Genome-Wide Analysis and Abiotic Stress-Responsive Patterns of COBRA-like Gene Family in Liriodendron chinense
Abstract
:1. Introduction
2. Results
2.1. Identification of the COBL Gene Family in the L. chinense Genome
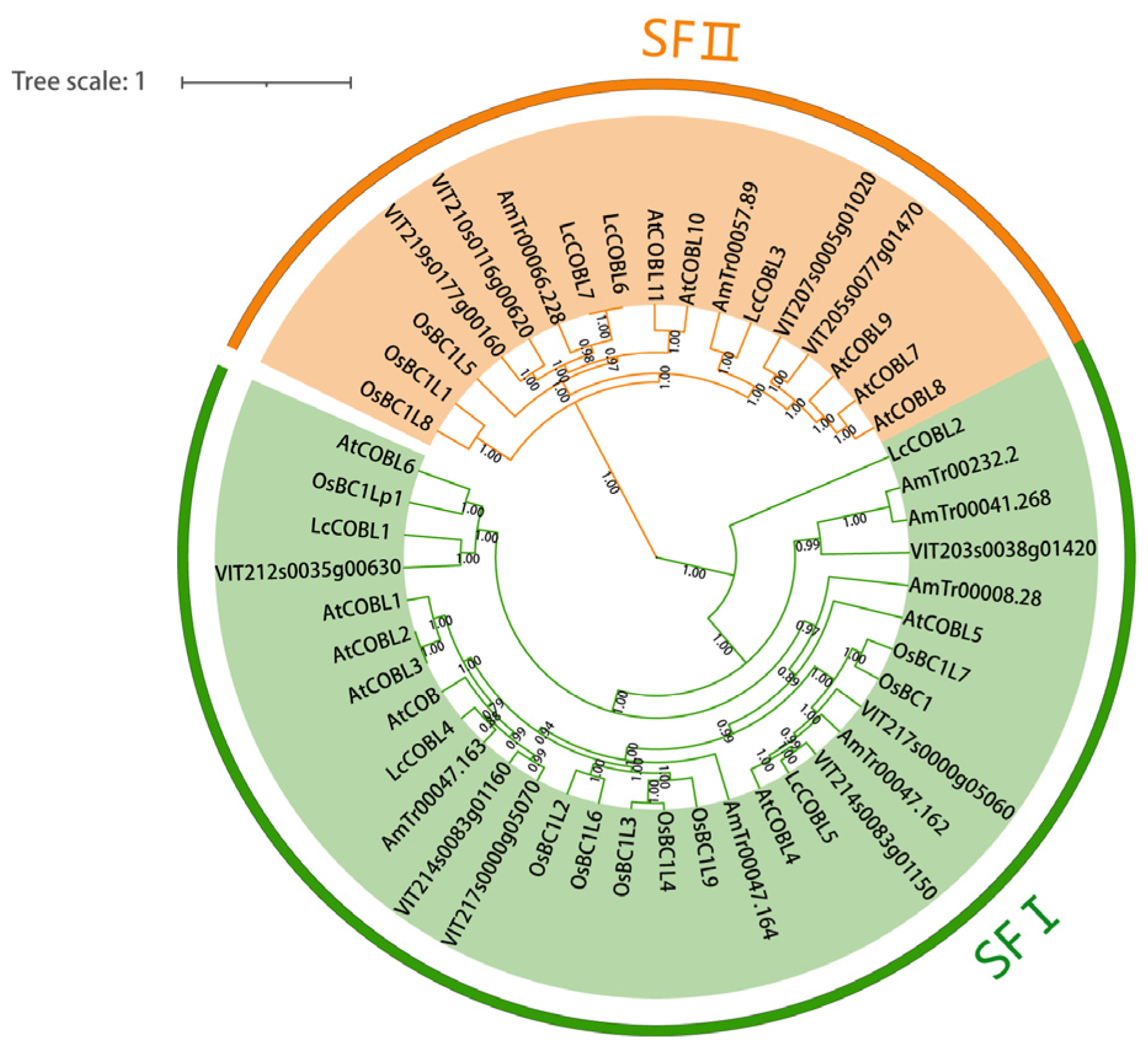
2.2. Phylogenetic Analysis of LcCOBL Proteins
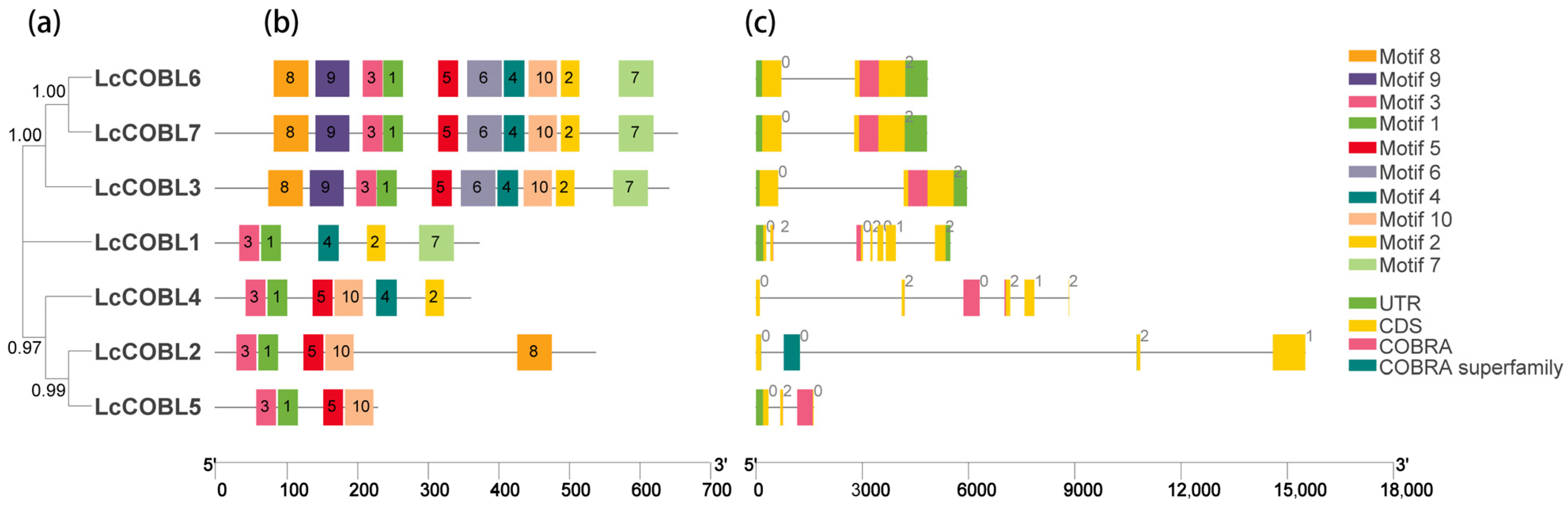
2.3. Analyses of Locations, Structures and Conserved Motifs of LcCOBL Genes
2.4. Cis-Element Analysis of LcCOBL Promoters
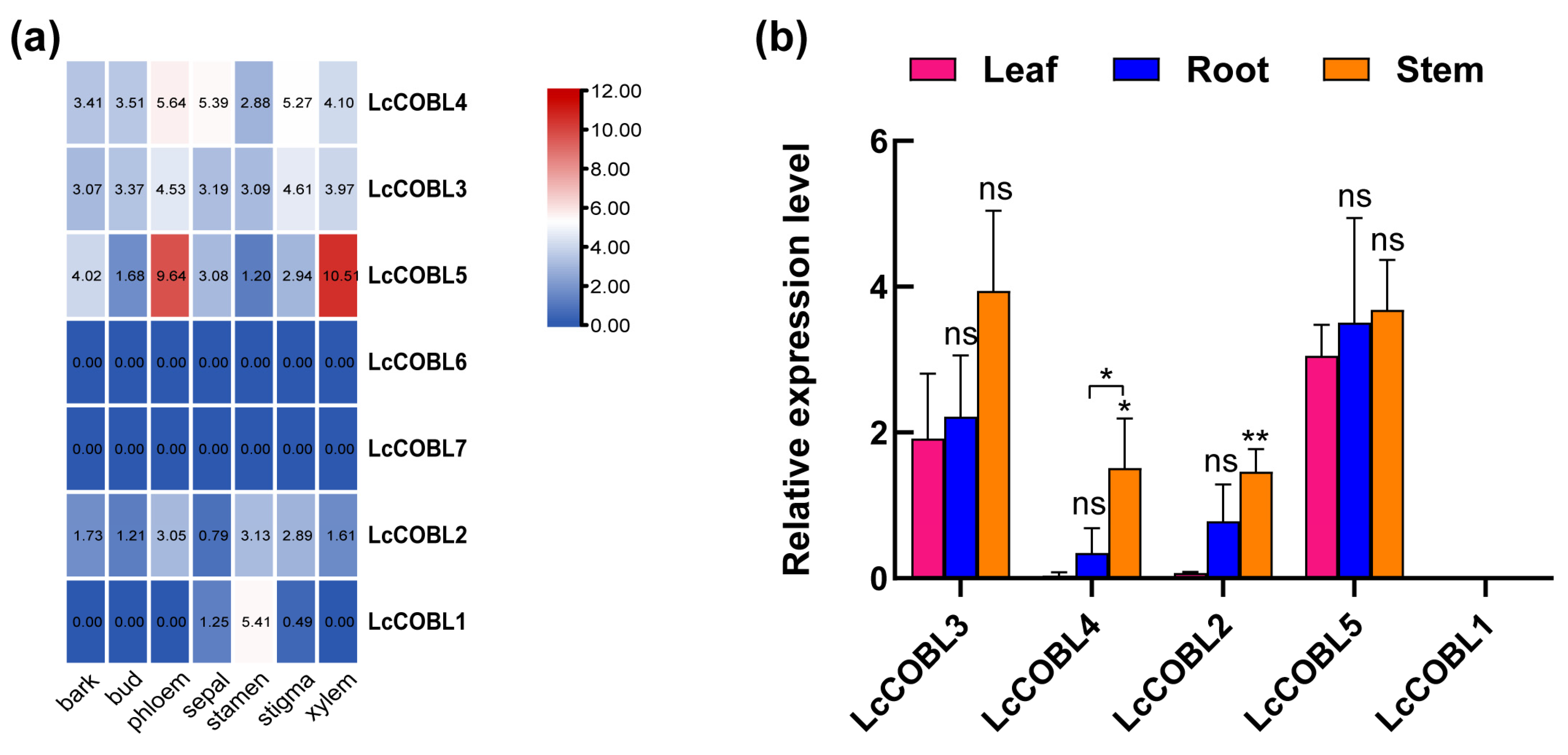
2.5. Organspecific Expression Patterns of LcCOBLs
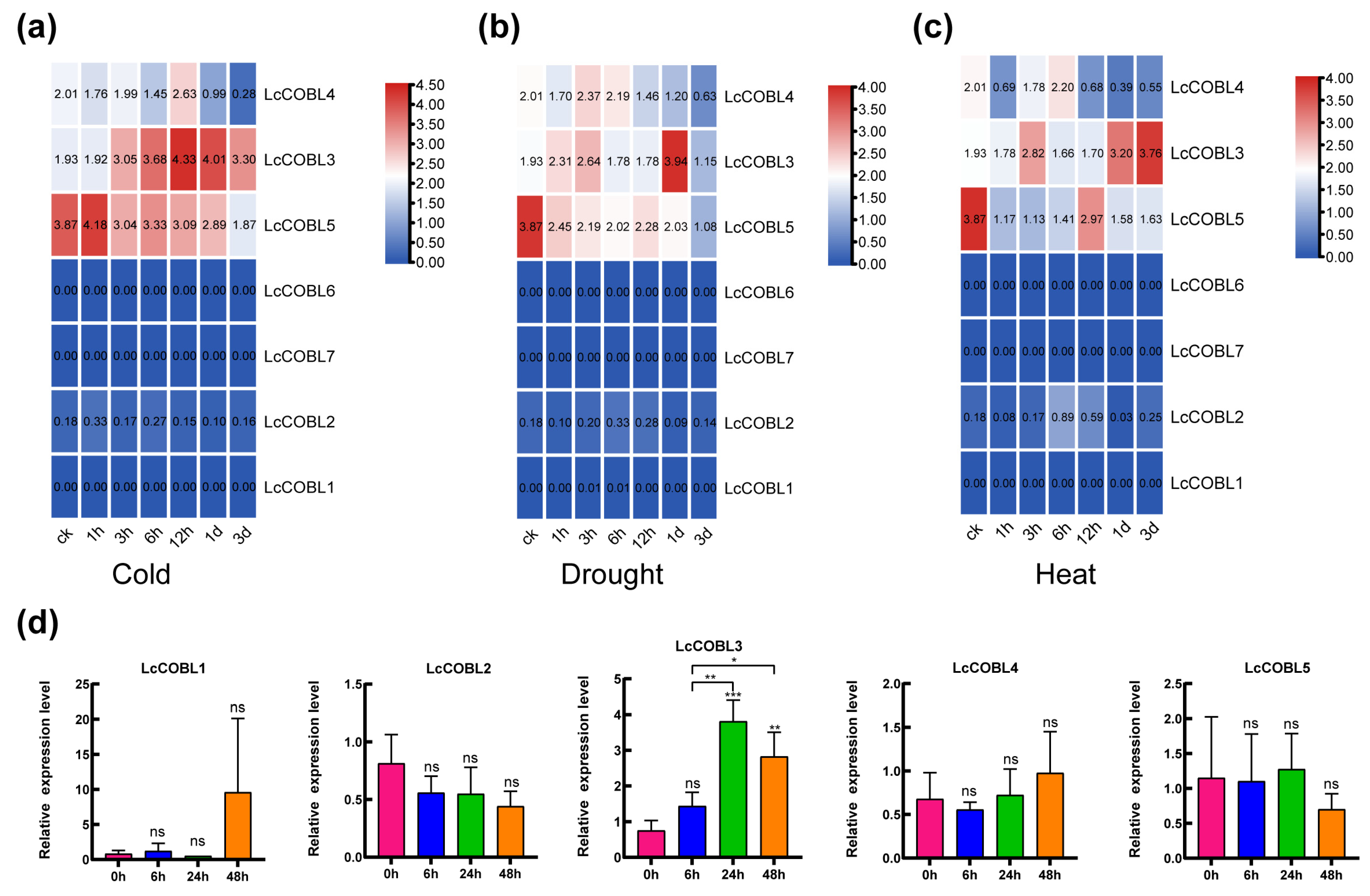
2.6. Expression Patterns of LcCOBLs in Response to Abiotic Stresses
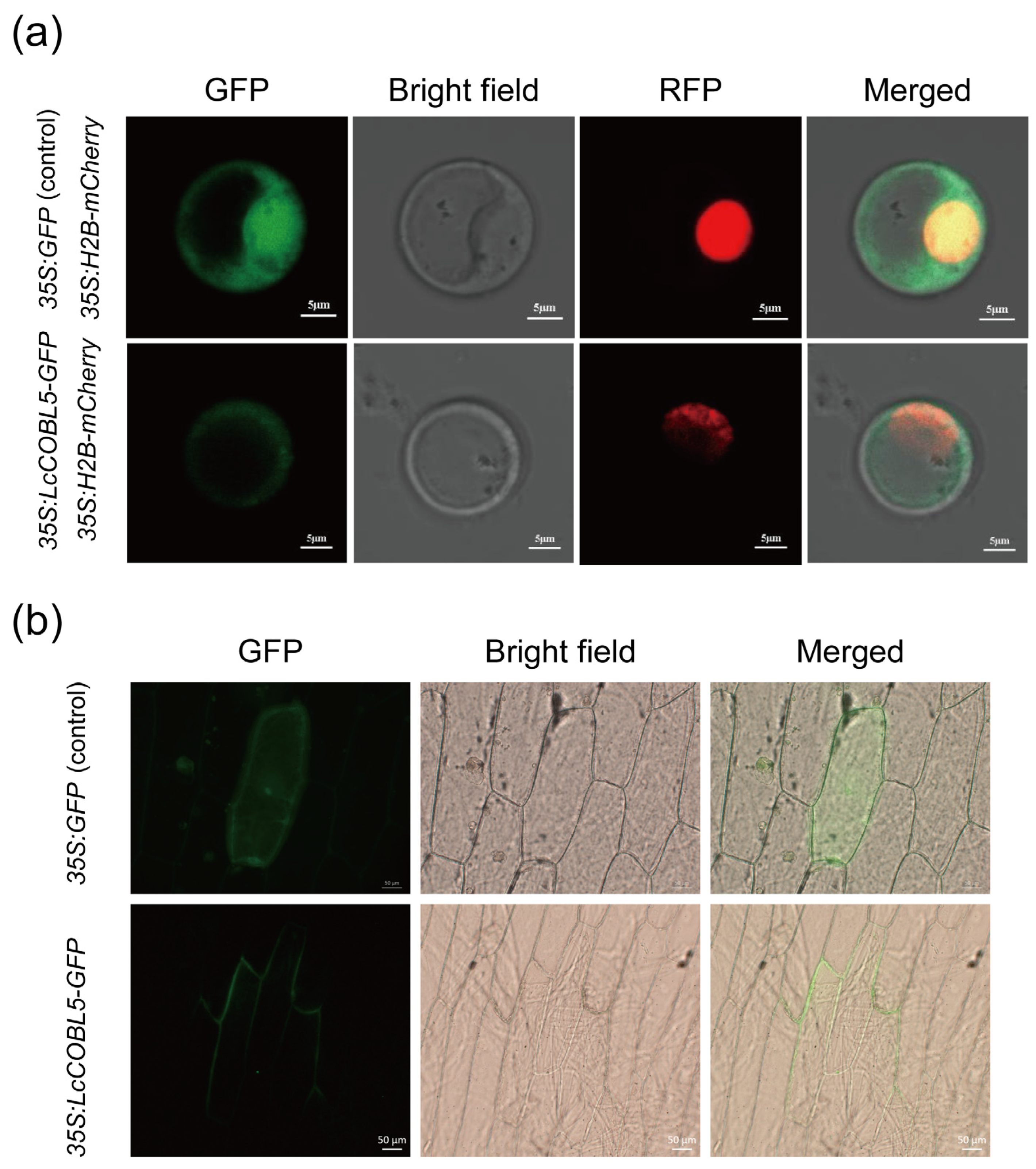
2.7. Subcellular Localization of LcCOBL Proteins
3. Discussion
3.1. Functions of LcCOBLs in Stem Development and Stress Responses
3.2. Subcellular Localization of LcCOBLs and Their Associations with Potential Functions
4. Materials and Methods
4.1. Identification of the LcCOBLs Family in the L. chinense Genome
4.2. Phylogenetic Analysis and Classification of the LcCOBL Family
4.3. Analyses of the Chromosomal Locations, Structures and Conserved Motifs of LcCOBL Genes
4.4. Plant Materials, RNA Extraction, qRT-PCR Analysis, and Transcriptome Sequencing Analysis of Abiotic Stress in L. chinense
4.5. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mikkelsen, M.D.; Harholt, J.; Ulvskov, P.; Johansen, I.E.; Fangel, J.U.; Doblin, M.S.; Bacic, A.; Willats, W.G. Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae. Ann. Bot. 2014, 114, 1217–1236. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xu, S.; Wang, Y.; Zhou, M.; Hong, L. Primary Cell Wall Modifying Proteins Regulate Wall Mechanics to Steer Plant Morphogenesis. Front. Plant Sci. 2021, 12, 751372. [Google Scholar] [CrossRef] [PubMed]
- Skrovankova, S.; Mlcek, J. Improvement of Postharvest Quality and Bioactive Compounds Content of Persimmon Fruits after Hydrocolloid-Based Edible Coating Application. Horticulturae 2022, 8, 1045. [Google Scholar]
- Handakumbura, P.P.; Hazen, S.P. Transcriptional Regulation of Grass Secondary Cell Wall Biosynthesis: Playing Catch-Up with Arabidopsis thaliana. Front. Plant Sci. 2012, 3, 74. [Google Scholar] [CrossRef] [Green Version]
- Xi, W.; Song, D.; Sun, J.; Shen, J.; Li, L. Formation of wood secondary cell wall may involve two type cellulose synthase complexes in Populus. Plant Mol. Biol. 2017, 93, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131, s207373. [Google Scholar] [CrossRef] [Green Version]
- Khan, G.A.; Persson, S. Cell Wall Biology: Dual Control of Cellulose Synthase Guidance. Curr. Biol. 2020, 30, 232–234. [Google Scholar] [CrossRef]
- Que, F.; Zha, R.F.; Wei, Q. Advances in research of cellulose synthase genes in plants. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2022, 46, 207–214. [Google Scholar]
- Houston, K.; Burton, R.A.; Sznajder, B. A genome-wide association study for culm cellulose content in barley reveals candidate genes co-expressed with members of the CELLULOSE SYNTHASE A gene family. PLoS ONE 2015, 10, e0130890. [Google Scholar] [CrossRef] [Green Version]
- Persson, S.; Wei, H.; Milne, J.; Page, G.P.; Somerville, C.R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 2005, 102, 8633–8638. [Google Scholar] [CrossRef] [Green Version]
- Schindelman, G.; Morikami, A.; Jung, J.; Baskin, T.I.; Carpita, N.C.; Derbyshire, P.; McCann, M.C.; Benfey, P.N. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001, 15, 1115–1127. [Google Scholar] [CrossRef] [Green Version]
- Brady, S.M.; Song, S.; Dhugga, K.S.; Rafalski, J.A.; Benfey, P.N. Combining Expression and Comparative Evolutionary Analysis. The COBRA Gene Family. Plant Physiol. 2007, 143, 172–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2007, 1771, 405–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borner, G.H.; Lilley, K.S.; Stevens, T.J.; Dupree, P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 2003, 132, 568–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Shang-Guan, K.; Zhang, B.; Liu, X.; Yan, M.; Zhang, L.; Shi, Y.; Zhang, M.; Qian, Q.; Li, J.; et al. Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet. 2013, 9, e1003704. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Schindelman, G.; DeSalle, R.; Benfey, P.N. The COBRA family of putative GPI-anchored proteins in Arabidopsis. A new fellowship in expansion. Plant Physiol. 2002, 130, 538–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Kang, B.-G.; Osburn, L.D.; Cheng, Z.-M. The COBRA gene family in Populus and gene expression in vegetative organs and in response to hormones and environmental stresses. Plant Growth Regul. 2009, 58, 211–223. [Google Scholar] [CrossRef]
- Dai, X.; You, C.; Wang, L.; Chen, G.; Zhang, Q.; Wu, C. Molecular characterization, expression pattern, and function analysis of the OsBC1L family in rice. Plant Mol. Biol. 2009, 71, 469–481. [Google Scholar] [CrossRef]
- Julius, B.T.; McCubbin, T.J.; Mertz, R.A.; Baert, N.; Knoblauch, J.; Grant, D.G.; Conner, K.; Bihmidine, S.; Chomet, P.; Wagner, R.; et al. Maize Brittle Stalk2-Like3, encoding a COBRA protein, functions in cell wall formation and carbohydrate partitioning. Plant Cell 2021, 33, 3348–3366. [Google Scholar] [CrossRef]
- Zhao, Y.; Su, X.; Wang, X.; Wang, M.; Feng, X.; Muhammad, A.M.; Cai, Y. Comparative genomic analysis of the COBRA genes in six Rosaceae species and expression analysis in Chinese white pear. Peer J. 2022, 10, e13723. [Google Scholar] [CrossRef]
- Benfey, P.N.; Linstead, P.J.; Roberts, K.; Schiefelbein, J.W.; Hauser, M.T.; Aeschbacher, R.A. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 1993, 119, 57–70. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Wen, T.-J.; Zimmermann, R.; Chimot-Marolle, P.; Silva, O.D.C.E.; Bruce, W.; Lamkey, K.R.; Wienand, U.; Schnable, P.S. The maize (Zea mays L.) roothairless3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J. 2008, 54, 888–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaheer, M.; Rehman, S.U.; Khan, S.H.; Shahid, S.; Rasheed, A.; Naz, R.; Sajjad, M. Characterization of new COBRA like (COBL) genes in wheat (Triticum aestivum) and their expression analysis under drought stress. Mol. Biol. Rep. 2022, 49, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, Y.; Tan, W.; Chen, J.; Zhu, M.; Lv, Y.; Liu, Y.; Yu, S.; Zhang, W.; Cai, H. Brittle Culm 1 encodes a COBRA-like protein involved in secondary cell wall cellulose biosynthesis in sorghum. Plant Cell Physiol. 2019, 60, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Tang, X.; Giovannoni, J.; Xiao, F.; Liu, Y. Functional characterization of a tomato COBRA-like gene functioning in fruit development and ripening. BMC Plant Biol. 2012, 12, 211. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Xiong, H.; Jiang, C.; Zhang, D.; Yang, Z.; Huang, Y.; Zhu, W.; Ma, S.; Duan, J.; Wang, X.; et al. Natural variation of DROT1 confers drought adaptation in upland rice. Nat. Commun. 2022, 13, 4265. [Google Scholar] [CrossRef]
- Kasirajan, L.; Valiyaparambth, R.; Kamaraj, K.; Sebastiar, S.; Hoang, N.V.; Athiappan, S.; Srinivasavedantham, V.; Subramanian, K. Deep sequencing of suppression subtractive library identifies differentially expressed transcripts of Saccharum spontaneum exposed to salinity stress. Physiol. Plant 2022, 174, e13645. [Google Scholar] [CrossRef]
- Ko, J.H.; Kim, J.H.; Jayanty, S.S.; Howe, G.A.; Han, K.H. Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defence responses in Arabidopsis thaliana. J. Exp. Bot. 2006, 57, 2923–2936. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Wang, P.; Long, X.; Wu, W.; Zhang, J.; Pan, Y.; Cheng, T.; Shi, J.; Chen, J. The PIN gene family in relic plant L. chinense: Genome-wide identification and gene expression profiling in different organizations and abiotic stress responses. Plant Physiol. Biochem. 2021, 162, 634–646. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.R. The current scenario and sustainable developmental strategies for genetically improved tree seed production bases in China. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2020, 44, 1–8. [Google Scholar]
- Thijs, G.; Marchal, K.; Lescot, M.; Rombauts, S.; De Moor, B.; Rouzé, P.; Moreau, Y. A Gibbs sampling method to detect over-represented motifs in the upstream regions of co-expressed genes. In Proceedings of the Fifth Annual International Conference on Computational Biology, Montreal, QC, Canada, 22–25 April 2001; pp. 305–312. [Google Scholar]
- Powell, R.V.; Willett, C.R.; Goertzen, L.R.; Rashotte, A.M. Lineage specific conservation of cis-regulatory elements in Cytokinin Response Factors. Sci. Rep. 2019, 9, 13387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidari, P.; Puresmaeli, F.; Mora-Poblete, F. Genome-wide identification and molecular evolution of the magnesium transporter (MGT) gene family in Citrullus lanatus and Cucumis sativus. Agronomy 2022, 12, 2253. [Google Scholar] [CrossRef]
- Dai, X.; You, C.; Chen, G.; Li, X.; Zhang, Q.; Wu, C. OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol. Biol. 2011, 75, 333–345. [Google Scholar] [CrossRef]
- Roudier, F.; Fernandez, A.G.; Fujita, M.; Himmelspach, R.; Borner, G.H.H.; Schindelman, G.; Song, S.; Baskin, T.I.; Dupree, P.; Wasteneys, G.; et al. COBRA, an Arabidopsis Extracellular Glycosyl-Phosphatidyl Inositol-Anchored Protein, Specifically Controls Highly Anisotropic Expansion through Its Involvement in Cellulose Microfibril Orientation. Plant Cell 2005, 17, 1749–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Wang, S.; Chen, H.; You, L.; Liu, F.; Liu, Z. Genome-wide identification and expression profiling of the COBRA-like genes reveal likely roles in stem strength in rapeseed (Brassica napus L.). PLoS ONE 2021, 16, e260268. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Song, W.; Huang, S.; Jiang, K.; Moriwaki, Y.; Wang, Y.; Men, Y.; Zhang, D.; Wen, X.; Han, Z.; et al. Extracellular pH sensing by plant cell-surface peptide-receptor complexes. Cell 2022, 185, 3341–3355. [Google Scholar] [CrossRef]
- Kneen, M.; Farinas, J.; Li, Y.; Verkman, A.S. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998, 74, 1591–1599. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Tang, M.; Chang, H.H.; Kanamala, M.; Davidson, A.J.; Wu, Z. Mannosylation of pH-sensitive liposomes promoted cytoplasmic delivery of protein to macrophages: Green fluorescent protein (GFP) performed as an endosomal escape tracer. Pharm. Dev. Technol. 2021, 26, 1000–1009. [Google Scholar] [CrossRef]
- Roberts, T.M.; Rudolf, F.; Meyer, A.; Pellaux, R.; Whitehead, E.; Panke, S.; Held, M. Identification and Characterisation of a pH-stable GFP. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Borner, G.H.; Sherrier, D.J.; Stevens, T.J.; Arkin, I.T.; Dupree, P. Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 2002, 129, 486–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Bian, L.; Shi, J.; Xu, J.; Xi, M.; Wang, G. Expression of a conifer COBRA-like gene ClCOBL1 from Chinese fir (Cunninghamia lanceolata) alters the leaf architecture in tobacco. Plant Physiol. Biochem. 2013, 70, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.S.; Jin, F.; Liu, Y.S. Functional Analysis of the GPI Transamidase Complex by Screening for Amino Acid Mutations in Each Subunit. Molecules 2021, 26, 5462. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Li, T.; Yuan, W.; Qiu, S.; Shi, J. Selection of reference genes for gene expression analysis in Liriodendron hybrids’ somatic embryogenesis and germinative tissues. Sci. Rep. 2021, 11, 4957. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Xu, L.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Hao, Z.; Lu, Y.; Yang, L.; et al. Genome-wide identification of the Liriodendron chinense WRKY gene family and its diverse roles in response to multiple abiotic stress. BMC Plant Biol. 2022, 22, 25. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Locus | MW (kDa) a | pI b | GRAVY c | N-Terminal Signal Peptide | GPI Modification Site | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| LcCOBL1 | Lchi01857 | Chr2 | 42.96 | 8.43 | −0.271 | Yes | Yes | Cell membrane |
| LcCOBL2 | Lchi02570 | Chr5 | 59.66 | 8.81 | −0.245 | Yes | No | Cell membrane; chloroplasts |
| LcCOBL3 | Lchi20746 | Chr10 | 70.30 | 5.53 | −0.059 | Yes | Yes | Cell membrane |
| LcCOBL4 | Lchi12311 | Chr13 | 40.58 | 8.95 | −0.199 | Yes | No | Cell membrane |
| LcCOBL5 | Lchi14677 | Chr16 | 25.30 | 7.53 | −0.239 | Yes | No | Cell membrane |
| LcCOBL6 | Lchi21892 | Chr19 | 73.59 | 8.47 | −0.28 | Yes | Yes | Cell membrane |
| LcCOBL7 | Lchi34460 | Contig2982 | 73.66 | 8.45 | −0.271 | Yes | Yes | Cell membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, C.; Chen, J.; Wu, W.; Liao, B.; Zheng, X.; Li, Y.; Huang, J.; Shi, J.; Hao, Z. Genome-Wide Analysis and Abiotic Stress-Responsive Patterns of COBRA-like Gene Family in Liriodendron chinense. Plants 2023, 12, 1616. https://doi.org/10.3390/plants12081616
Qiu C, Chen J, Wu W, Liao B, Zheng X, Li Y, Huang J, Shi J, Hao Z. Genome-Wide Analysis and Abiotic Stress-Responsive Patterns of COBRA-like Gene Family in Liriodendron chinense. Plants. 2023; 12(8):1616. https://doi.org/10.3390/plants12081616
Chicago/Turabian StyleQiu, Chen, Jinhui Chen, Weihuang Wu, Bojun Liao, Xueyan Zheng, Yong Li, Jing Huang, Jisen Shi, and Zhaodong Hao. 2023. "Genome-Wide Analysis and Abiotic Stress-Responsive Patterns of COBRA-like Gene Family in Liriodendron chinense" Plants 12, no. 8: 1616. https://doi.org/10.3390/plants12081616
APA StyleQiu, C., Chen, J., Wu, W., Liao, B., Zheng, X., Li, Y., Huang, J., Shi, J., & Hao, Z. (2023). Genome-Wide Analysis and Abiotic Stress-Responsive Patterns of COBRA-like Gene Family in Liriodendron chinense. Plants, 12(8), 1616. https://doi.org/10.3390/plants12081616






