Expression Dynamics of lpa1 Gene and Accumulation Pattern of Phytate in Maize Genotypes Possessing opaque2 and crtRB1 Genes at Different Stages of Kernel Development
Abstract
1. Introduction
2. Results
2.1. Genetic Variation for Nutritional Quality Traits
2.2. PA and Associated Traits during Different Stages of Kernel Development
2.3. Lysine and Tryptophan during Different Stages of Kernel Development
2.4. Provitamin-A during Different Stages of Kernel Development
2.5. Variation in Expression of Genes
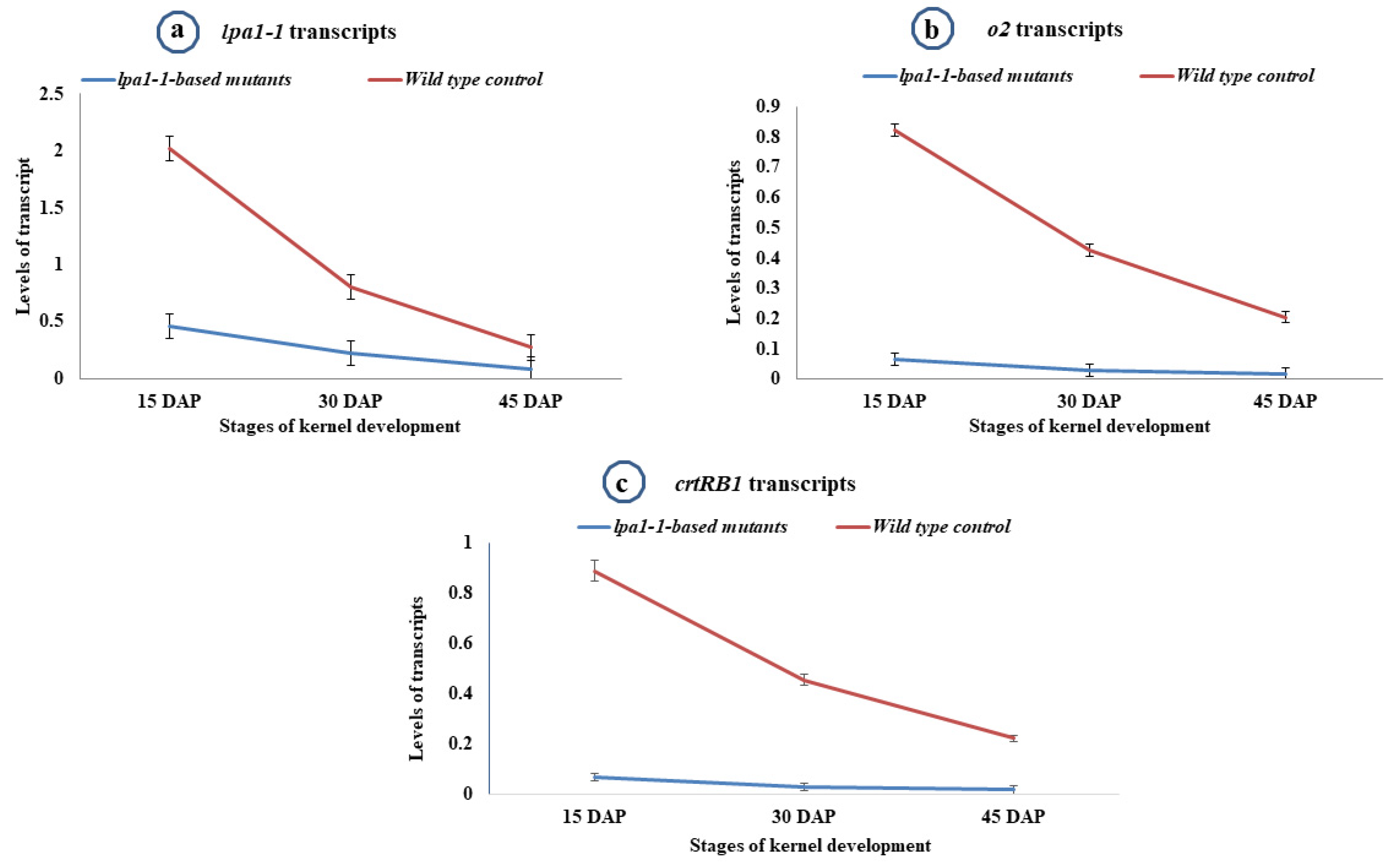
2.6. Expression Pattern of lpa-1-1 Gene during Kernel Development Stages
2.7. Expression Pattern of o2 Gene during Kernel Development Stages
2.8. Expression Pattern of crtRB1 Gene during Different Stages of Kernel Development
2.9. Correlation between Transcript Levels and Accumulation of Nutrients
3. Discussion
3.1. Expression Pattern of lpa1-1 and Its Effect on Kernel Phytate Accumulation
3.2. Expression Pattern of o2 and Its Effect on Lysine and Tryptophan
3.3. Expression Pattern of crtRB1 and Its Effect on proA Accumulation
4. Materials and Methods
4.1. Plant Materials
4.2. Field Experiments
4.3. Isolation of RNA and cDNA Synthesis
4.4. Designing of Primers for the Expression Studies
4.5. Gene Expression Analysis for lpa1-1, o2 and crtRB1 Genes
4.6. Samples Preparation for Biochemical Analysis
4.7. Determination of Phytic Acid (PA) and Inorganic Phosphorous (iP)
4.8. Determination of Lysine, Tryptophan and Provitamin-A
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Nutrition Report. 2021 Global Nutrition Report: The State of Global Nutrition; Development Initiatives: Bristol, UK, 2022; Available online: https://globalnutritionreport.org (accessed on 15 February 2023).
- Bouis, H.E.; Saltzman, A.; Birol, E. Improving nutrition through biofortification. In Agriculture for Improved Nutrition: Seizing the Momentum; Fan, S., Yosef, S., Pandya-Lorch, R., Eds.; International Food Policy Research Institute (IFPRI) and CABI: Wallingford, UK, 2019; pp. 47–57. [Google Scholar] [CrossRef]
- Tsakirpaloglou, N.; Bueno-Mota, G.M.; Soriano, J.C.; Arcillas, E.; Arines, F.M.; Yu, S.M.; Stangoulis, J.; Trijatmiko, K.R.; Reinke, R.; Tohme, J.; et al. Proof of concept and early development stage of market-oriented high iron and zinc rice expressing dicot ferritin and rice nicotianamine synthase genes. Sci. Rep. 2023, 13, 676. [Google Scholar] [CrossRef]
- McAuliffe, S.; Ray, S.; Fallon, E.; Bradfield, J.; Eden, T.; Kohlmeier, M. Dietary micronutrients in the wake of COVID-19: An appraisal of evidence with a focus on high-risk groups and preventative healthcare. BMJ Nutr. Prev. Health 2020, 3, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Yadava, D.K.; Hossain, F.; Mohapatra, T. Nutritional security through crop biofortification in India: Status and future prospects. Indian J. Med. Res. 2018, 148, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Neeraja, C.N.; Hossain, F.; Hariprasanna, K.; Sewa, R.; Satyawathi, C.T.; Longvah, T.; Raghu, P.; Voleti, S.R.; Sundaram, R.M. Towards nutrition security of India with biofortified cereal varieties. Curr. Sci. 2022, 123, 271. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, B.; Raigond, P.; Sahu, C.; Mishra, U.N.; Sharma, S.; Lal, M.K. Phytic acid: Blessing in disguise, a prime compound required for both plant and human nutrition. Food Res. Int. 2021, 142, 110193. [Google Scholar] [CrossRef] [PubMed]
- Raboy, V.; Young, K.A.; Dorsch, J.A.; Cook, A. Genetics and breeding of seed phosphorus and phytic acid. J. Plant Physiol. 2001, 158, 489–497. [Google Scholar] [CrossRef]
- Prasanna, B.M.; Palacios-Rojas, N.; Hossain, F.; Muthusamy, V.; Menkir, A.; Dhliwayo, T.; Ndhlela, T.; San Vicente, F.; Nair, S.K.; Vivek, B.S.; et al. Molecular breeding for nutritionally enriched maize: Status and prospects. Front. Genet. 2020, 10, 1392. [Google Scholar] [CrossRef]
- Bohn, L.; Meyer, A.S.; Rasmussen, S.K. Phytate: Impact on environment and human nutrition. A challenge for molecular breeding. J. Zhejiang Univ.-Sci. B 2008, 9, 165–191. [Google Scholar] [CrossRef]
- Gupta, H.S.; Hossain, F.; Muthusamy, V. Biofortification of maize: An Indian perspective. Indian J. Genet. Plant Breed. 2015, 75, 1–22. [Google Scholar] [CrossRef]
- Cromwell, G.L.; Coffey, R.D. Phosphorus: A key essential nutrient, yet a possible major pollutant. Its central role in animal nutrition. In Biotechnology in the Feed Industry; Alltech Tech Publishers: Nicholasville, KY, USA, 1991; pp. 133–145. [Google Scholar]
- Puppala, K.R.; Buddhiwant, P.G.; Agawane, S.B.; Kadam, A.S.; Mote, C.S.; Lonkar, V.D.; Khire, J.M.; Dharne, M.S. Performance of Aspergillus niger (NCIM 563) phytase based feed supplement for broiler growth and phosphorus excretion. Biocatal. Agric. Biotechnol. 2021, 31, 101887. [Google Scholar] [CrossRef]
- Ragi, S.; Muthusamy, V.; Zunjare, R.U.; Bhatt, V.; Katral, A.; Abhijith, K.P.; Chand, G.; Mishra, S.J.; Sekhar, J.C.; Yadava, D.K.; et al. Genetic variation for grain phytate and molecular characterization of low phytic acid-2 (lpa2) gene-based maize (Zea mays L.) inbreds. Plant Breed. 2022, 141, 212–222. [Google Scholar] [CrossRef]
- Erenstein, O.; Jaleta, M.; Sonder, K.; Mottaleb, K.; Prasanna, B.M. Global maize production, consumption and trade: Trends and R&D implications. Food Secur. 2022, 14, 1295–1319. [Google Scholar] [CrossRef]
- Schlemmer, U.; Frølich, W.; Prieto, R.M.; Grases, F. Phytate in foods and significance for humans: Food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 2009, 53, S330–S375. [Google Scholar] [CrossRef] [PubMed]
- Hossain, F.; Zunjare, R.U.; Muthusamy, V.; Bhat, J.S.; Mehta, B.K.; Sharma, D.; Talukder, Z.A.; Chhabra, R.; Katral, A.; Dutta, S.; et al. Biofortification of Maize for Nutritional Security. In Biofortification of Staple Crops; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Raboy, V.; Gerbasi, P.F.; Young, K.A.; Stoneberg, S.D.; Pickett, S.G.; Bauman, A.T.; Murthy PP, N.; Sheridan, W.F.; Ertl, D.S. Origin and seed phenotype of maize low phytic acid 1–1 and low phytic acid 2–1. Plant Physiol. 2000, 124, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Pilu, R.; Panzeri, D.; Gavazzi, G.; Rasmussen, S.K.; Consonni, G.; Nielsen, E. Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (lpa241). Theor. Appl. Genet. 2003, 107, 980–987. [Google Scholar] [CrossRef]
- Shi, J.; Wang, H.; Wu, Y.; Hazebroek, J.; Meeley, R.B.; Ertl, D.S. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003, 131, 507–515. [Google Scholar] [CrossRef]
- Badone, F.C.; Amelotti, M.; Cassani, E.; Pilu, R. Study of low phytic acid1-7 (lpa1-7), a new ZmMRP4 mutation in maize. J. Hered. 2012, 103, 598–605. [Google Scholar] [CrossRef]
- Raboy, V. Low phytic acid Crops: Observations Based on Four Decades of Research. Plants 2020, 9, 140. [Google Scholar] [CrossRef]
- Pilu, R.; Landoni, M.; Cassani, E.; Doria, E.; Nielsen, E. The maize lpa241 mutation causes a remarkable variability of expression and some pleiotropic effects. Crop Sci. 2005, 45, 2096–2105. [Google Scholar] [CrossRef]
- Abhijith, K.P.; Muthusamy, V.; Chhabra, R.; Dosad, S.; Bhatt, V.; Chand, G.; Jaiswal, S.K.; Zunjare, R.U.; Vasudev, S.; Yadava, D.K.; et al. Development and validation of breeder-friendly gene-based marker for lpa1-1 and lpa2-1 genes conferring low phytic acid in maize kernel. 3 Biotech 2020, 10, 121. [Google Scholar] [CrossRef]
- Ragi, S.; Muthusamy, V.; Zunjare, R.U.; Bhatt, V.; Katral, A.; Abhijith, K.P.; Kasana, R.; Gain, N.; Sekhar, J.C.; Yadava, D.K.; et al. Genetic and molecular characterization of sub-tropically adapted low phytate genotypes for utilization in mineral biofortification of maize (Zea mays L.). Crop Pasture Sci. 2021, 73, 104–115. [Google Scholar] [CrossRef]
- Baveja, A.; Chhabra, R.; Panda, K.K.; Muthusamy, V.; Zunjare, R.U.; Hossain, F. Development and validation of multiplex-PCR assay to simultaneously detect favourable alleles of shrunken2, opaque2, crtRB1 and lcyE genes in marker-assisted selection for maize biofortification. J. Plant Biochem. Biotechnol. 2020, 30, 265–274. [Google Scholar] [CrossRef]
- Zunjare, R.U.; Hossain, F.; Muthusamy, V.; Baveja, A.; Chauhan, H.S.; Bhat, J.S.; Thirunavukkarasu, N.; Saha, S.; Gupta, H.S. Development of biofortified maize hybrids through markerassisted stacking of β-carotene hydroxylase, lycopene-ε-cyclase and opaque2 genes. Front. Plant Sci. 2018, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, W.; Zhao, Q.; Zhou, X.; Jiang, L.; Ma, S.; Liu, X.; Li, Y.; Zhang, C.; Fan, Y.; et al. Analysis of weighted co-regulatory networks in maize provides insights into new genes and regulatory mechanisms related to inositol phosphate metabolism. BMC Genom. 2016, 17, 129. [Google Scholar] [CrossRef]
- Vallabhaneni, R.; Wurtzel, E.T. Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol. 2009, 150, 562–572. [Google Scholar] [CrossRef]
- Zhan, J.; Li, G.; Ryu, C.H.; Ma, C.; Zhang, S.; Lloyd, A.; Hunter, B.G.; Larkins, B.A.; Drews, G.N.; Wang, X.; et al. Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell 2018, 30, 2425–2446. [Google Scholar] [CrossRef]
- Shi, J.; Wang, H.; Schellin, K.; Li, B.; Faller, M.; Stoop, J.M.; Meeley, R.B.; Ertl, D.S.; Ranch, J.P.; Glassman, K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 2007, 25, 930–937. [Google Scholar] [CrossRef]
- Mehta, B.K.; Chhabra, R.; Muthusamy, V.; Zunjare, R.U.; Baveja, A.; Chauhan, H.S.; Prakash, N.R.; Chalam, V.C.; Singh, A.K.; Hossain, F. Expression analysis of β-carotene hydroxylase1 and opaque2 genes governing accumulation of provitamin-A, lysine and tryptophan during kernel development in biofortified sweet corn. 3 Biotech 2021, 11, 325. [Google Scholar] [CrossRef]
- Hossain, F.; Muthusamy, V.; Pandey, N.; Vishwakarma, A.K.; Baveja, A.; Zunjare, R.U.; Thirunavukkarasu, N.; Saha, S.; Manjaiah, K.M.M.; Prasanna, B.M.; et al. Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J. Genet. 2018, 97, 287–298. [Google Scholar] [CrossRef]
- Baveja, A.; Chhabra, R.; Panda, K.K.; Muthusamy, V.; Mehta, B.K.; Mishra, S.J.; Zunjare, R.U.; Hossain, F. Expression analysis of opaque2, crtRB1 and shrunken2 genes during different stages of kernel development in biofortified sweet corn. J. Cereal Sci. 2022, 105, 103466. [Google Scholar] [CrossRef]
- Feng, F.; Wang, Q.; Zhang, J.; Yang, R.; Li, X. Assessment of carotenoid and tocopherol level in sweet corn inbred lines during kernel development stages. Indian J. Genet. Plant Breed. 2015, 75, 196–200. [Google Scholar] [CrossRef]
- Yan, J.; Kandianis, C.B.; Harjes, C.E.; Bai, L.; Kim, E.H.; Yang, X.; Skinner, D.J.; Fu, Z.; Mitchell, S.; Li, Q.; et al. Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 2010, 42, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.S.; Chhabra, R.; Rashmi, T.; Muthusamy, V.; Zunjare, R.U.; Mishra, S.J.; Gain, N.; Mehta, B.K.; Singh, A.K.; Gupta, H.S.; et al. Impact of vte4 and crtRB1 genes on composition of vitamin-E and provitamin-A carotenoids during kernel-stages in sweet corn. J. Food Compos. Anal. 2022, 105, 104264. [Google Scholar] [CrossRef]
- Dutta, S.; Muthusamy, V.; Chhabra, R.; Baveja, A.; Zunjare, R.U.; Mondal, T.K.; Yadava, D.K.; Hossain, F. Low expression of carotenoids cleavage dioxygenase 1(ccd1) gene improves the retention of provitamin-A in maize grains during storage. Mol. Genet. Genom. 2021, 296, 141–153. [Google Scholar] [CrossRef]
- Ueda, T.; Waverczak, W.; Ward, K.; Sher, M.; Ketudat, M.; Schmidt, R.J.; Messing, J. Mutation of 22 and 27-kD zein promoters affect transactivation by the opaque-2 protein. Plant Cell 1992, 4, 701–709. [Google Scholar]
- Dennis, E.S.; Gerlach, W.L.; Pryor, A.J.; Bennetzen, J.L.; Inglis, A.; Llewellyn, D.; Sachs, M.M.; Ferl, R.J.; Peacock, W.J. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic. Acids. Res. 1984, 12, 3983–4000. [Google Scholar] [CrossRef]
- Mehta, B.K.; Muthusamy, V.; Baveja, A.; Chauhan, H.S.; Chhabra, R.; Bhatt, V.; Chand, G.; Zunjare, R.U.; Singh, A.K.; Hossain, F. Composition analysis of lysine, tryptophan and provitamin-A during different stages of kernel development in biofortified sweet corn. J. Food Compos. Anal. 2020, 94, 103625. [Google Scholar] [CrossRef]
- Lorenz, A.J.; Scott, M.P.; Lamkey, K.R. Quantitative determination of phytate and inorganic phosphorus for maize breeding. Crop Sci. 2007, 47, 600–604. [Google Scholar] [CrossRef]
- Muthusamy, V.; Hossain, F.; Thirunavukkarasu, N.; Choudhary, M.; Saha, S.; Bhat, J.S.; Prasanna, B.M.; Gupta, H.S. Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 2014, 9, e113583. [Google Scholar] [CrossRef]
- Sarika, K.; Hossain, F.; Muthusamy, V.; Zunjare, R.U.; Baveja, A.; Goswami, R.; Bhat, J.S.; Saha, S.; Gupta, H.S. Marker-assisted pyramiding of opaque2 and novel opaque16 genes for further enrichment of lysine and tryptophan in sub-tropical maize. Plant Sci. 2018, 272, 142–152. [Google Scholar] [CrossRef]
- Kurilich, A.C.; Juvik, J.A. Quantification of carotenoid and tocopherol antioxidants in Zea mays. J. Agric. Food Chem. 1999, 47, 1948–1955. [Google Scholar] [CrossRef] [PubMed]
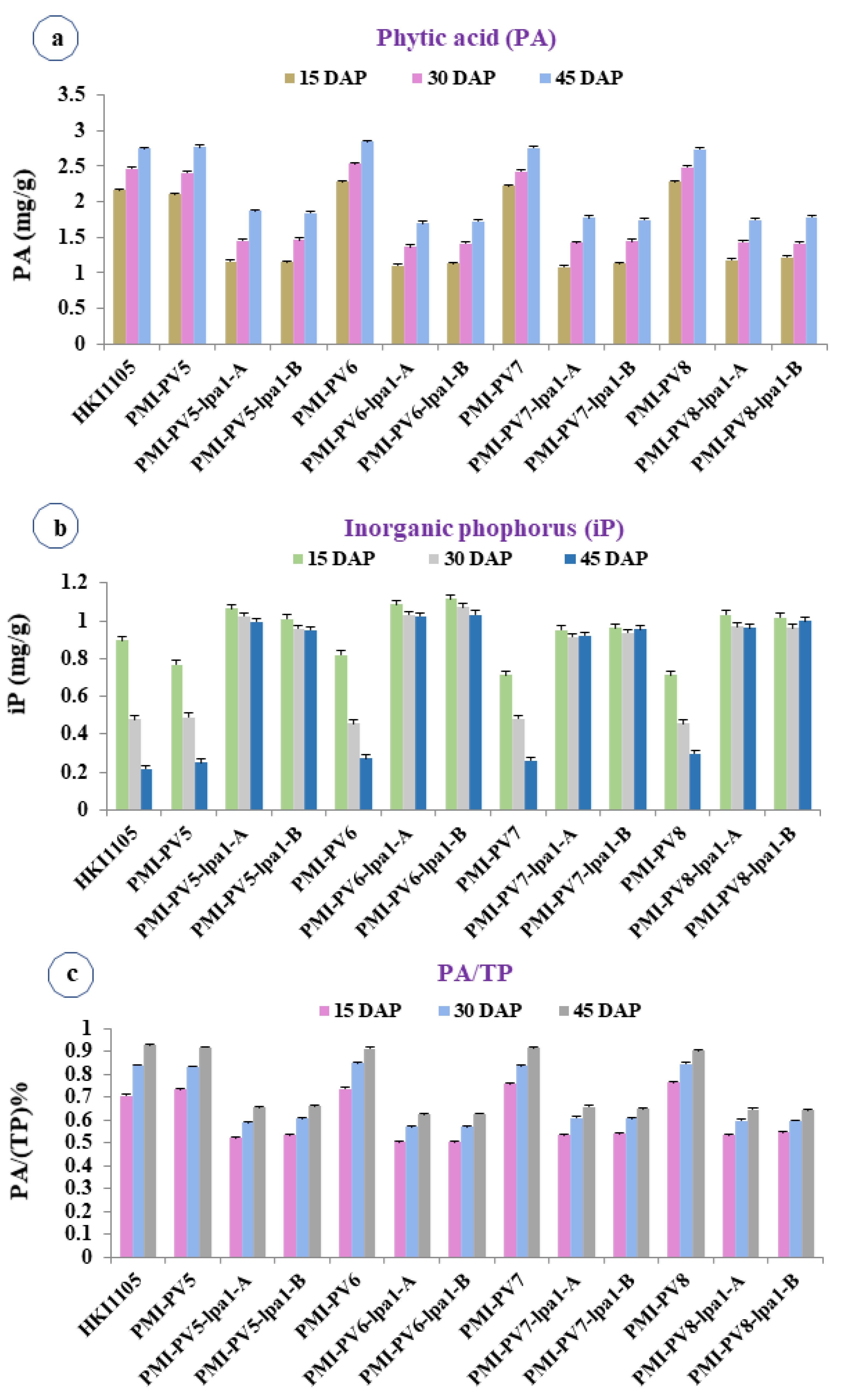

| Source of Variation | df | PA | iP | TP | PA/TP | Lysine | Tryptophan | ProA |
|---|---|---|---|---|---|---|---|---|
| Replication | 2 | 0.00090 | 0.00023 | 0.00158 | 0.00001 | 0.00006 | 0.00001 | 0.00100 |
| Genotype (G) | 12 | 2.47800 ** | 0.57529 ** | 0.69676 ** | 0.13798 ** | 0.04815 ** | 0.00305 ** | 75.62000 ** |
| Days after pollination (DAP) | 2 | 3.55780 ** | 0.53805 ** | 1.39162 ** | 0.19262 ** | 0.22726 ** | 0.03695 ** | 259.14900 ** |
| G × DAP | 24 | 0.00470 ** | 0.04631** | 0.06084 ** | 0.00099 ** | 0.00092 ** | 0.00010 ** | 0.33000 ** |
| Error | 76 | 0.00170 | 0.00118 | 0.00352 | 0.00009 | 0.00003 | 0.00002 | 0.00800 |
| Source of Variation | df | lpa1-1 | crtRB1 | o2 |
|---|---|---|---|---|
| Replication | 2 | 0.16210 | 0.00062 | 0.00002 |
| Genotype (G) | 12 | 1.45110 ** | 0.05380 ** | 0.13870 ** |
| Days after pollination (DAP) | 2 | 8.40920 ** | 0.00901** | 0.09144 ** |
| Genotype × DAP | 24 | 0.39270 ** | 0.00151 ** | 0.01889 ** |
| Error | 76 | 0.05520 | 0.00061 | 0.00044 |
| Genotypes | lpa1-1 | crtRB1 | o2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 15 DAP | 30 DAP | 45 DAP | 15 DAP | 30 DAP | 45 DAP | 15 DAP | 30 DAP | 45 DAP | |
| PMI-PV5 | 2.091 | 0.880 | 0.276 | 0.032 | 0.025 | 0.016 | 0.061 | 0.027 | 0.012 |
| PMI-PV5-lpa1-A | 0.464 | 0.214 | 0.099 | 0.030 | 0.026 | 0.017 | 0.062 | 0.027 | 0.016 |
| PMI-PV5-lpa1-B | 0.473 | 0.222 | 0.075 | 0.037 | 0.028 | 0.014 | 0.065 | 0.024 | 0.012 |
| PMI-PV6 | 1.816 | 0.715 | 0.252 | 0.035 | 0.025 | 0.013 | 0.083 | 0.029 | 0.019 |
| PMI-PV6-lpa1-A | 0.421 | 0.225 | 0.091 | 0.030 | 0.027 | 0.016 | 0.079 | 0.029 | 0.017 |
| PMI-PV6-lpa1-B | 0.469 | 0.241 | 0.087 | 0.032 | 0.026 | 0.010 | 0.077 | 0.031 | 0.018 |
| PMI-PV7 | 2.045 | 0.836 | 0.264 | 0.033 | 0.026 | 0.013 | 0.056 | 0.028 | 0.011 |
| PMI-PV7-lpa1-A | 0.429 | 0.205 | 0.064 | 0.031 | 0.028 | 0.011 | 0.062 | 0.026 | 0.019 |
| PMI-PV7-lpa1-B | 0.451 | 0.219 | 0.074 | 0.030 | 0.029 | 0.018 | 0.057 | 0.027 | 0.016 |
| PMI-PV8 | 2.121 | 0.792 | 0.295 | 0.029 | 0.028 | 0.018 | 0.067 | 0.025 | 0.020 |
| PMI-PV8-lpa1-A | 0.494 | 0.238 | 0.085 | 0.033 | 0.028 | 0.015 | 0.069 | 0.030 | 0.019 |
| PMI-PV8-lpa1-B | 0.475 | 0.202 | 0.073 | 0.032 | 0.029 | 0.014 | 0.066 | 0.027 | 0.017 |
| HKI1105 | 2.105 | 0.870 | 0.288 | 0.387 | 0.314 | 0.209 | 0.821 | 0.427 | 0.205 |
| CD at 5% | 0.599 | 0.289 | 0.086 | 0.578 | 0.044 | 0.007 | 0.041 | 0.014 | 0.019 |
| SE | 0.205 | 0.099 | 0.029 | 0.020 | 0.015 | 0.002 | 0.014 | 0.040 | 0.006 |
| S. No. | Traits | DAP | Correlation Coefficient (r) |
|---|---|---|---|
| lpa1-1 | |||
| 1 | PA | 15 DAP | 0.99 *** |
| 2 | PA | 30 DAP | 0.98 *** |
| 3 | PA | 45 DAP | 0.98 *** |
| 4 | iP | 15 DAP | −0.90 *** |
| 5 | iP | 30 DAP | −0.97 *** |
| 6 | iP | 45 DAP | −0.98 *** |
| 7 | PA/TP | 15 DAP | 0.98 *** |
| 8 | PA/TP | 30 DAP | 0.98 *** |
| 9 | PA/TP | 45 DAP | 0.98 *** |
| o2 | |||
| 1 | Lysine | 15 DAP | −0.84 *** |
| 2 | Lysine | 30 DAP | −0.91 *** |
| 3 | Lysine | 45 DAP | −0.91 *** |
| 4 | Tryptophan | 15 DAP | −0.92 *** |
| 5 | Tryptophan | 30 DAP | −0.85 *** |
| 6 | Tryptophan | 45 DAP | −0.91 *** |
| crtRB1 | |||
| 7 | ProA | 15 DAP | −0.94 *** |
| 8 | ProA | 30 DAP | −0.94 *** |
| 9 | ProA | 45 DAP | −0.94 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatt, V.; Muthusamy, V.; Panda, K.K.; Katral, A.; Chhabra, R.; Mishra, S.J.; Gopinath, I.; Zunjare, R.U.; Neeraja, C.N.; Rakshit, S.; et al. Expression Dynamics of lpa1 Gene and Accumulation Pattern of Phytate in Maize Genotypes Possessing opaque2 and crtRB1 Genes at Different Stages of Kernel Development. Plants 2023, 12, 1745. https://doi.org/10.3390/plants12091745
Bhatt V, Muthusamy V, Panda KK, Katral A, Chhabra R, Mishra SJ, Gopinath I, Zunjare RU, Neeraja CN, Rakshit S, et al. Expression Dynamics of lpa1 Gene and Accumulation Pattern of Phytate in Maize Genotypes Possessing opaque2 and crtRB1 Genes at Different Stages of Kernel Development. Plants. 2023; 12(9):1745. https://doi.org/10.3390/plants12091745
Chicago/Turabian StyleBhatt, Vinay, Vignesh Muthusamy, Kusuma Kumari Panda, Ashvinkumar Katral, Rashmi Chhabra, Subhra J. Mishra, Ikkurti Gopinath, Rajkumar U. Zunjare, Chirravuri Naga Neeraja, Sujay Rakshit, and et al. 2023. "Expression Dynamics of lpa1 Gene and Accumulation Pattern of Phytate in Maize Genotypes Possessing opaque2 and crtRB1 Genes at Different Stages of Kernel Development" Plants 12, no. 9: 1745. https://doi.org/10.3390/plants12091745
APA StyleBhatt, V., Muthusamy, V., Panda, K. K., Katral, A., Chhabra, R., Mishra, S. J., Gopinath, I., Zunjare, R. U., Neeraja, C. N., Rakshit, S., Yadava, D. K., & Hossain, F. (2023). Expression Dynamics of lpa1 Gene and Accumulation Pattern of Phytate in Maize Genotypes Possessing opaque2 and crtRB1 Genes at Different Stages of Kernel Development. Plants, 12(9), 1745. https://doi.org/10.3390/plants12091745






