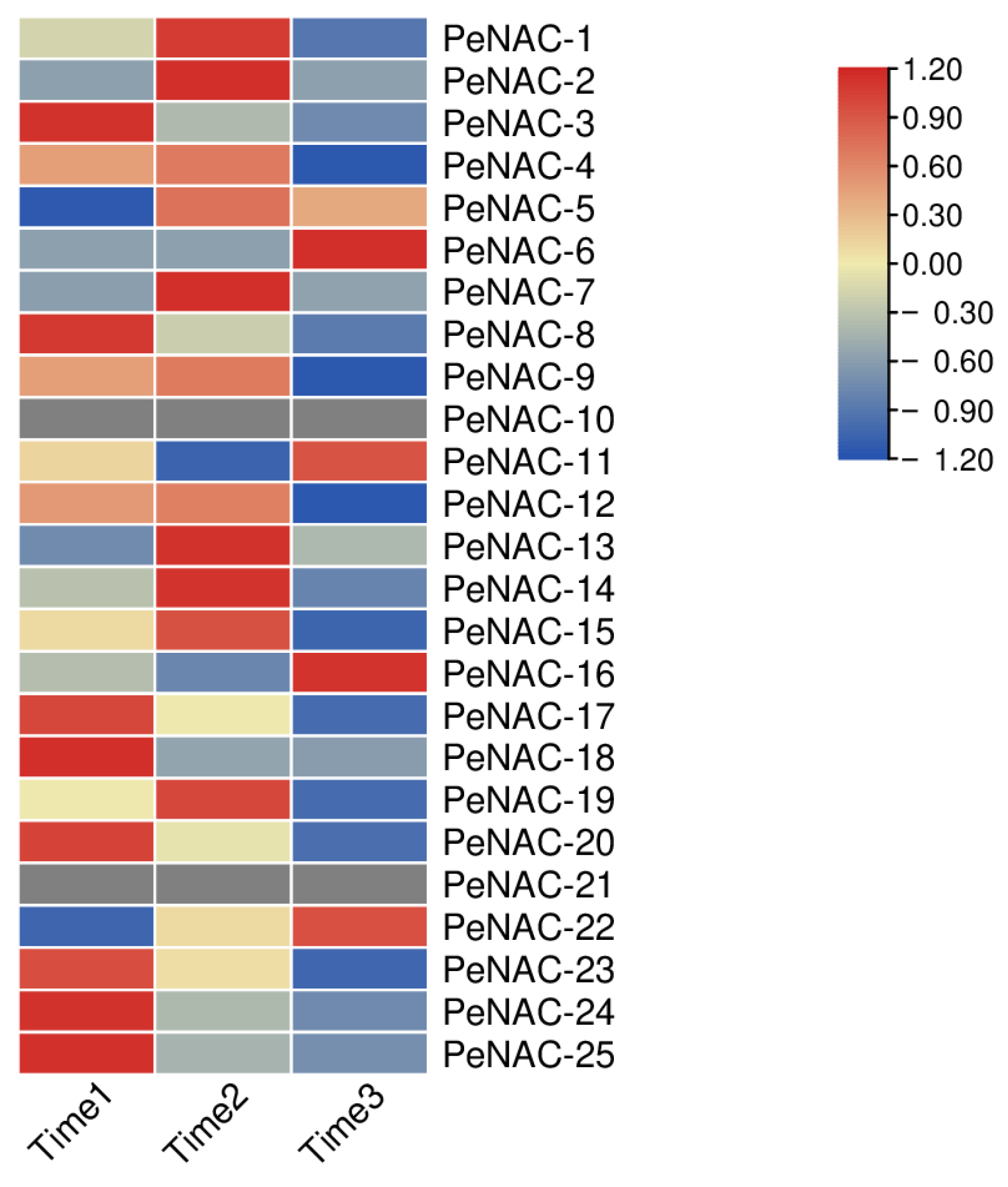
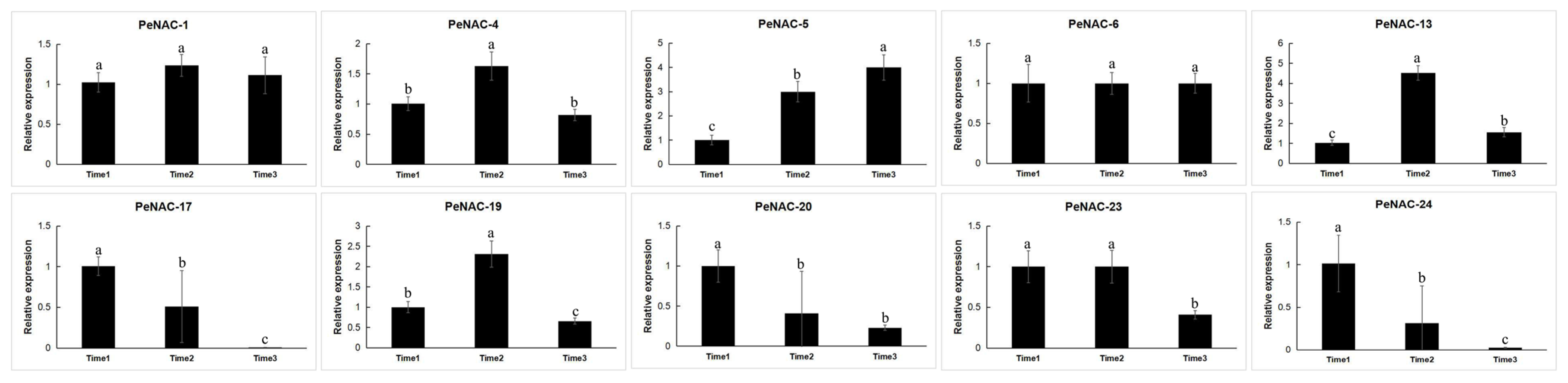
We performed transcript sequencing during three periods: the seventh day before fruit harvest (Time1), the harvest period (Time2) and the seventh day after fruit harvest (Time3) in passion fruit [37], wherein we analyzed the expression levels of the PeNACs (Figure 4). The results show that most of the genes demonstrated the highest expression levels at Time1, and their expression decreased as the fruit matured, with the lowest expression in the third period, such as PeNAC3/8/17/20/23/24/25. Among them, PeNAC22 is most expressed in the third period of fruit maturity. Some genes were verified by qRT-PCR and the results were consistent with the transcript sequencing (Figure 5). This indicated that this gene may be associated with fruit maturity.
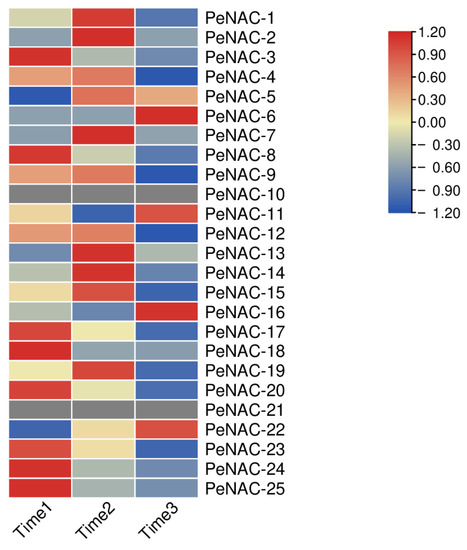
Figure 4.
Expression profiles of PeNACs genes during three fruit-ripening periods (Time1: the 7th day before fruit harvest, Time2: the harvest period, Time3: the 7th day after fruit harvest). The details are shown in Table S3. A low expression level is shown in blue and a high expression level is shown in red. The heat map was generated using TBtools.
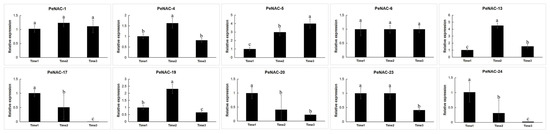
Figure 5.
Expression analysis of 10 PeNACs during three fruit-ripening periods in the passion fruit. The experimental materials are identical to those used for RNA-seq during three fruit-ripening periods. The details are shown in Table S4. Data are means ± SD of n = 3 biological replicates. Means denoted by the same letter are not significantly different at p < 0.05 as determined by Duncan’s multiple range test.
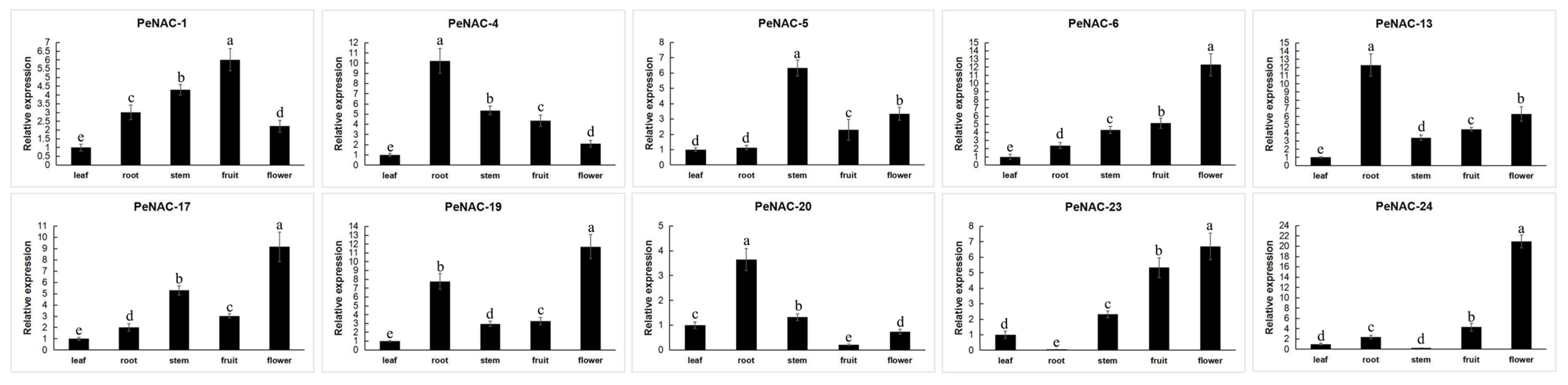
The expression of some PeNACs in different tissues of passion fruit has been analyzed (Figure 6). Among them, four were mainly expressed in the flower (PeNAC-6, PeNAC-17, PeNAC-19, PeNAC-23). PeNAC-1 was mainly expressed in the fruit. Additionally, PeNAC-4/13/20 were mainly expressed in the root. The results show that the gene was expressed in different parts.
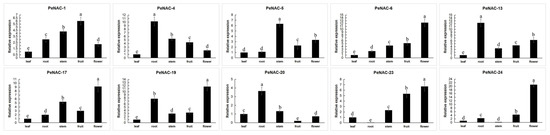
Figure 6.
Expression analysis of 10 PeNACs in the leaf, root, stem, fruit and flower of the passion fruit. The leaf, root, stem, fruit, flower and fruits were obtained from the healthy passion fruit (Passiflora edulis). The details are shown in Table S5. Data are means ± SD of n = 3 biological replicates. Means denoted by the same letter are not significantly different at p < 0.05 as determined by Duncan’s multiple range test.
2.4. Cold Stress Analysis in Transgenic Tobacco
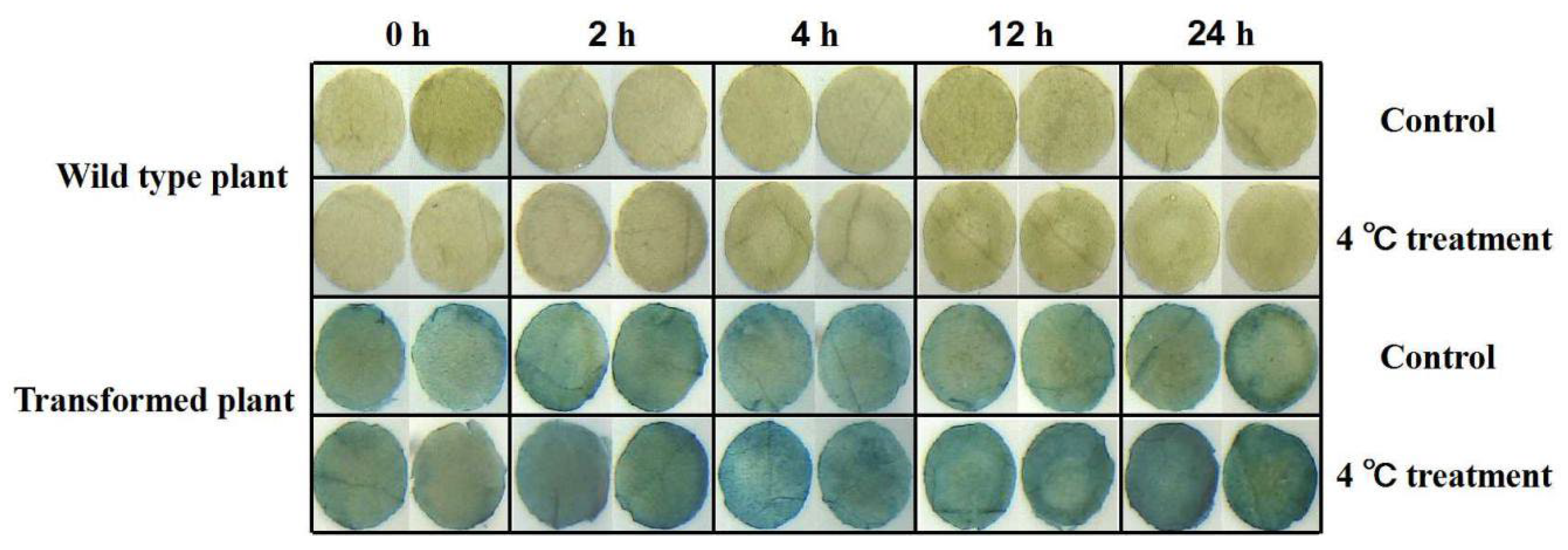
We further transiently transformed tobacco with the promoter of PeNAC-19 (Figure 7). The PeNAC-19p-transformed tobacco and control were subjected to low-temperature stress treatment at 4 °C. The results show that the gene was highly induced during 2 h and 24 h treatments.
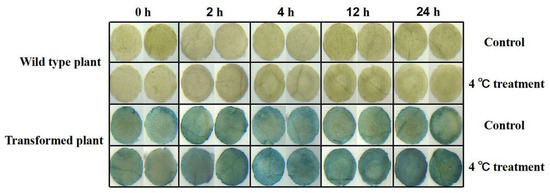
Figure 7.
Gus staining was performed on transgenic tobacco under normal and cold treatment conditions. Tobacco was treated for 0, 2, 4, 12 and 24 h at low temperature (3 replicates per treatment) and then processed into discs (diameter 0.5 cm). The leaf discs that floated under normal growth temperature (25 °C) were used as control.
2.5. Functional Complementation Validation of PeNAC-19
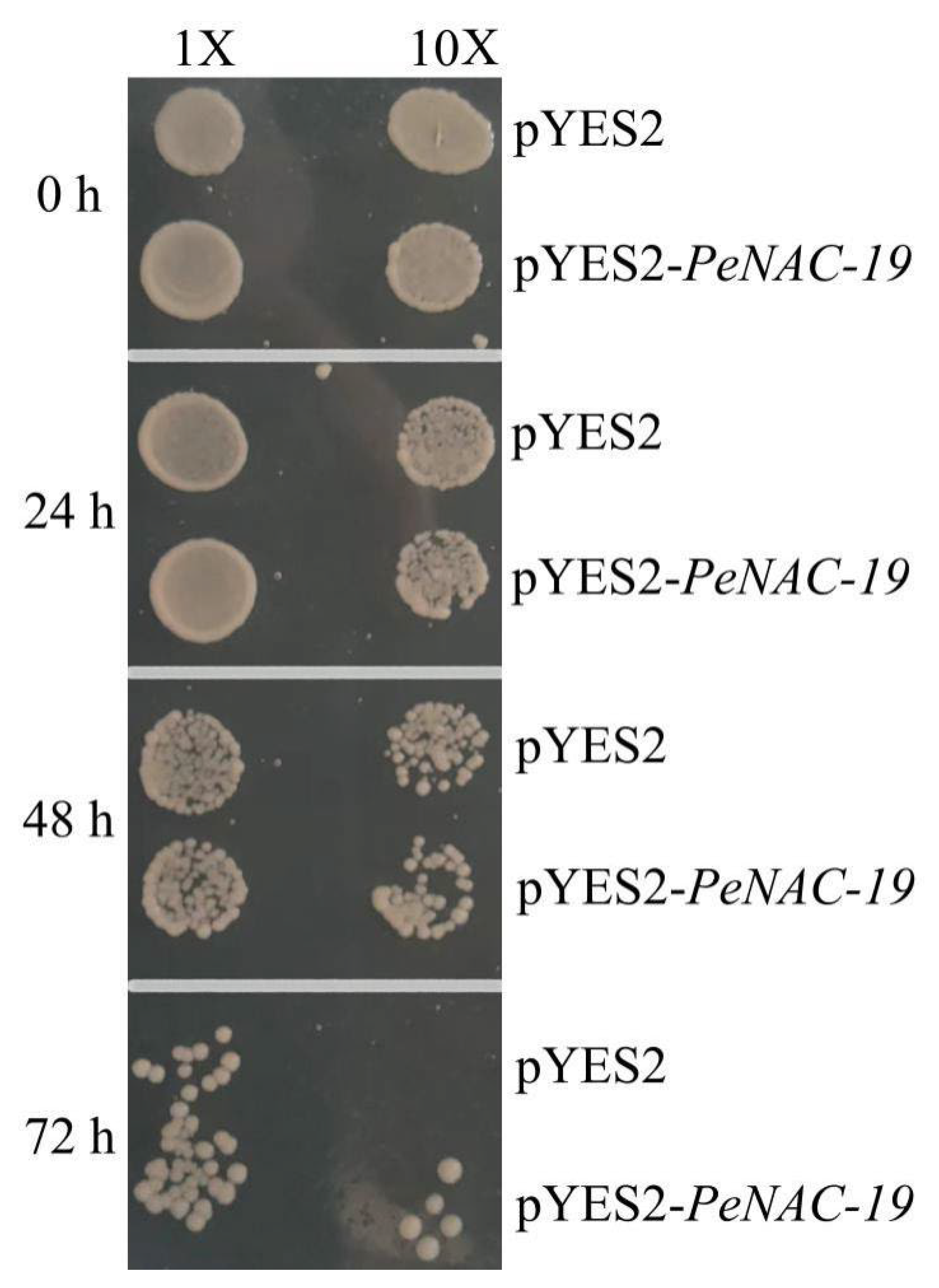
In Figure 8, the pYES2-PeNAC-19 and the pYES2 empty vector (control) were transformed into INVSC1 (Saccharomyces cerevisiae) for the cold-stress experiment (−20 °C for 0 h, 24 h, 48 h and 72 h). The results indicate that the growth of both the empty vector and the transgenic yeast is more and more restricted with increasing treatment time. When treated at −20 °C for 72 h, the PeNAC-19-transformed yeast grew better than the control. This suggests that PeNAC-19 plays a certain role under the cold stress.
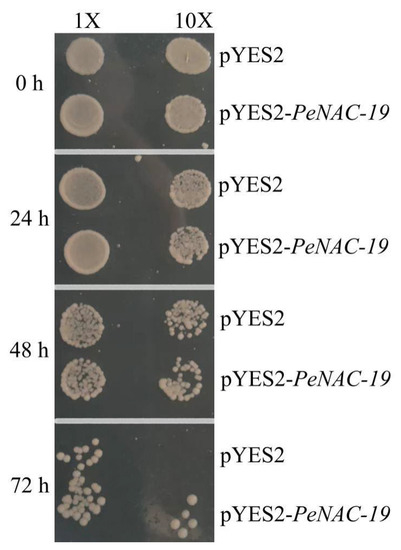
Figure 8.
Growth status of the Saccharomyces cerevisiae INVSc1 strain expressing pYES2–PeNAC-19 and pYES2 (control) under cold stress (−20 °C). 1× means the original bacterial fluid, 10× is the fluid diluted 10 times.
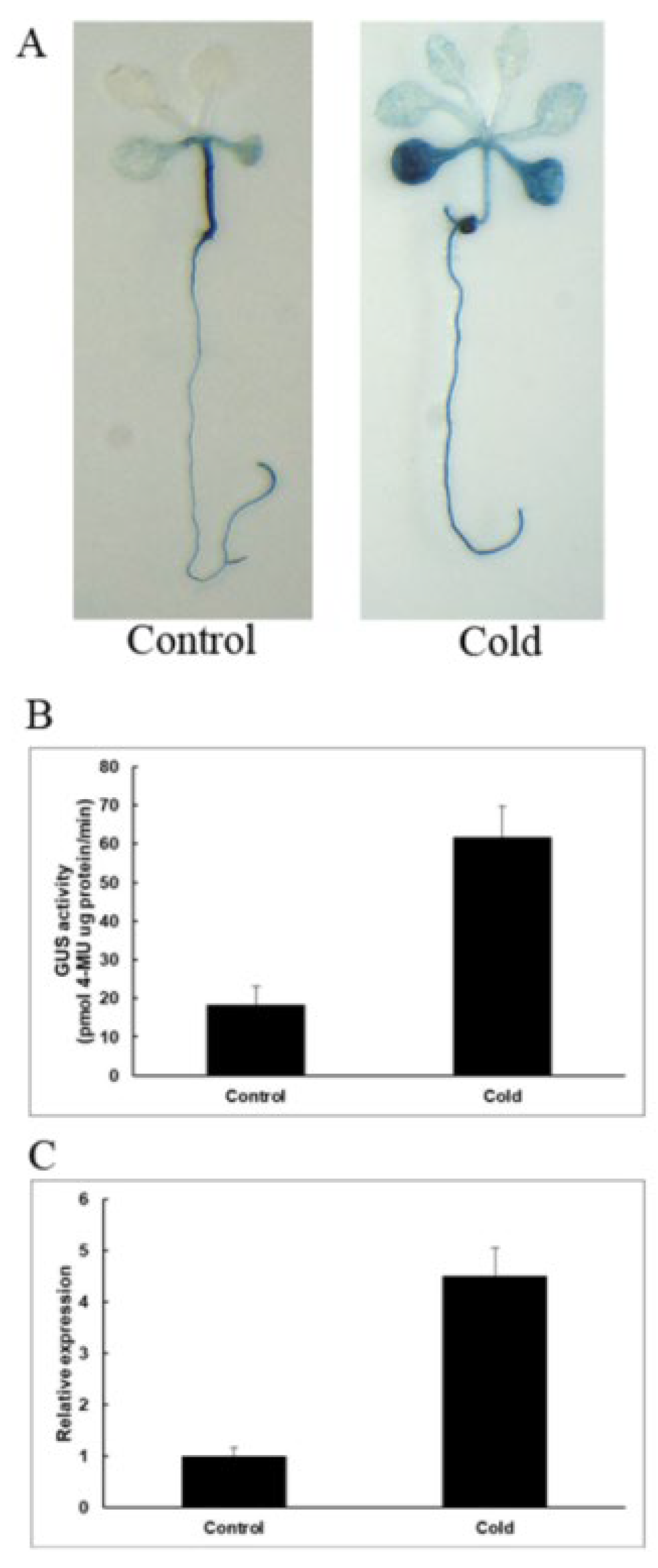
2.6. Response of Transgenic Arabidopsis to Low-Temperature Stress
As shown in Figure 9A, under normal growth conditions, GUS staining was mainly concentrated in stems, and under low-temperature stress, GUS staining was enhanced and mainly concentrated in the leaves and roots of the seedling. The GUS enzyme activity test was carried out, and we found that GUS activities are 3.4-fold higher than the control. The results show that PeNAC-19 was induced by low-temperature stress.
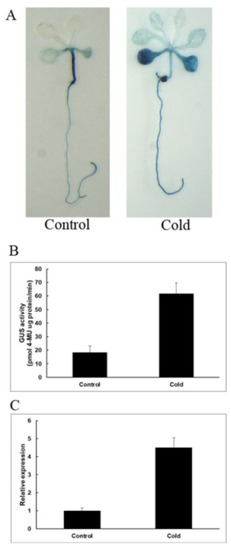
Figure 9.
Induction and expression pattern of PeNAC-19 under low temperature (4 °C for 36 h). (A) GUS staining of transgenic Arabidopsis. (B) GUS enzyme activity analysis of transgenic Arabidopsis thaliana. (C) Expression of PeNAC-19 under the cold stress. Means denoted by the same letter are not significantly different at p < 0.05 as determined by Duncan’s multiple range test.
3. Materials and Methods
3.1. Identification of NAC Genes in Passion Fruit
The passion fruit genome data were downloaded from the NGDC database (https://ngdc.cncb.ac.cn/gwh/Genome/557/show, accessed on 2 April 2021). Additionally, the PeNACs were initially screened and identified using hmmsearch and local blast (Output E value: 1 × 10−5). In addition, analysis was performed using the NAC with the highest comparison value to further identify possible NACs in the passion fruit database. The protein sequences identified by the two methods described above were integrated and resolved to remove redundancy. NAC protein sequences in Arabidopsis and rice were downloaded from databases (http://plants.ensembl.org/Arabidopsis_thaliana/Info/Index, accessed on 2 June 2022, http://plants.ensembl.org/Oryza_sativa/Info/Index, accessed on 3 June 2022). The phylogenetic trees were constructed using MEGA 7.0 software (https://www.megasoftware.net/, accessed on 12 June 2022) [40]. The members of PeNACs were finally obtained through the screening of the above methods.
3.2. Gene Identification
The physical and chemical properties of protein were calculated using the ProtParam website (http://www.expasy.org/tools/protparam.html, accessed on 15 June 2022) (https://meme-suite.org/meme/tools/meme, accessed on 20 June 2022), NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/, accessed on 22 June 2022) and WoLF PSORT 0.2 software (https://www.genscript.com/wolf-psort.html, accessed on 29 June 2022).
3.3. Plant Materials and Growth Conditions
For the abiotic stress treatment, the two-months-old healthy passion fruit seedlings (Passiflora edulis) were chosen. They were grown in soil under a growth chamber (30 °C; 200 µ mol·m−2·s−1 light intensity; 12 h light/12 h dark cycle; 70% relative humidity) to a height of about 1 m and with 8–10 functional leaves, which were subjected to various abiotic stress treatments [36]. For the sampling of fruit during ripening, the fruits were chosen in the 7th day before fruit harvest (Time1), the harvest period (Time2), and the 7th day after fruit harvest (Time3) in passion fruit [37]. Each treatment was repeated three times with 10 plants/fruits at a time, and the material was RNA extracted and RNAseq.
3.4. Heat Map Construction
Transcriptome data used for heat map construction are shown in Tables S1 and S3. The analysis was conducted using TBtools software (https://bio.tools/tbtools, accessed on 10 July 2022) [40].
3.5. Cloning and Vector Construction of PeNAC-19 and the Promoter
The full-length cDNA of PeNAC-19 and a 2000 bp DNA fragment before the start codon of the PeNAC-19 was amplified from the passion fruit (Passiflora edulis). Healthy passion fruit seedlings were used to construct a single-stranded cDNA template.
To examine whether PeNAC-19 responds to low-temperature stress, the expression vectors have been constructed. The PCR products of the PeNAC-19 ORF were inserted into the pCAMBIA1304 (Cambia, Australia) expression vector, which is called pCAMBIA1304-PeNAC-19. The PCR products of the PeNAC-19 promoter were cloned into the pCAMBIA1304 vector called pCAMBIA1304-PeNAC-19p. The vectors were transferred into the Agrobacterium strain EHA105.
3.6. Cold Stress Treatment in Wild-Type and Transgenic Plants
The two-month-old tobacco leaves were used for transient expression experiments. The Agrobacterium transformed with pCAMBIA1304-PeNAC-19p was shaken at 28 °C to OD 600 = 0.8–1.0, and the bacterial solution was infected with an equal volume of MES buffer (10 mM MES; 10 mM MgCl2; 200 mM MAS) and then kept in the dark at room temperature for 2–3 h. After that, the solution had been injected into the back of the tobacco which was left to culture for 2–3 d. The plants were treated at 4 °C and 25 °C for 0, 2, 4, 12 and 24 h. The leaf discs with a diameter of 0.5 cm were cut out for further staining testing.
The Agrobacterium transformed with pCAMBIA1304-PeNAC-19 and pCAMBIA1304-PeNAC19p were shaken at 28 °C in YEB medium, and then added to 1/2 MS solution until OD 600 = 0.8–1.0. The transformation of Arabidopsis thaliana was conducted using the inflorescence dip method. The seeds of Arabidopsis thaliana were disinfected with 75% ethanol and spreading on the medium (1/2MS, 25 μg/mL hygromycin B). After culturing in the dark at 4 °C for 3 days, the seeds were transferred to a culture at 23 °C for growth. After ten days, the normal growth seedlings were the T1 generation. Additionally, the T2 generation grown in the selection medium were for further experimental analysis. The 14-day-old transgenic Arabidopsis with about 6–8 leaves was treated in an incubator at 4 °C and 23 °C for 36 h.
3.7. GUS Activity Detection
The fresh transgenic tobacco and Arabidopsis samples under the normal and cold conditions were placed in X-Gluc solution (GBT, St. Louis, MO, USA) [41] for histochemical analysis. Gus enzyme activity was determined by 4-methyl umbelliferate glucuronide fluorescence method [42].
3.8. Functional Complementation of PeNAC-19 in Yeast
The pYES2–PeNAC-19 vector was constructed using the full-length cDNA of PeNAC-19. Then, the pYES2–PeNAC-19 and pYES2 vector (control) were transfected into the INVSc1 Strain (Saccharomyces cerevisiae). To perform the yeast complementation assays, the yeast liquid was cultured in SD-Ura liquid medium at 30 °C until OD600 = 1.0 and treated under the cold stress [43]. The experiment was repeated three times.
3.9. RNA Extraction, Transcriptome Sequencing and qRT-PCR
Total RNA was extracted from frozen samples with plant RNA isolation kit (Fuji, China, Chengdu), and three biological replicates were performed. The cDNA was used for transcriptome sequencing analysis and quantitative real-time polymerase chain reaction (qRT-PCR). The SYBR® Premix Ex Taq™ kit (TaKaRa, Tokyo, Japan) was used to detect in qRT-PCR. Relative expression levels were calculated using the 2−∆∆CT method and normalized to the PeNACs.
4. Discussion
Passion fruit (Passiflora edulis Sim), a perennial vine, is rich in nutrients and contains a variety of amino acids and vitamins. Due to its short growth cycle and good economic value, it is very popular [36,37]. However, only a few studies have investigated its function [44]. Previous studies have shown that transcription factors can regulate plant growth and development and enhance plant responses to abiotic stresses [45]. The NAC family, which is one of the largest transcription factor families, plays an important role in plant growth, development and abiotic stress responses [46].
In this study, we have reported 25 PeNACs in passion fruit, and performed the expression patterns analysis under different abiotic stresses and during different fruit-ripening stages. At the same time, a low-temperature stress-inducible gene, PeNAC-19, was screened for transient expression in tobacco, transgenic Arabidopsis and in vivo in yeast. These results provide evidence for the response of PeNAC members under cold stress. In physical and chemical property terms, the isoelectric point of the PeNAC protein ranges from 3.84 to 10.44, with an average of 6.38, which is consistent with BjuNAC [47]. All PeNAC family members are subcellularly localized in the nucleus by prediction. This was consistent with the localization of most transcription factors.
In general, the expression level of a gene determines its function [48]. Transcription factors usually play a key role in regulating the expression of tissue-specific genes [49]. In this study, we have selected some PeNAC genes for qRT-PCR analysis of each tissue, and the results show that some genes exhibited higher expression in the roots, such as PeNAC-4/13/20. Similar results were also shown in other plants, such as orchardgrass (Dactylis glomerata L.) [45], Fagopyrum tataricum [50], Panicum miliaceum [51] and Triticum aestivum [52]. In orchards, DgNAC046/087/103 had the highest expression in the stems, which may play an important role in stem development. In this research, the expression of PeNAC-6/19/17/23 is higher in flowers, which may be related to the development of floral organs. In addition, the overexpression of the tissue’s specifically expressed NAC gene, poplar NAC15, was able to promote wood formation [53]. In orchardgrass, DgNAC genes are extensively involved in tissue development [45]. The NAC domain transcription factor PdWND3A affects lignin biosynthesis [54].
The relationship between NAC family members and plant abiotic stress has also been reported in many species. In B. juncea var. tumida, some NAC genes such as BjuNAC112, BjuNAC178, BjuNAC184 and BjuNAC240 can respond to high-temperature stress [55]. In addition, in tobacco, the overexpression of LpNAC13 in Lilium pumilum reduced the tolerance to drought stress, but positively regulated the response to salt stress [47]. In Cerasus humilis, ChNAC1 positively regulates the expression of abscisic acid (ABA)-responsive genes, and its overexpressed plants exhibited higher drought tolerance [25,56]. Arabidopsis AtNTL6 expressed the highest expression at low temperature for 18 h [57]. OsNAC6/SNAC2 overexpression could improve the drought, salt and low-temperature tolerance of rice [23,24]. SlNAC1 from Suaeda liaotungensis and VvNAC1 from Vitis vinifera L. can positively regulate the cold resistance of transgenic plants [58,59]. In wheat (Triticum aestivum L.), TaNAC2/47/67 can improve cold tolerance in transgenic plants [60,61,62]. GmNAC20 transgenic rice has stronger salt tolerance and cold tolerance by regulating the expression of abiotic stress-response genes [63].
In this research, one member of the PeNACs, PeNAC-19, was induced in the different abiotic stresses. PeNAC-19 was induced by four various abiotic stresses. It may be a candidate gene for stress-resistant breeding. At present, low temperatures have seriously endangered the development of passion fruit cultivation. Therefore, we first focus on the relationship between PeNAC-19 and cold stress. The results show that PeNAC-19 responded to cold stress significantly in tobacco and Arabidopsis, and could improve the low-temperature tolerance of yeast. This study lays a foundation for the functional study of PeNAC gene family members under fruit ripening and abiotic stress.
5. Conclusions
In this study, the NAC family members in passion fruit were identified and analyzed, and the transcriptome results at different fruit-ripening stages and abiotic stresses were verified by qRT-PCR. The expression of one of the NAC genes (PeNAC-19) was induced by cold stress. Further verification of this gene showed that it could enhance the ability of transgenic tobacco, Arabidopsis and yeast to resist cold stress. The results lay a good foundation for further studies on the ability of the passion fruit to resist to abiotic stresses.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants12061393/s1, Table S1. The transcriptome data of passion fruit under the drought, salt, cold and high-temperature treatment. Table S2. qPCR data of PeNACs under the drought, salt, cold and high-temperature treatment. Table S3. The transcriptome data of passion fruit in the three stages of fruit ripening. Table S4. qPCR data of PeNACs in the three stages of fruit ripening. Table S5. qPCR data of PeNACs in different tissues. Table S6. A list of the oligo primers of PeNACs used for qRT-PCR.
Author Contributions
Experiments were performed by P.L., F.M., W.X., D.H., B.W. and P.S. who analyzed the data; Y.X. and S.S. drafted the manuscript; F.M. and B.X. supervised the experiments and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was sponsored by National Natural Science Foundation of China (32260737), Project of Sanya Yazhou Bay Science and Technology City (SCKJ-JYRC-2022-93, SCKJ-JYRC-2022-84) and Hainan Provincial Natural Science Foundation of China (321RC1088, 321MS091).
Data Availability Statement
The raw sequence data have been deposited in the National Genomics Data Center (NGDC), and the link is https://ngdc.cncb.ac.cn/gwh/Genome/557/show accessed on 2 April 2021. All datasets are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Riaño-Pachón, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jin, J.; Tang, L.; Zhao, Y.; Gu, X.; Gao, G.; Luo, J. PlantTFDB 2.0: Updateand improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011, 39 (Suppl. S1), D1114–D1117. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.; Samaha, R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Wray, G.A.; Hahn, M.W.; Abouheif, E.; Balhoff, J.P.; Pizer, M.; Rockman, M.V.; Romano, L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristemgene of Petunia is required for pattern formation in embryos and flowersand is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Kikuchi, K.; Ueguchi-Tanaka, M.; Yoshida, K.; Nagato, Y.; Matsusoka, M.; Hirano, H.-Y. Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 2000, 262, 1047–1051. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Welner, D.; Deeba, F.; Lo Leggio, L.; Skriver, K. NAC transcription factors: From structure to function in stress-associated networks. In Plant Transcription Factors; Gonzalez, D.H., Ed.; Academic Press: Boston, MA, USA, 2015; pp. 199–212. [Google Scholar]
- Jensen, M.K.; Skriver, K. NAC transcription factor gene regulatory and protein–protein interaction networks in plant stress responses and senescence. IUBMB Life 2014, 66, 156–166. [Google Scholar] [CrossRef]
- Mathew, I.E.; Agarwal, P. May the fittest protein evolve: Favoring the plant-specific origin and expansion of NAC transcription factors. BioEssays 2018, 40, 1800018. [Google Scholar] [CrossRef]
- Zhu, G.; Chen, G.; Zhu, J.; Zhu, Y.; Lu, X.; Li, X.; Hu, Y.; Yan, Y. Molecular characterization and expression profiling of NAC transcription factors in Brachypodium distachyon L. PLoS ONE 2015, 10, e0139794. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fan, R.Y.; Yang, Q.S.; Hu, C.H.; Sheng, O.; Deng, G.M.; Dong, T.; Li, C.Y.; Peng, X.X.; Bi, F.C.; et al. Genome-Wide Identification and Characterization of the NAC Transcription Factor Family in Musa Acuminata and Expression Analysis during Fruit Ripening. Int. J. Mol. Sci. 2020, 21, 634. [Google Scholar] [CrossRef] [PubMed]
- Min, X.Y.; Jin, X.Y.; Zhang, Z.S.; Wei, X.Y.; Ndayambaza, B.; Wang, Y.R.; Liu, W.X. Genome-Wide Identification of NAC Transcription Factor Family and Functional Analysis of the Abiotic Stress-Responsive Genes in Medicago sativa L. J. Plant Growth Regul. 2020, 39, 324–337. [Google Scholar] [CrossRef]
- Nigarish, M.; Chen, Y.K.; Chen, X.H.; Azher, N.M.; Junaid, I.; Muhammad, R.H.; Shen, X.; Lin, Y.L.; Xu, X.H.; Lai, Z.X. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression patterns during somatic embryogenesis in Dimocarpus longan Lour. Plant Physiol. Biochem. 2020, 157, 169–184. [Google Scholar]
- Feng, J.D.; Qiang, J.Z.; Hui, F.H.; Chen, L.; Liao, G.L.; He, Y.Q.; Huang, C.H.; Xu, X.B. Genome-wide identification and comprehensive analysis of NAC family genes involved in fruit development in kiwifruit (Actinidia). BMC Plant Biol. 2021, 21, 44. [Google Scholar]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zheng, Y.; Xin, H.; Fang, L.; Li, S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Yan, X.; Li, J.; Yang, Q.; Jamil, W.; Teng, Y.; Bai, S. Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biol. 2018, 18, 214. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Choi, Y.D.; Kim, M.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Han, D.G.; Du, M.; Zhou, Z.Y.; Wang, S.; Li, T.M.; Han, J.X.; Xu, T.L.; Yang, G.H. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef]
- Jin, C.; Li, K.Q.; Xu, X.Y.; Zhang, H.P.; Chen, H.X.; Chen, Y.H.; Hao, J.; Wang, Y.; Huang, X.S.; Zhang, S.L. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef]
- Li, X.D.; Zhuang, K.Y.; Liu, Z.M.; Yang, D.Y.; Ma, N.N.; Meng, Q.W. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco. J. Plant Physiol. 2016, 204, 54–65. [Google Scholar] [CrossRef]
- Giovannoni, J. Molecular Biology of Fruit Maturation and Ripening. Annu. Rev. Plant Biol. 2001, 52, 725–749. [Google Scholar] [CrossRef]
- Wang, J.F.; Wang, Y.P.; Zhang, J.; Ren, Y.; Li, M.Y.; Tian, S.W.; Yu, Y.T.; Zuo, Y.; Gong, G.Y.; Zhang, H.Y.; et al. The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 2021, 8, 214. [Google Scholar] [CrossRef]
- Kerstin, D.; Johanna, W.J.; Miguel, N.G.; Almuth, H.; Karl, L.; Ines, E.; Malin, E. Overexpression of PaNAC03; a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biol. 2017, 17, 6. [Google Scholar]
- Zhang, S.L.; Dong, R.Z.; Wang, Y.W.; Li, X.M.; Ji, M.M.; Wang, X.P. NAC domain gene VvNAC26 interacts with VvMADS9 and influences seed and fruit development. Plant Physiol. Biochem. 2021, 164, 63–72. [Google Scholar] [CrossRef]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef]
- Carrasco-Orellana, C.; Stappung, Y.; Mendez-Yañez, A.; Allan, A.C.; Espley, R.V.; Plunkett, B.J.; Moya-Leon, M.A.; Herrera, R. Characterization of a ripeningrelated transcription factor FcNAC1 from Fragaria chiloensis fruit. Sci. Rep. 2018, 8, 10524. [Google Scholar] [CrossRef]
- Huang, D.; Xu, Y.; Wu, B.; Ma, F.N.; Song, S. Comparative analysis of basic quality of passion fruits (Passiflora edulis sims) in Guangxi, Guizhou and Fujian, China. Banglandesh J. Bot. 2019, 48, 901–906. [Google Scholar]
- Costa, J.L.; Jesus, O.N.D.; Oliverira, G.A.F.; Oliverira, E.J.D. Effect of selection on genetic variability in yellow passion fruit. Crop Breed. Appl. Biotechnol. 2012, 12, 253–260. [Google Scholar] [CrossRef]
- Song, S.; Zhang, D.; Ma, F.; Xing, W.; Huang, D.; Wu, B.; Chen, J.; Chen, D.; Xu, B.; Xu, Y. Genome-Wide Identification and Expression Analyses of the Aquaporin Gene Family in Passion Fruit (Passiflora edulis), Revealing PeTIP3-2 to Be Involved in Drought Stress. Int. J. Mol. Sci. 2022, 23, 5720. [Google Scholar] [CrossRef]
- Xia, Z.; Huang, D.; Zhang, S.; Wang, W.; Ma, F.; Wu, B.; Xu, Y.; Xu, B.; Chen, D.; Zou, M.; et al. Chromosome-scale genome assembly provides insights into the evolution and flflavor synthesis of passion fruit (Passiflflora edulis Sims). Hortic. Res. 2021, 8, 14. [Google Scholar] [CrossRef]
- Yang, Q.; Li, B.; Rizwan, H.M.; Sun, K.; Zeng, J.; Shi, M.; Guo, T.; Chen, F. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression analysis under Fusarium kyushuense and drought stress conditions in Passiflora edulis. Front. Plant Sci. 2022, 13, 972734. [Google Scholar] [CrossRef]
- Ma, D.; Dong, S.; Zhang, S.; Wei, X.; Xie, Q.; Ding, Q. Chromosome-level reference genome assembly provides insights into aroma biosynthesis in passion fruit (Passiflora edulis). Mol. Ecol. Resour. 2021, 21, 955–968. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4, molecular evolutionary genetics analysis MEGA software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatilegene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Xu, Y.; Jin, Z.Q.; Xu, B.Y.; Li, J.Y.; Li, Y.J.; Wang, X.Y.; Wang, A.B.; Hu, W.; Huang, D.M.; Wei, Q.; et al. Identification of transcription factors interacting with a 1274bp promoter of MaPIP1;1 which confers high-level gene expression and drought stress Inducibility in transgenic Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 278. [Google Scholar] [CrossRef]
- Wang, B.F.; Wang, Y.C.; Zhang, D.W.; Li, H.Y.; Yang, C.P. Verification of the resistance of a LEA gene from Tamarix expression in Saccharomyces cerevisiae to abiotic stresses. J. For. Res. 2008, 19, 58–62. [Google Scholar] [CrossRef]
- Li, X.; Cai, K.; Pei, X.; Li, Y.; Hu, Y.; Meng, F.; Song, X.; Tigabu, M.; Ding, C.; Zhao, X. Genome-Wide Identification of NAC Transcription Factor Family in Juglans mandshurica and Their Expression Analysis during the Fruit Development and Ripening. Int. J. Mol. Sci. 2021, 22, 12414. [Google Scholar] [CrossRef]
- Yang, Z.F.; Nie, G.; Feng, G.Y.; Han, J.T.; Huang, L.K.; Zhang, X.Q. Genome-wide identification, characterization and expression analysis of the NAC transcription factor family in orchardgrass (Dactylis glomerata L.). BMC Genom. 2021, 22, 178. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, S.J.; Guan, C.J.; Kong, X.; Wang, Y.P.; Cui, Y.; Liu, B.; Zhou, Y.W.; Zhang, Y.N. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, S.; Chen, F.; Liu, B.; Wu, L.; Li, F.; Zhang, J.; Bao, M.; Liu, G. Genome-wide identification and characterization of the SBP-box gene family in Petunia. BMC Genom. 2018, 19, 193. [Google Scholar] [CrossRef]
- Naya, F.J.; Stellrecht, C.; Tsai, M.J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995, 9, 1009–1019. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef]
- Shan, Z.; Jiang, Y.; Li, H.; Guo, J.; Dong, M.; Zhang, J.; Liu, G. Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genom. 2020, 21, 96. [Google Scholar] [CrossRef]
- Guerin, C.; Roche, J.; Allard, V.; Ravel, C.; Mouzeyar, S.; Bouzidi, M.F. Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. aestivum L.). PLoS ONE 2019, 14, e0213390. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Zhang, D.; Zhou, B.; Wang, J.; Li, R.; Jiang, T. Over-expression of poplar NAC15 gene enhances wood formation in transgenic tobacco. BMC Plant Biol. 2020, 20, 12. [Google Scholar] [CrossRef]
- Yang, Y.; Yoo, C.G.; Rottmann, W.; Winkeler, K.A.; Collins, C.M.; Gunter, L.E.; Jawdy, S.S.; Yang, X.; Pu, Y.; Ragauskas, A.J. PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in Populus. BMC Plant Biol. 2019, 19, 486. [Google Scholar] [CrossRef]
- Jiang, L.; Sun, Q.; Wang, Y.; Chang, P.; Kong, H.; Luo, C.; He, X. Genome-wide identification and characterization of NAC genes in Brassica juncea var tumida. PeerJ 2021, 9, e11212. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, J.W.; Sun, L.J.; Song, X.S. The molecular cloning and functional characterization of ChNAC1; a NAC transcription factor in Cerasus humilis. Plant Growth Regul. 2019, 89, 331–343. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, C.M. A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal. Behav. 2010, 5, 481–483. [Google Scholar] [CrossRef]
- Li, X.L.; Yang, X.; Hu, Y.X.; Yu, X.D.; Li, Q.L. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014, 33, 767–778. [Google Scholar] [CrossRef]
- Le, H.G.; Profizi, C.; Courteaux, B.; Rabenoelina, F.; Gérard, C.; Clément, C.; Baillieul, F.; Cordelier, S.; Dhondtcordelier, S. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 2013, 64, 4877–4893. [Google Scholar]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Jia, J.; Kong, X. The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 2015, 6, 1174. [Google Scholar] [CrossRef]
- Mao, X.; Chen, S.; Li, A.; Zhai, C.; Jing, R. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 2014, 9, e84359. [Google Scholar] [CrossRef] [PubMed]
- Yarra, R.; Wei, W. The NAC-type transcription factor GmNAC20 improves cold, salinity toleranced lateral root formation in transgenic rice plants. Funct. Integr. Genom. 2021, 21, 473–487. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).