Molecular Cloning and Expression Analysis of the Typical Class III Chitinase Genes from Three Mangrove Species under Heavy Metal Stress
Abstract
1. Introduction
2. Results
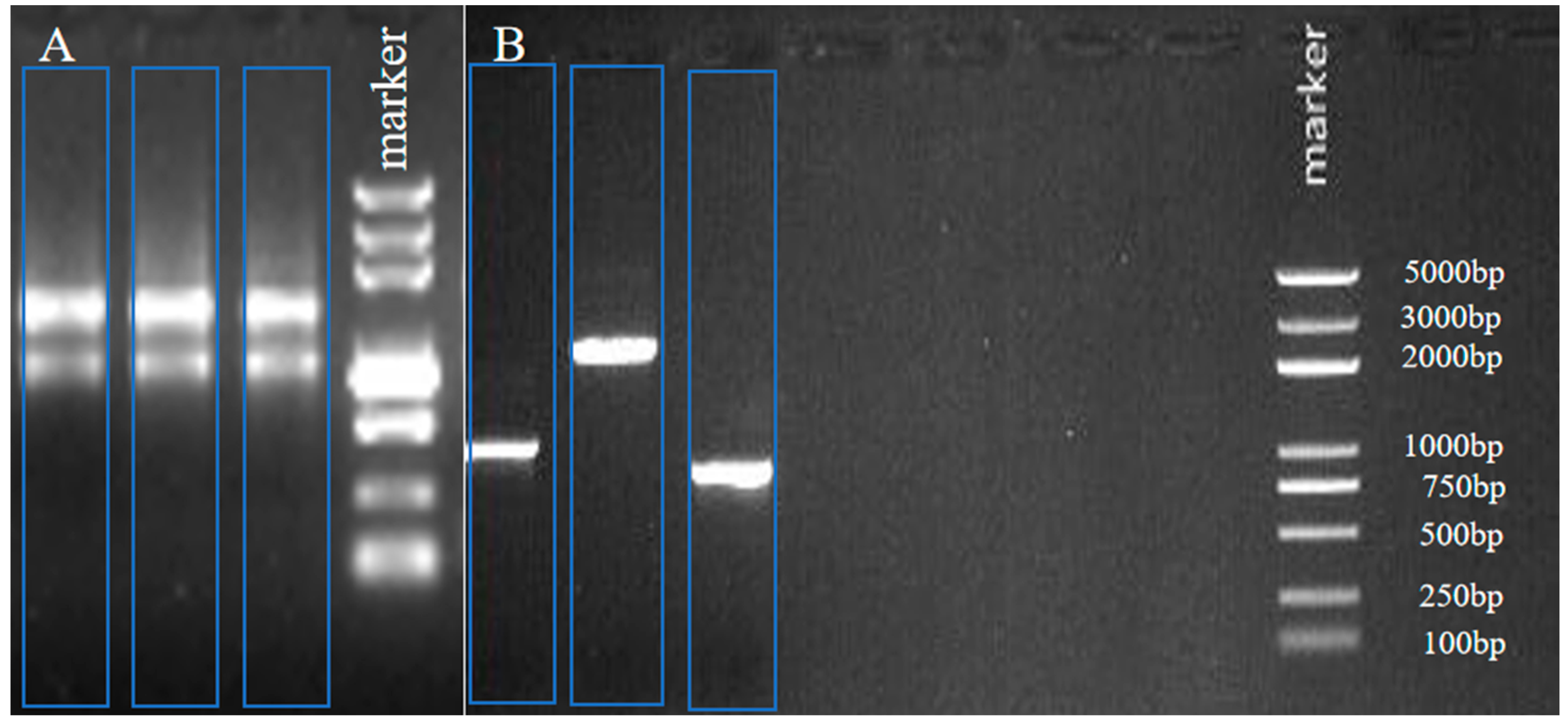
2.1. The Full-Length cDNA of the CHI III Gene Cloning
2.2. The Effect on Mangrove under Heavy Metal Treatment
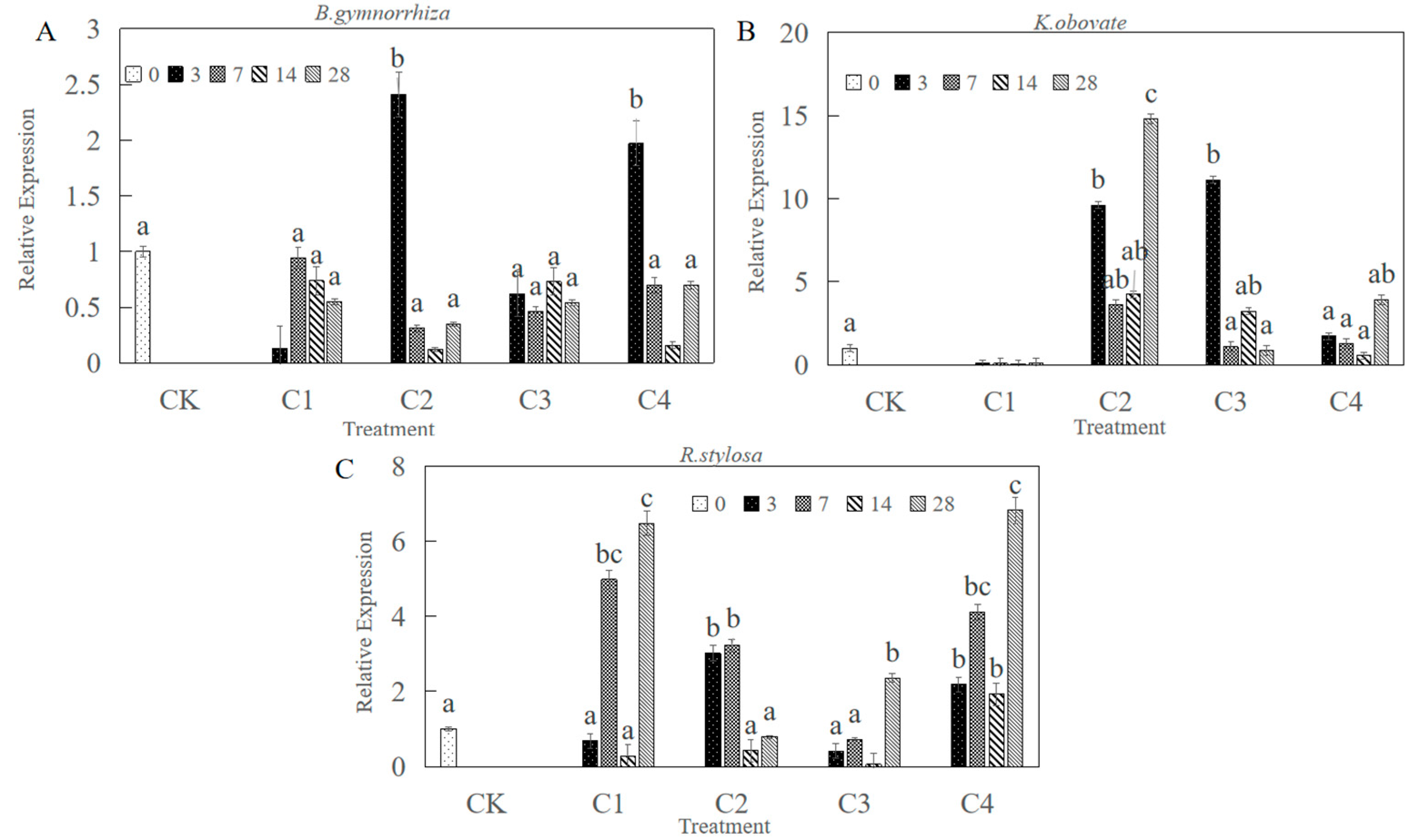
2.3. CHI III mRNA Expression in Leaf in Response to Heavy Metals
3. Discussion
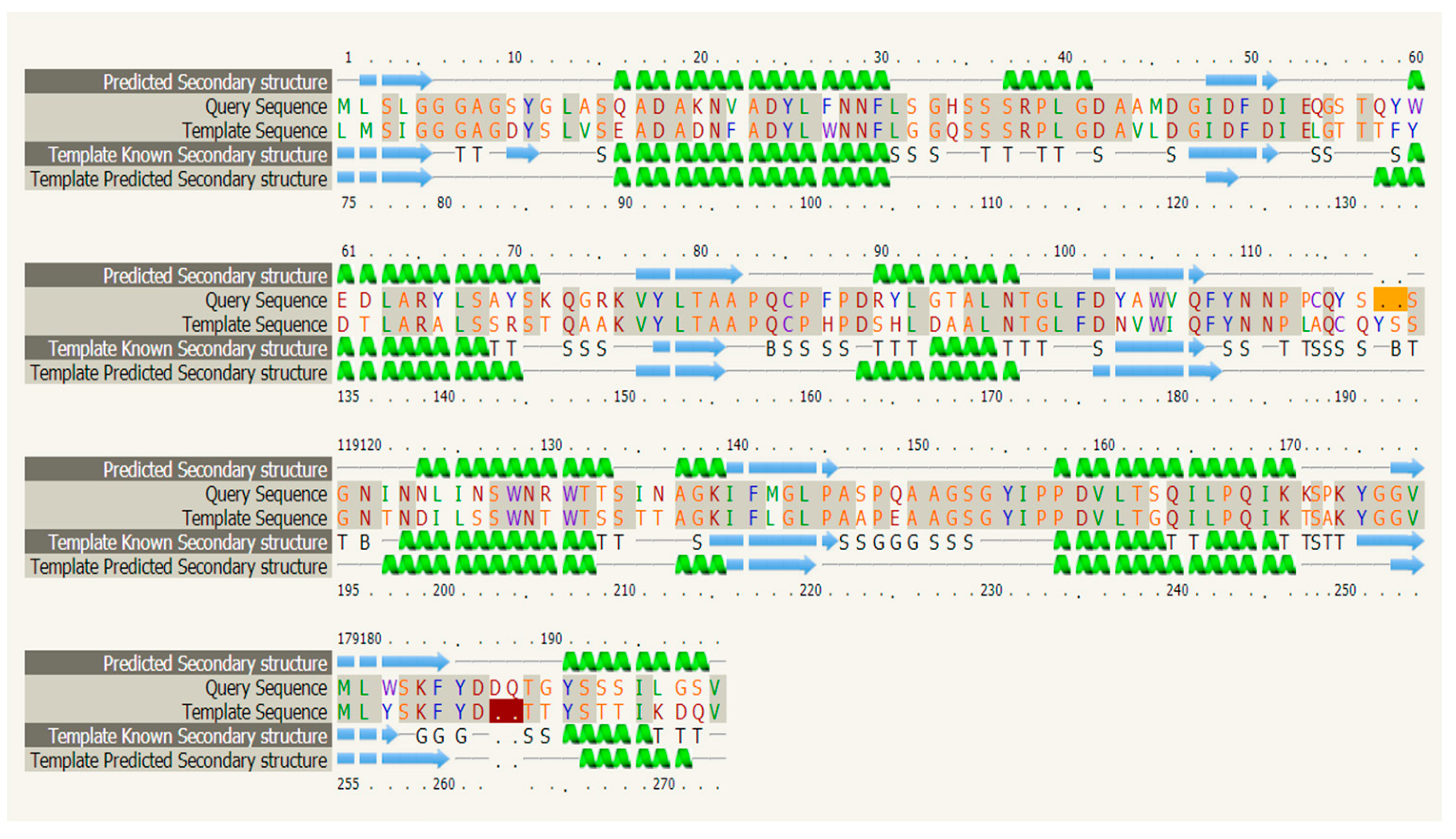
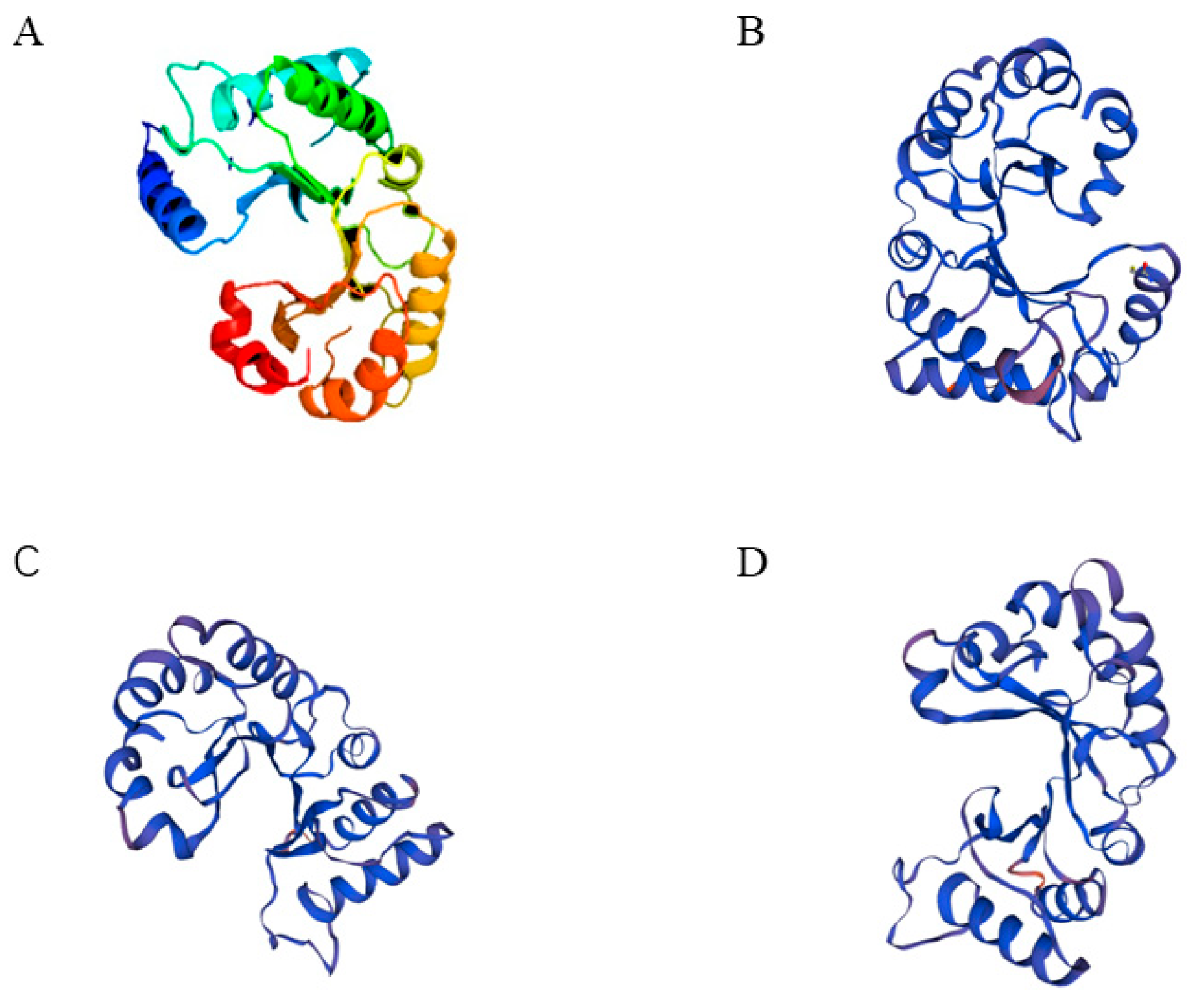
3.1. Cloning and Structural Characterization Analysis of CHI III
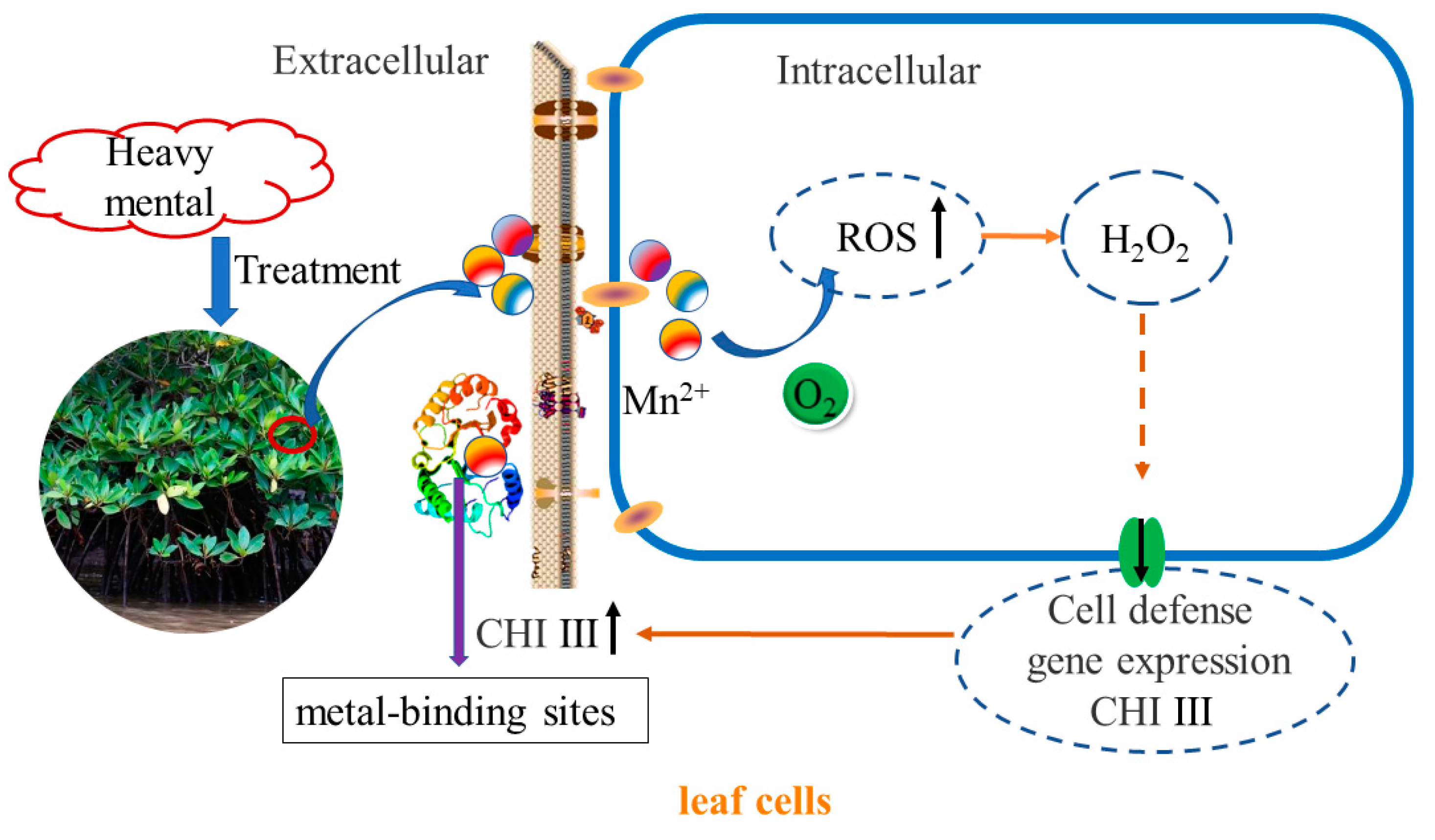
3.2. Expression of CHI III in Leaves in Response to Heavy Metal
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Extraction of the Total RNA
4.3. Primer Designs and Sequence Comparison Analysis
4.4. Cloning the Full-Length cDNA of the Chitinase Gene
4.5. Bioinformatic Analysis
4.6. Analysis of Gene Expression using Real-Time Quantitative PCR
4.7. Statistics Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.S. Restoration and Evaluation Technology of Mangrove Ecosystem; The Science Publishing Company: Beijing, China, 2013; pp. 1–20. [Google Scholar]
- Wang, Y.S. Molecular Ecology of Mangroves; The Science Publishing Company: Beijing, China, 2019; pp. 3–15. [Google Scholar]
- Wang, Y.S.; Gu, J.D. Ecological responses, adaptation and mechanisms of mangrove wetland ecosystem to the global climate change and anthropogenic activities. Int. Biodeterior. Biodegrad. 2021, 162, 105–248. [Google Scholar] [CrossRef]
- Valls, M.; Lorenzo, V.D. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol. Rev. 2002, 26, 327–338. [Google Scholar] [CrossRef]
- Kamala, K.S.; Batvari, B.; Lee, K.J.; Kannan, N.; Krishnamoorthy, R.; Shanthi, K.; Jayaprakash, M. Assessment of heavy metals (Cd, Cr, and Pb) in water, sediment, and seaweed (Ulva lactuca) in the Pulicat Lake, Southeast India. Chemosphere 2008, 71, 1233–1240. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Bhargava, P.; Rai, L.C. Salinity and copper-induced oxidative damage and changes in the antioxidative defense system of Anabaena doliolum. World J. Microbiol. Biotechnol. 2005, 21, 1291–1298. [Google Scholar] [CrossRef]
- Dong, L.L.; Xiao, M.L.; Ze, Y.P. Flavanol derivatives from Rhizophora stylosa and their DPPH radical scavenging activity. Molecules 2007, 12, 1163–1169. [Google Scholar]
- He, Y.J.; Sun, M.H.; Shen, Y.T. Progress in interaction mechanism and application between super enriched plants and heavy metals. Rock Min. Test 2020, 39, 639–657. [Google Scholar]
- Zhang, F.Q.; Wang, Y.S.; Lou, Z.P.; Dong, J.D. Effect of heavy metal stress on antioxidative enzymes and lipid peroxidation in leaves and roots of two mangrove plant seedlings (Kandelia candel and Bruguiera gymnorrhiza). Chemosphere 2007, 67, 44–50. [Google Scholar] [CrossRef]
- Zhang, F.Q.; Wang, C.C.; Luo, Z.P.; Dong, J.D. A novel metallothionein gene from a mangrove plant Kandelia candel. Ecotoxicology 2012, 21, 1633–1641. [Google Scholar] [CrossRef]
- Wu, Z.G.; Zhu, X.F. Molecular biology characterization and application in transgenic plant for chitinase. Chin. Bull. Life Sci. 2002, 14, 117–121. [Google Scholar]
- Peumans, W.J.; Proost, P.; Swennen, R.L.; Damme, V. The abundant class III chitinase homolog in young developing banana fruits behaves as a transient vegetative storage protein and most probably serves as an important supply of amino acids for the synthesis of ripening-associated proteins. Plant Physiol. 2002, 130, 1063–1072. [Google Scholar] [CrossRef]
- Kasprzewska, A. Plant chitinases-regulation and function. Cell Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar]
- Vellicce, G.R.; Ricci, J.C.D.; Hernández, L. Enhanced resistance to Botrytis cinerea, mediated by the transgenic expression of the chitinase gene ch5b in strawberry. Transgenic Res. 2006, 15, 57–68. [Google Scholar] [CrossRef]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef]
- Melchers, L.S.; Apotheker-de Groot, M.; van der Knaap, J.A.; Ponstein, A.S.; Linthorst, H.J.M. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 1994, 5, 469–480. [Google Scholar] [CrossRef]
- Arimori, T.; Tamada, T. Structure and function of a novel chitinase belonging to GH family 23. X-rays 2014, 56, 201–206. [Google Scholar]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active Enzymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Li, D.C. Review of fungal chitinases. Mycopathologia 2006, 161, 345–360. [Google Scholar]
- Arakane, Y.; Koga, D. Purification and characterization of a novel chitinase isozyme from yam tuber. Biosci. Biotech. Bioch. 1999, 63, 1895–1901. [Google Scholar] [CrossRef]
- Graham, L.S.; Sticklen, M.B. Plant chitinases. Can. J. Bot. 1994, 72, 1057–1083. [Google Scholar] [CrossRef]
- Berglund, L.; Brunstedt, J.; Nielsen, K.K.; Chen, Z.C.; Mikkelsen, J.D.; Marcker, K.A. A proline-rich chitinase from Beta vulgaris. Plant Mol. Biol. 1995, 27, 211–216. [Google Scholar] [CrossRef]
- Tang, C.M.; Chye, M.L.; Ramalingam, S.; Ouyang, S.W.; Zhao, K.J.; Ubhayasekera, W.; Mowbray, S.L. Functional analyses of the chitin-binding domains and the catalytic domain of Brassica juncea chitinase BjCHI1. Plant Mol. Biol. 2004, 56, 285–298. [Google Scholar] [CrossRef]
- Loon, L.C.V.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- van Keulen, H.; Wei, R.; Cutright, T.J. Arsenate-induced expression of a class III chitinase in the dwarf sunflower Helianthus annuus. Environ. Exp. Bot. 2008, 63, 281–288. [Google Scholar] [CrossRef]
- Su, Y.C.; Xu, L.P.; Fu, Z.W.; Yang, Y.T.; Guo, J.L.; Wang, S.S.; Que, Y.X. ScChi encoding an acidic class III chitinase of sugarcane, confers positive responses to biotic and abiotic stresses in sugarcane. Int. J. Mol. Sci. 2014, 15, 2738–2760. [Google Scholar] [CrossRef]
- Békésiová, B.; Hraška, Š.; Libantová, J.; Moravcíková, J.; Matušíková, I. Heavy-metal stress induced accumulation of chitinase isoforms in plants. Mol. Biol. Rep. 2008, 35, 579–588. [Google Scholar] [CrossRef]
- Mészáros, P.; Rybanský, L.; Spieß, N.; Socha, P.; Kuna, R.; Libantová, J.; Moravčíková, J.; Piršelová, B.; Hauptvogel, P.; Matušíková, I. Plant chitinase responses to different metal-type stresses reveal specificity. Plant Cell Rep. 2014, 33, 1789–1799. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, Q.J.; Zhang, C.Z.; Luo, J.M.; Shen, Y.B.; Wang, M. Characterization of antifungal chitinase from Bacillus licheniformis TCCC10016. Lect. Notes Electr. Eng. 2014, 249, 597–607. [Google Scholar]
- Wang, L.Y.; Wang, Y.S.; Zhang, J.P.; Gu, J.D. Molecular cloning of class III chitinase gene from Avicennia marina and its expression analysis in response to cadmium and lead stress. Ecotoxicology 2015, 24, 1697–1704. [Google Scholar] [CrossRef]
- Wang, L.Y.; Wang, Y.S.; Cheng, H.; Zhang, J.P.; Foong, S.Y. Cloning of the Aegiceras corniculatum class I chitinase gene (AcCHI I) and the response of AcCHI I mRNA expression to cadmium stress. Ecotoxicology 2015, 24, 1705–1713. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Meins, F.J.; Bernard, F.; Linthorst, H.U.M. Plant chitinase genes. Plant Mol. Biol. Report. 1994, 12, 22–28. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef]
- Takenaka, Y.; Nakano, S.; Tamoi, M.; Sakuda, S.; Fukamizo, T. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: Chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotech. Bioch. 2009, 73, 1066–1071. [Google Scholar] [CrossRef]
- Chandra, S.; Dutta, A.K.; Chandrashekara, K.N.; Acharya, K. In silico characterization, homology modeling of Camellia sinensis chitinase and its evolutionary analyses with other plant chitinases. Proc. Natl. Acad. Sci. USA 2015, 87, 685–695. [Google Scholar] [CrossRef]
- Nagpure, A.; Choudhary, B.; Gupta, R.K. Chitinases: In agriculture and human healthcare. Crit. Rev. Biotechnol. 2013, 34, 215–232. [Google Scholar] [CrossRef]
- Tapia, G.; Morales-Quintana, L.; Inostroza, L. Molecular characterisation of Ltchi7, a gene encoding a class III endochitinase induced by drought stress in Lotus spp. Plant Biol. 2011, 13, 69–77. [Google Scholar] [CrossRef]
- Yang, Y.W.; Zhang, B.L.; Ni, W.C.; Shen, X.L.; Zhang, X.G.; Xu, Y.J. Molecular cloning and expression analysis of two chitinase in upland cotton. Mianhua Xuebao 2008, 20, 88–93. [Google Scholar]
- Zhou, J.; Huang, J. Cloning and Functional Identification of Chitinase Gene SlChi in Salix. Mol. Plant Breed. 2018, 16, 8013–8021. [Google Scholar]
- De las Mercedes, D.M.; Pintor-Toro, J.A.; Cubero, B. Transgenic tobacco plants over expressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 2006, 142, 722–730. [Google Scholar]
- Rivera-Becerril, F.; Metwally, A.; Martin-Laurent, F.; Van Tuinen, D.; Dietz, K.J.; Gianinazzi, S.; Gianinazzi-Pearson, V. Molecular responses to cadmium in roots of Pisum sativum L. Water Air Soil Pollut. 2005, 168, 171–186. [Google Scholar] [CrossRef]
- Rivera-Becerril, F.; van Tuinen, D.; Martin-Laurent, F.; Metwally, A.; Dietz, K.J.; Gianinazzi, S.; Gianinazzi-Pearson, V. Molecular changes in Pisum sativum L. roots during arbuscular mycorrhiza buffering of cadmium stress. Mycorrhiza 2005, 16, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Maksymiec, W. Effect of copper on cellular processes in higher plants. Photosynthetica 1997, 34, 321–342. [Google Scholar] [CrossRef]
- Siedlecka, A.; Tukendorf, A.; Skorzynska-Polit, E.; Maksymiec, W.; Wojcik, M.; Baszynski, T.; Krupa, Z. Angiosperms (Asteraceae, Convolvulaceae, Fabaceae and Poaceae; other than Brassicaceae). In Metals in the Environment; Marcel Dekker Inc.: New York, NY, USA, 2001; pp. 171–215. [Google Scholar]
- Fang, W.C.; Kao, C.H. Enhanced peroxidase activity in rice leaves in response to excess iron, copper and zinc. Plant Sci. 2000, 158, 71–76. [Google Scholar] [CrossRef]
- Tewari, R.K.; Kumar, P.; Sharma, P.N.; Bisht, S.S. Modulation of oxidative stress responsive enzymes by excess cobalt. Plant Sci. 2002, 162, 381–388. [Google Scholar] [CrossRef]
- Allan, A.C.; Fluhr, R. Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 1997, 9, 1559–1572. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef]
- Zhang, R. Oxidative burst and H2O2 signal transduction in plant cells. Plant Physiol. Lett. 2000, 36, 376–383. [Google Scholar]
- Mészáros, P.; Rybanský, L.; Hauptvogel, P.; Kuna, R.; Libantová, J.; Moravčíková, J.; Piršelová, B.; Tirpaková, A.; Matušíková, I. Cultivar-specific kinetics of chitinase induction in soybean roots during exposure to arsenic. Mol. Biol. Rep. 2013, 40, 2127–2138. [Google Scholar] [CrossRef]
- Corrales, I.; Poschenrieder, C.; Barcelo, J. Boron-induced amelioration of aluminium toxicity in a monocot and a dicot species. J. Plant Physiol. 2008, 165, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Yang, Y.; Chen, P.C.; Li, C.; Wu, J.; Li, S. Ammonium Molybdate Spectrophotometric Determination of Hydrogen Peroxide Residues in Water and Hair Products. Mod. Prev. Med. 2008, 8, 1556–1558. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



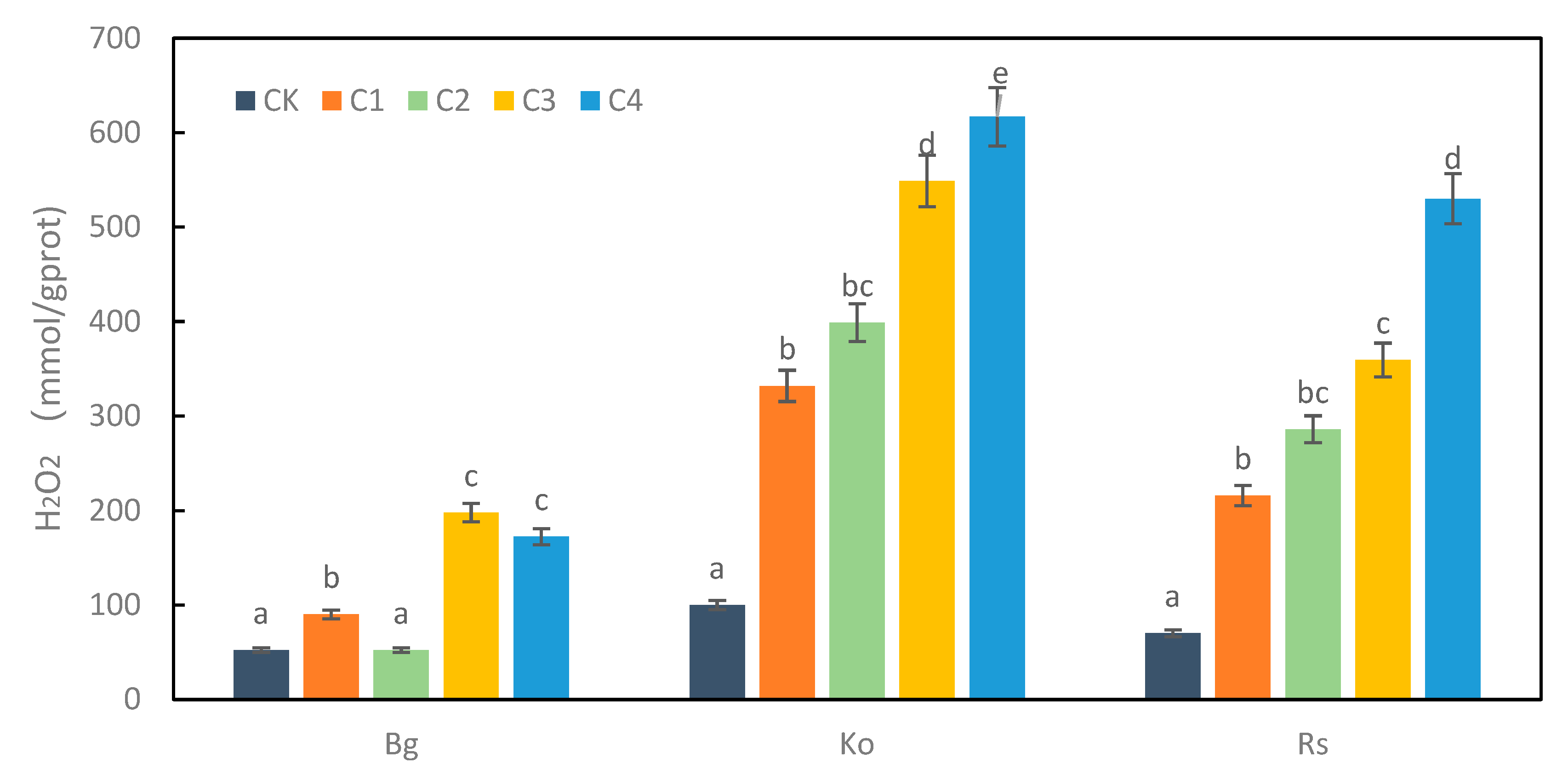
| Heavy Metal (mg/L) | Control Group (CK) | C1 | C2 | C3 | C4 |
|---|---|---|---|---|---|
| Cu2+ | 0 | 5 | 25 | 50 | 75 |
| Pb2+ | 0 | 1 | 5 | 10 | 15 |
| Cd2+ | 0 | 0.2 | 1 | 2 | 3 |
| Primers | Sequence (5′–3′) |
|---|---|
| F1 | TTGCCATCTACTGGGGTCA |
| R1 | CGTACTTTGGGGACTTCTTTAT |
| UPS | CTAATACGACTCACTATAGGGC |
| GSP-3′ | GATTACGCCAAGCTTGGGTCAAAACGGCAACGA |
| GSP-5′ | GATTACGCCAAGCTTGTAAAACGACGGCCAGTG |
| NGSP-3′ | GATTACGCCAAGCTTTACGGGTCTGTTTGACTATG |
| NGSP-5′ | GATTACGCCAAGCTTATCTGCGGAAGGATTTGA |
| F | GGACATCAAAAGTTGCCAGAG |
| R | GCATCACCTAAAGGACGAGAA |
| Bg18S(F) | CGGGGGCATTCGTATTTC |
| Bg18S(R) | CCTGGTCGGCATCGTTTAT |
| Ko18S(F) | CCTGAGAAACGGCTACCACATC |
| Ko18S(R) | ACCCATCCCAAGGTCCAACTAC |
| Rs18S(F) | ACCATAAACGATGCCGACC |
| Rs18S(R) | CCTTGCGACCATACTCCC |
| Function | Tool |
|---|---|
| Spliced sequence alignment | ApE software |
| Predict the open reading frame | ORFFinder (https://www.ncbi.nlm.nih.gov/orffinder/ accessed on 15 October 2021) |
| Predict the molecular weight, theoretical pI and hydrophilia | ExPASy-Compute pl/Mw (http://web.expasy.org/compute_pi/ accessed on 15 October 2021) |
| Physical and chemical properties of protein | ExPASy-ProtParam (http://web.expasy.org/protparam/ accessed on 15 October 2021)) |
| Hydrophilia | ExPASy-ProtScale (http://web.expasy.org/protscale/ accessed on 15 October 2021) |
| Functional site prediction of amino acid sequence | SoftBerryPSITE (http://www.softberry.com/berry.phtml?topic=psite&group=programs&subgroup=proloc/ accessed on 15 October 2021) |
| Protein transmembrane domain analysis | TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM accessed on 15 October 2021) |
| Predict potential signal peptide cleavage site | SignalP4.0-Server (http://www.cbs.dtu.dk/services/SignalP-4.0 accessed on 15 October 2021) |
| Analysis of protein structure and function domain | SMART (http://smart.embl-heidelberg.de/ accessed on 15 October 2021) |
| Prediction of protein secondary structure | Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index accessed on 15 October 2021) |
| Automated 3D structure building | ExPASy-SWISS-MODEL (https://www.swissmodel.expasy.org/ accessed on 15 October 2021) |
| Multiple sequence alignment | ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/ accessed on 15 October 2021) |
| Reconstruct phylogenetic tree | MEGA6 |
| Predict the subcellular localization | Plant-PLoc (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc/ accessed on 15 October 2021) |
| Amplification Program | Ingredient | Volume | |
|---|---|---|---|
| 95 °C 10 min | 2xqPCRmix | 10 μL | |
| 95 °C 10 s | 40 cycles | F primer (10 pmol/uL) | 0.5 μL |
| 60 °C 60 s | R primer (10 pmol/uL) | 0.5 μL | |
| 95 °C 5 s | DNA template | 2 μL | |
| 60 °C 5 s | Test once every 0.5 °C temperature rise | ddH2O | 7 μL |
| 95 °C 5 s | total | 20 μL | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.-Y.; Wang, Y.-S.; Sun, C.-C. Molecular Cloning and Expression Analysis of the Typical Class III Chitinase Genes from Three Mangrove Species under Heavy Metal Stress. Plants 2023, 12, 1681. https://doi.org/10.3390/plants12081681
Zhou Y-Y, Wang Y-S, Sun C-C. Molecular Cloning and Expression Analysis of the Typical Class III Chitinase Genes from Three Mangrove Species under Heavy Metal Stress. Plants. 2023; 12(8):1681. https://doi.org/10.3390/plants12081681
Chicago/Turabian StyleZhou, Yue-Yue, You-Shao Wang, and Cui-Ci Sun. 2023. "Molecular Cloning and Expression Analysis of the Typical Class III Chitinase Genes from Three Mangrove Species under Heavy Metal Stress" Plants 12, no. 8: 1681. https://doi.org/10.3390/plants12081681
APA StyleZhou, Y.-Y., Wang, Y.-S., & Sun, C.-C. (2023). Molecular Cloning and Expression Analysis of the Typical Class III Chitinase Genes from Three Mangrove Species under Heavy Metal Stress. Plants, 12(8), 1681. https://doi.org/10.3390/plants12081681






