Abstract
Glucose-6-phosphate dehydrogenase (G6PD) catalyzes the first committed step of the oxidative pentose phosphate pathway (OPPP). Our recent phosphoproteomics study revealed that the cytosolic G6PD6 isozyme became hyperphosphorylated at Ser12, Thr13 and Ser18, 48 h following phosphate (Pi) resupply to Pi-starved (–Pi) Arabidopsis thaliana cell cultures. The aim of the present study was to assess whether G6PD6 phosphorylation also occurs in shoots or roots following Pi resupply to –Pi Arabidopsis seedlings, and to investigate its relationship with G6PD activity. Interrogation of phosphoproteomic databases indicated that N-terminal, multi-site phosphorylation of G6PD6 and its orthologs is quite prevalent. However, the functions of these phosphorylation events remain unknown. Immunoblotting with an anti-(pSer18 phosphosite-specific G6PD6) antibody confirmed that G6PD6 from Pi-resupplied, but not –Pi, Arabidopsis cell cultures or seedlings (i.e., roots) was phosphorylated at Ser18; this correlated with a significant increase in extractable G6PD activity, and biomass accumulation. Peptide kinase assays of Pi-resupplied cell culture extracts indicated that G6PD6 phosphorylation at Ser18 is catalyzed by a Ca2+-dependent protein kinase (CDPK), which correlates with the ‘CDPK-like’ targeting motif that flanks Ser18. Our results support the hypothesis that N-terminal phosphorylation activates G6PD6 to enhance OPPP flux and thus the production of reducing power (i.e., NADPH) and C-skeletons needed to establish the rapid resumption of growth that ensues Pi-resupply to –Pi Arabidopsis.
1. Introduction
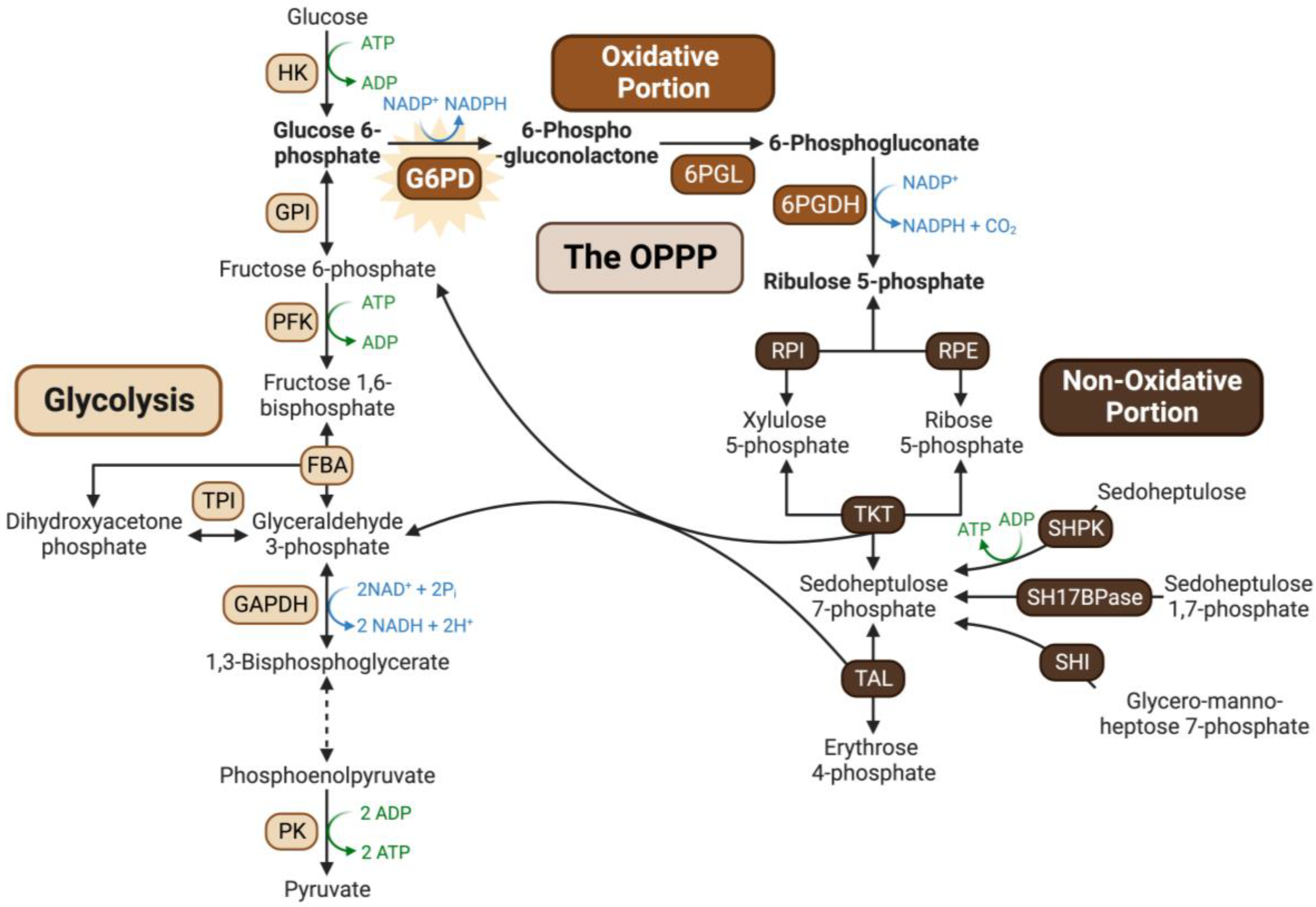
Glucose-6-phosphate dehydrogenase (G6PD) catalyzes the first committed step of the oxidative pentose–phosphate pathway (OPPP) by irreversibly converting glucose-6-phosphate (G6P) and NADP+ into 6-phosphogluconolactone and NADPH. The OPPP’s principal functions are to produce: (i) NADPH needed for ‘reductive’ biosynthetic reactions and reactive oxygen species (ROS) detoxification, and (ii) anabolic precursors (e.g., ribose-5-phosphate and erythrose-4-phosphate) required for nucleotide, aromatic amino acid, and co-factor synthesis (Figure 1) [1].
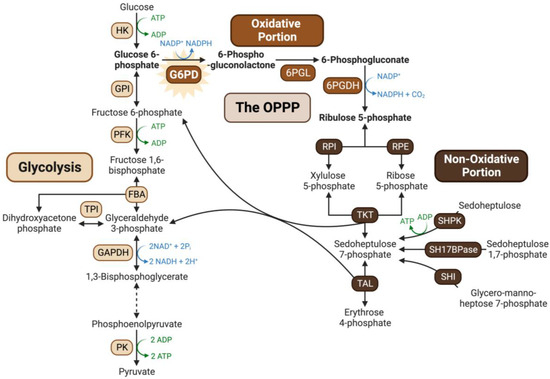
Figure 1.
Overview of the oxidative pentose phosphate pathway. The OPPP integrates with the upper portion of the glycolytic pathway, providing a bypass for G6P oxidation to triose-phosphates while generating reducing power in the form of NADPH, and anabolic precursors such as ribose-5-phosphate and erythrose-4-phosphate required for nucleotide synthesis and the shikimate pathway, respectively. Abbreviations: 6PGL, 6-phosphogluconolactonase; 6PGDH, 6-phosphogluconate dehydrogenase; RPI, ribose 5-phosphate isomerase; RPE, ribulose 5-phosphate epimerase; TKT, transketolase; TAL, transaldolase; SHI, sedoheptulose 7-phosphate isomerase; SH17BPase, sedoheptulose 1,7-biphosphatase; SHPK, sedoheptulokinase; HK, hexokinase; GPI, glucose phosphate isomerase; PFK, phosphofructokinase; FBA, fructose biphosphate aldolase; TPI, triosephosphate isomerase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PK, pyruvate kinase. Figure created with Biorender.com.
Plant G6PDs are derived from a small multigene family that encode distinct cytosolic and plastidic isozymes (G6PDc and G6PDp, respectively) that exhibit marked differences in their respective molecular and kinetic properties. G6PDp isozymes are primarily expressed in green tissues, where they are reversibly inactivated in a light-dependent manner by a thioredoxin-mediated, dithiol–disulfide interconversion mechanism that helps to maintain photosynthetic efficiency [1]. G6PDc is believed to have a more central role in regulating basal metabolism, as it contributes the majority (~80%) of G6PD activity in plant tissues [2,3]. However, the functions and regulation of G6PDc are less understood relative to G6PDp. G6PDc isozymes lack the conserved redox-sensitive cysteine residues found in G6PDp and appear to be primarily controlled by the intracellular NADPH/NADP+ ratio [4,5]. The Arabidopsis G6PD gene family encodes four plastidic (G6PD1-4) and two cytosolic (G6PD5 and G6PD6) isozymes, as indicated by genome annotation and transit peptide sequence analysis [4]. Interestingly, alternative splice variants of G6PD5 localize at the cytosolic face of the endoplasmic reticulum, along with other metabolically sequential OPPP enzymes (i.e., 6-phosphogluconolactonase and 6-phosphogluconate (6PG) dehydrogenase) [6]. These endoplasmic-reticulum-associated G6PD5 isoforms likely operate within a membrane-bound metabolon to provide NADPH for various metabolic pathways under stressful conditions that require enhanced NADPH production via the OPPP.
Apart from supporting anabolism and cell growth, G6PDc and the OPPP play a crucial role in mitigating oxidative stress through the generation of NADPH, an essential co-factor of cellular antioxidant systems [1]. For example, to attenuate salinity-induced oxidative damage, salt-stressed plants require an enhanced supply of NADPH for osmolyte synthesis, ROS detoxification, and the maintenance of ionic balance [7]. G6PDc has been identified as a key player during the acclimation of wheat, barley, soybean, and Arabidopsis thaliana to salinity stress [7,8,9,10]. Salt-stressed Arabidopsis seedlings lacking G6PDc (i.e., g6pd5/6 double mutants) displayed reduced root growth, low germination rates and enhanced accumulation of ROS [11]. Moreover, the salt-stress-induced expression of ascorbate peroxidase and glutathione reductase was attenuated in the atg6pd5/6 mutant, suggesting that G6PDc plays a key role in mediating ROS scavenging during salinity stress [11]. It is also notable that g6pd5/6 knockout plants were more sensitive to salt stress than the corresponding single mutants, which displayed only slight phenotypic impairments [11]. Nevertheless, the high abundance of G6PD6 mRNA and in vivo activity throughout the plant indicates that G6PD6 is the predominant G6PDc isozyme expressed in Arabidopsis [4]. G6PD5 and G6PD6 are likely coordinated in their role of cytosolic NADPH production within various Arabidopsis tissues, and appear to be post-transcriptionally regulated by an unknown mechanism, given the low correlation between G6PD5 and G6PD6 mRNA abundance and corresponding G6PD activities or amounts [4,12].
Inorganic phosphate (Pi; HPO42−) is an indispensable but environmentally limiting macronutrient that has crucial functions in energy transduction and metabolic regulation while also serving as a structural constituent of essential biomolecules such as nucleic acids, phospholipids, sugar-phosphates, and adenylates. Pi also controls the activity of many plant proteins via its covalent attachment to phosphoproteins and/or by acting as an allosteric activator or inhibitor of key regulatory enzymes of central metabolism [13]. However, Pi is one of the least available macronutrients in many terrestrial and aquatic ecosystems. Plants have therefore evolved an impressive array of adaptive strategies known as the ‘Pi starvation response’ (PSR) that facilitate their acclimation to the suboptimal levels of soluble Pi that characterize most soils. The PSR is regulated by Pi-starvation inducible genes, as well as various post-transcriptional and post-translational strategies that reprioritize internal Pi use while maximizing root Pi acquisition [14]. For example, –Pi plants optimize root Pi uptake via the following processes: (i) induction of high-affinity Pi transporters that actively assimilate Pi against a steep concentration gradient; and (ii) exudation of Pi-mobilizing compounds such as organic acid anions (e.g., malate, citrate), nucleases, and purple acid phosphatases [14]. Protein post-translational modifications (PTMs) such as phosphorylation and ubiquitination play a central role in plant PSR signal transduction pathways and the control of phosphate-starvation-inducible gene expression and gene products. For example, during Pi deprivation, root phosphoenolpyruvate carboxylase (PEPC), a tightly regulated cytosolic enzyme situated at a pivotal branchpoint of central metabolism, is activated by protein-kinase-mediated phosphorylation at a conserved N-terminal serine residue [15,16]. This enhances anaplerotic partitioning of imported sucrose into Pi-mobilizing organic acid anions [16,17].
We recently reported that proteomic remodeling occurring 48 h following the resupply of 2 mM Pi to heterotrophic, –Pi Arabidopsis cell cultures occurred largely at the level of protein phosphorylation, with limited fluctuations in protein abundance [18]. It is notable that G6PD6 was one of the most differentially phosphorylated of the over 500 proteins whose phosphorylation status significantly changed following Pi resupply. Multiple N-terminal residues of G6PD6 (i.e., Ser12, Thr13 and Ser18) were hyperphosphorylated following Pi resupply [18], highlighting a potential link between Pi nutrition and a novel post-translational G6PD6 regulatory mechanism via reversible phosphorylation. The initial aim of the present study was to interrogate phosphoproteomic datasets to assess the prevalence of N-terminal phosphorylation of G6PD6 and its orthologs. We then developed an anti-(pSer18 phosphosite-specific G6PD6) antibody (anti-pSer18) for immunoblot assessment of the impact of Pi nutrition on G6PD6’s in vivo Ser18 phosphorylation status in Arabidopsis cell cultures and seedlings. Our results are consistent with the hypothesis that this PTM activates G6PD6 to mediate increased cytosolic OPPP flux, and thus enhance the provision of NADPH and biosynthetic precursors needed to support the resumption of cell growth that rapidly occurs following Pi resupply.
2. Results
2.1. Multisite N-Terminal Phosphorylation of Arabidopsis G6PD6 and Its Orthologs Is Quite Prevalent
A multiple sequence alignment was conducted to evaluate the conservation of G6PD6’s Ser3, Ser12, Thr13, and Ser18 phosphosites [18,19,20] amongst the N-termini of other G6PDs (Figure 2). These residues are highly conserved between the two Arabidopsis G6PDc paralogs, G6PD5 and G6PD6 (≈90% sequence identity), as well as relatively well conserved between G6PDc orthologs from other vascular plant species (i.e., soybean, potato, tomato, barley, rice). By contrast, these sites are not evident in Arabidopsis G6PDp isozymes (i.e., G6PD1-4), which share much lower overall sequence identities with G6PD6 (≤45%) (Figure 2).

Figure 2.
Multiple sequence alignment of Arabidopsis G6PD6’s N-terminus with N-termini of other G6PDs. G6PD6 phosphosites that were mapped via LC-MS/MS following 48 h of Pi resupply to –Pi Arabidopsis cell cultures [18] are colored in red and underlined, other experimentally determined phosphosites (Table 1) are colored in red, and corresponding conserved residues within other G6PDs are colored in blue. Arabidopsis G6PD6 is marked with an arrow. N-termini sequences are highlighted as follows: green = Arabidopsis G6PDp; pink = plant G6PDc; yellow = animal G6PD; blue = fungal G6PD; purple = bacterial G6PD. The abbreviations are as follows: An, Aspergillus niger (common mold); At, Arabidopsis thaliana (thale cress); Ce, Caenorhabditis elegans (nematode); Cv, Chlorella vulgaris (green alga); Ec, Escherichia coli; Gm, Glycine max (soybean); Hs, Homo sapiens (human); Hv, Hordeum vulgare (barley); Mm, Mus musculus (mouse); Np, Nostoc punctiforme (cyanobacterium); Os, Oryza sativa (rice); Pp, Physcomitrium patens (earthmoss); Rn, Rattus norvegicus (rat); Sl, Solanum lycopersicum (tomato); St, Solanum tuberosum (potato). Protein accession numbers are listed in Supplementary Table S1.
Interrogation of various phosphoproteomic datasets indicates that N-terminal phosphorylation of Arabidopsis G6PD6 and its orthologs is relatively common (Figure 2 and Table 1). Of the two Arabidopsis G6PDc isozymes, N-terminal phosphorylation of G6PD5 appears to occur less frequently (Table 1), implying that G6PD6 may be the primary G6PDc isozyme subjected to this PTM. Similar N-terminal phosphosites have also been identified in vertebrate (Ser8/Thr10 for human and mouse) and invertebrate (Ser7 for Caenorhabditis elegans) animal G6PDs (Figure 2) despite their phylogenetic distance from Arabidopsis G6PD6 (≤48% sequence identity).

Table 1.
Summary of vascular plant G6PDc phosphosites mapped during various phosphoproteomic studies.
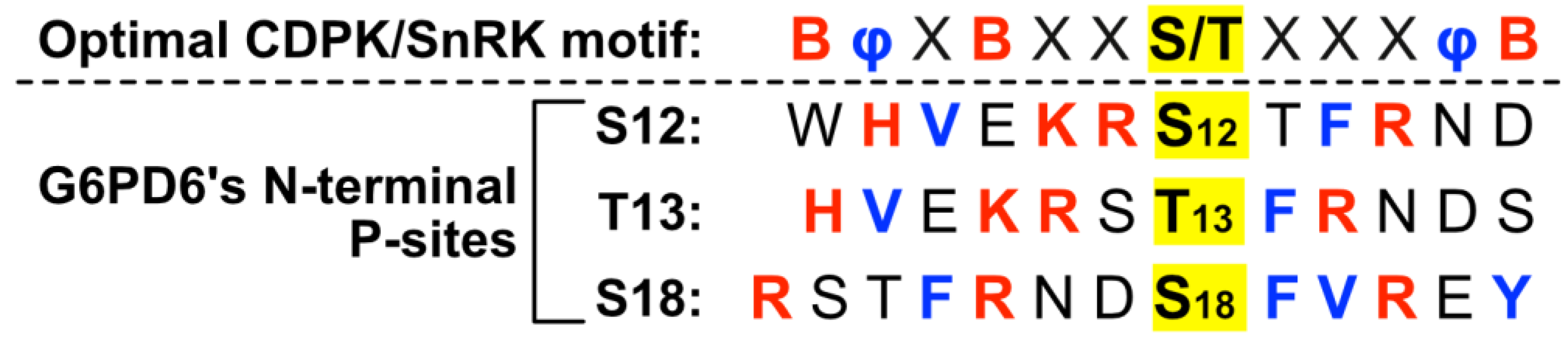
2.2. Residues Flanking G6PD6’s N-Terminal Phosphosites Are Representative of a Basophilic Motif
Residues neighboring G6PD6’s N-terminal phosphosites comprise an S- or T-type motif [40], with basic (i.e., Lys, Arg or His) residues occurring at positions −1, −2, −5 and +3 for Ser12; −2, −3, −6 and +2 for Thr13; and −3, −7 and +3 for Ser18 (Figure 3). Basophilic motifs are commonly recognized by Ca2+-dependent protein kinases (CDPKs), SNF1-related protein kinases (SnRKs), and Ca2+/calmodulin-dependent protein kinases [40,41,42]. In particular, G6PD6’s N-terminal phosphosites present several motifs reminiscent of the optimal recognition motif for plant CDPKs and SnRKs, i.e., containing basic residues at positions −6, −3 and +5, and hydrophobic residues at positions −5 and +4 (Figure 3) [40,41,42]. N-terminal phosphosites of G6PDc isozymes from other plant species possess similar flanking residues (Figure 2), indicating a potentially conserved mechanism of regulation.
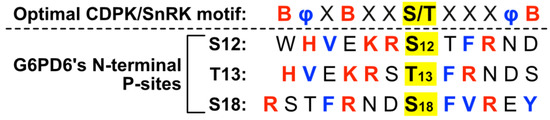
Figure 3.
Motif analysis of phosphorylated Arabidopsis G6PD6 residues. Sequence alignment of amino acids flanking G6PD6 Ser12, Thr13, and Ser18 residues that were phosphorylated 48 h following Pi resupply to –Pi Arabidopsis cell cultures [18]; basic and hydrophobic residues are colored red and blue, respectively. The optimal CDPK/SnRK recognition motif is located above the alignment to highlight residues of interest, where φ represents a hydrophobic amino acid, B represents a basic amino acid, and X represents any amino acid.
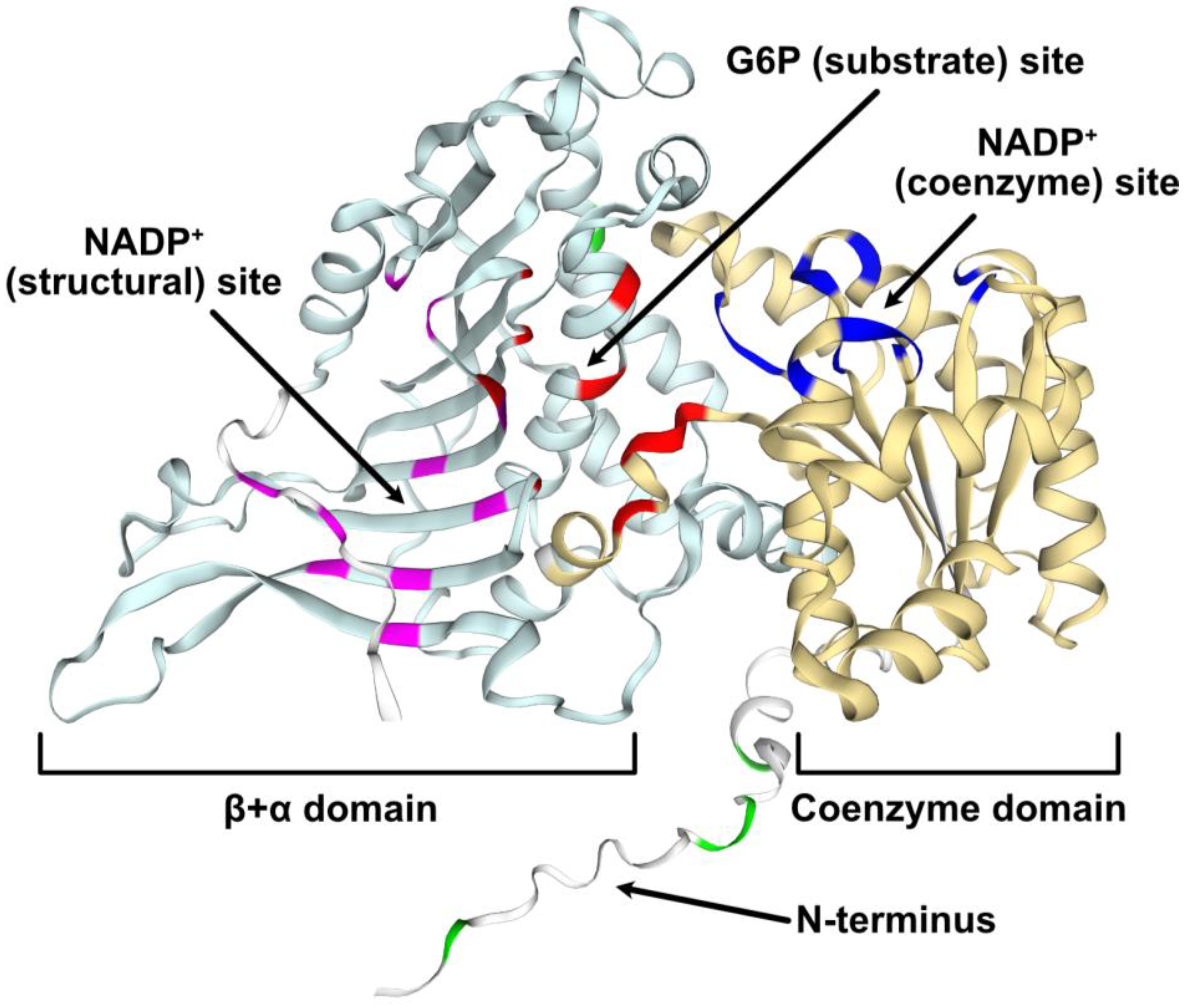
2.3. Structural Modeling of G6PD6 Indicates That Its N-Terminal Region Is Intrinsically Disordered
The Arabidopsis G6PD6 gene encodes a 515 amino acid polypeptide with a predicted size of 59 kD. The first 22 N-terminal residues of G6PD6 appear to be intrinsically disordered as predicted via MobiDB and structural modeling (Figure 4).
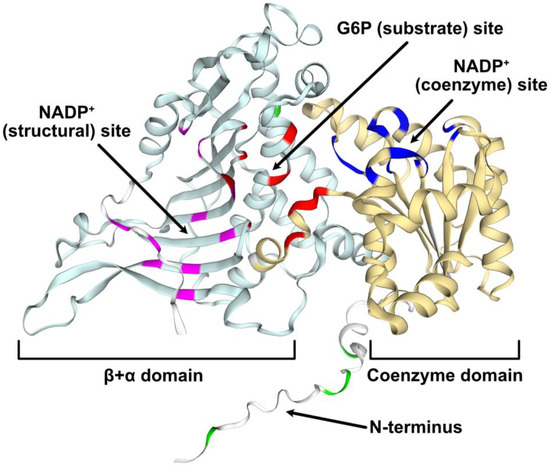
Figure 4.
Structural model of Arabidopsis G6PD6. The predicted G6PD6 protein model (AF-Q9FJI5-F1) was obtained from AlphaFold and annotated in the SWISS-MODEL workspace. Based on pairwise sequence alignment with human G6PD, conserved key residues that interact with G6P or NADP+ (deduced from crystallized structures of human G6PD) are annotated as follows: red = G6P (substrate); dark blue = NADP+ (coenzyme); magenta = NADP+ (structural) [43]. Phosphorylated residues are annotated in green. Domains were retrieved from InterPro (coenzyme (yellow) = IPR036291, β+α (light blue) = IPR022675) and binding sites are annotated accordingly.
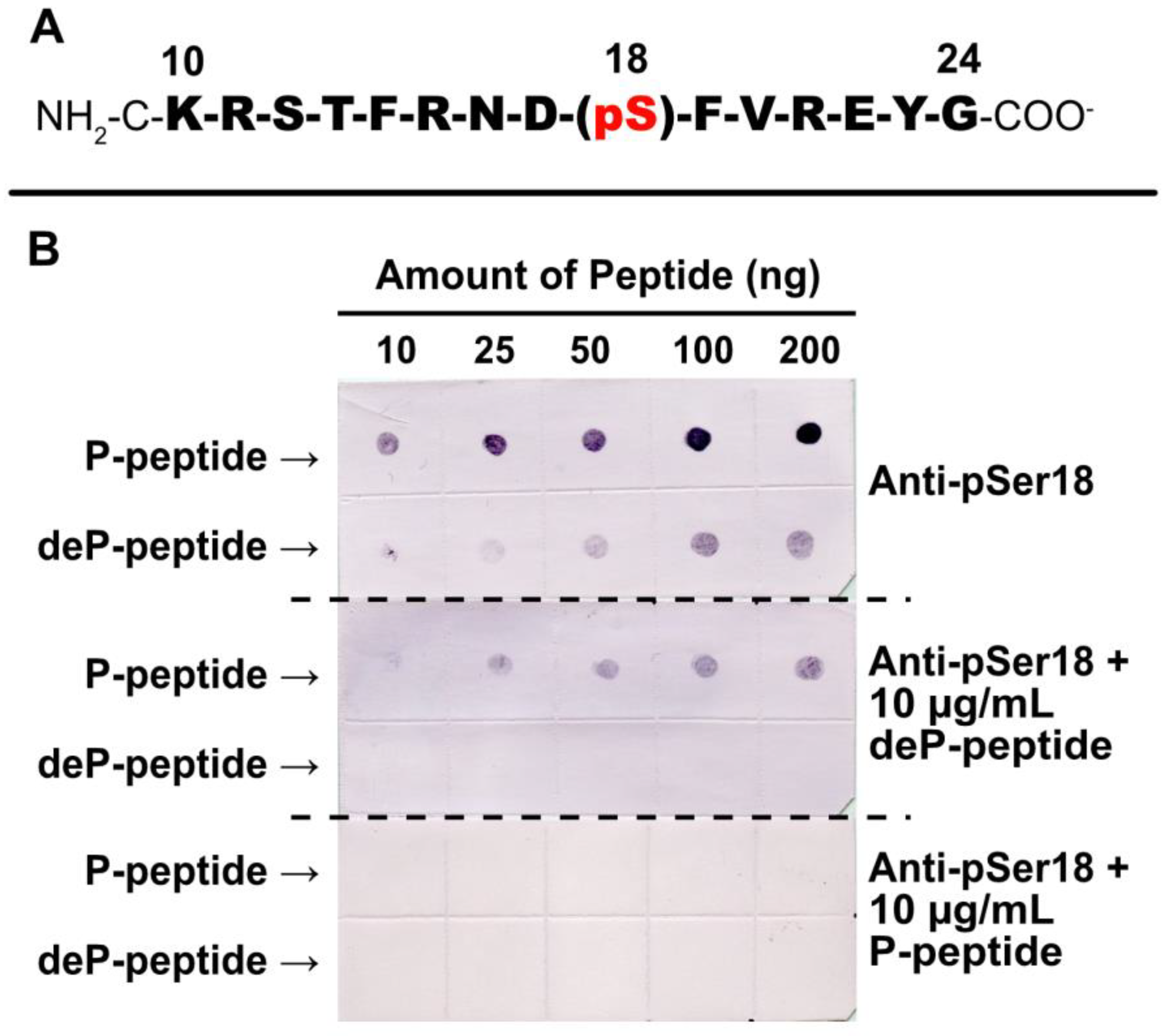
2.4. Specificity of Anti-pSer18 Immune Serum
Rabbit-phosphosite-specific antiserum was raised against a synthetic phospho- (P-) peptide corresponding to residues 10–24 of G6PD6 with phosphorylated Ser18 (Figure 5A). pSer18 was selected (as opposed to pSer12 or pThr13) as (i) it is the most frequently documented P-site of G6PD6 (and its orthologs) in various phosphoproteomic studies (Table 1), and (ii) Mehta et al. [18] determined that G6PD6’s Ser18 residue exhibited the greatest log fold-increase in phosphorylation 48 h following Pi resupply to –Pi Arabidopsis cells.
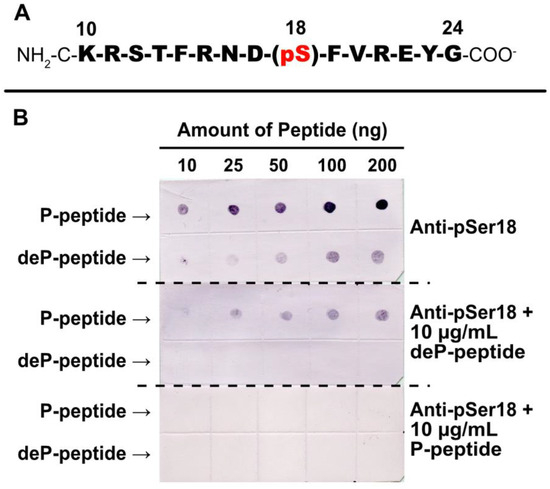
Figure 5.
Specificity of phosphosite-specific antibody raised against pSer18 of Arabidopsis G6PD6. (A) Sequence of phosphorylated synthetic peptide that was covalently coupled to keyhole limpet hemocyanin (KLH) and used for rabbit immunization. The sequence numbering represents amino acid position relative to G6PD6’s N-terminus. The Ser18 phosphorylation site is indicated. The peptide was synthesized with an extra N-terminal Cys residue to facilitate its conjugation to KLH. (B) Anti-pSer18 immune serum (±10 μg/mL of phospho- (P-) or dephospho- (deP-) peptide) was used to probe immunodot blots of varying amounts of the pSer18 P-peptide and corresponding deP-peptide.
Immunodot blotting of synthetic P- and dephospho- (deP-) peptides revealed that a 1:500 dilution of anti-pSer18 antiserum cross-reacted with as little as 10 ng of the P-peptide, but also weakly cross-reacted with higher amounts of the corresponding deP-peptide (Figure 5B). However, the inclusion of 10 µg/mL deP-peptide with the diluted antiserum abolished this non-specific cross-reaction. Therefore, all subsequent anti-pSer18 immunoblots were probed in the presence of 10 µg/mL deP-peptide unless otherwise indicated. By contrast, incubation with 10 µg/mL of blocking P-peptide eliminated any cross-reaction of anti-pSer18 with either P- or deP-peptide (Figure 5B).
2.5. G6PD6 Phosphorylation at Ser18 Is Influenced by Pi Nutrition in Arabidopsis Suspension Cells and Roots
The use of anti-pSer18 for examining G6PD6 phosphorylation via immunoblotting was complemented with rabbit antiserum raised against purified pea G6PDc (anti-pea G6PDc) [44]; pea G6PDc shares 81.4% sequence identity with Arabidopsis G6PD6. The anti-(pea G6PDc), which detects both P- and deP-G6PD6, allowed for the normalization of total G6PD6 on immunoblots. Clarified protein extracts of –Pi and 48 h Pi-resupplied Arabidopsis suspension cells and seedlings (i.e., roots and shoots) cultivated in sterile liquid media were subjected to SDS-PAGE followed by immunoblotting with anti-pSer18 and anti-(pea G6PDc) (Figure 6 and Figure S1).
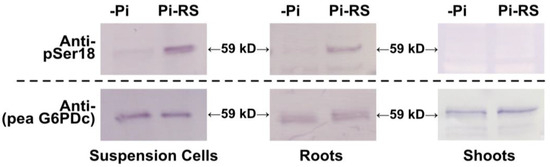
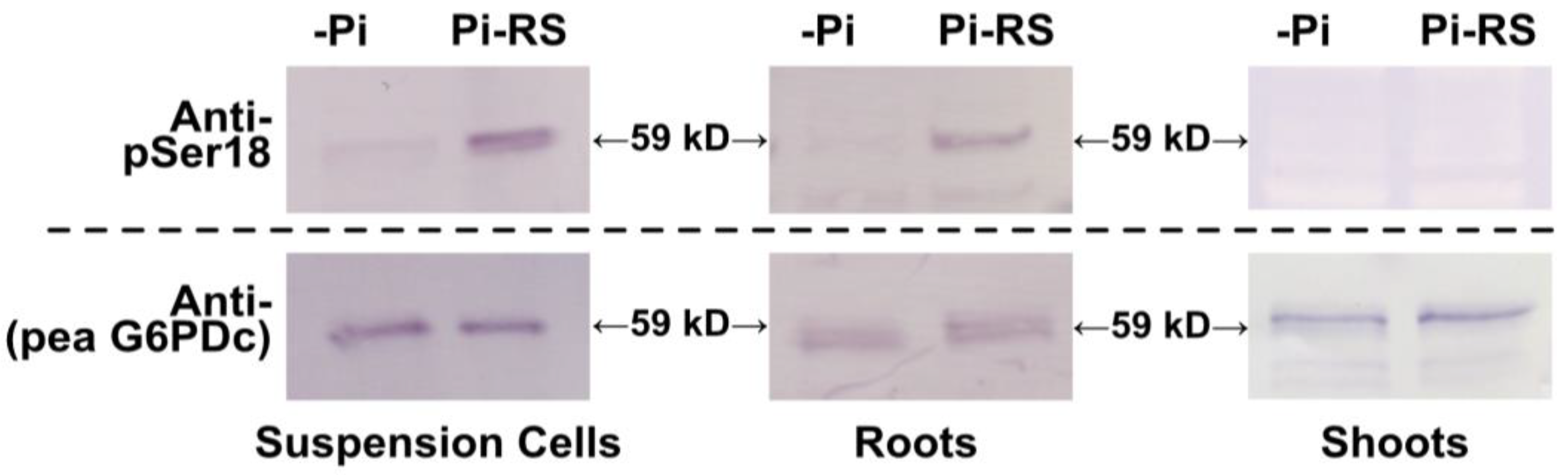
Figure 6.
G6PD6 is in vivo phosphorylated at Ser18, 48 h following Pi resupply to Pi-starved Arabidopsis suspension cells and seedlings. Clarified extracts from –Pi and 48 h Pi-resupplied (Pi-RS) cells or seedlings (shoots and roots) were subjected to SDS/PAGE followed by immunoblotting with anti-pSer18 (+10 μg/mL dephospho- (deP)-peptide) or anti-(pea G6PDc). Approximately 8 and 1 µg of protein were loaded into each lane of the anti-pSer18 and anti-(pea G6PDc) immunoblots, respectively.
Phosphorylation of 59 kD G6PD6 polypeptides at Ser18 was evident on immunoblots of Pi-resupplied cells and root extracts probed with anti-pSer18 in the presence of 10 µg/mL corresponding deP-peptide, with no apparent change in the abundance of 59 kD G6PD6 polypeptides (based upon anti-(pea G6PDc) immunoblotting) (Figure 6). By contrast, no cross-reaction of anti-pSer18 with 59 kD G6PD6 polypeptides occurred when immunoblots of the Pi-resupplied suspension cell or root extracts were probed in the presence of 10 µg/mL of the blocking P-peptide (Supplementary Figure S2); this further attests to the specificity of anti-pSer18 in exclusively cross-reacting with 59 kD G6PD6 polypeptides phosphorylated at Ser18. The collective results confirm that in vivo G6PD6 phosphorylation at Ser18 occurs in Arabidopsis cell cultures and seedlings and is dependent on nutritional Pi status. It is also notable that Ser18 phosphorylation of G6PD6 in response to Pi resupply appears to be root- and cell-culture-specific as it was not detected on immunoblots of shoot extracts (Figure 6 and Figure S1). The anti-(pea G6PDc) immunoblots of extracts prepared from –Pi and/or Pi-resupplied cell culture and roots also detected an immunoreactive 70 kD polypeptide that did not cross-react with anti-pSer18 (Supplementary Figures S1 and S2). This is likely due to a non-specific cross-reaction since the predicted size of Arabidopsis G6PD1-6 polypeptides ranges from 59 to 65 kD [4].
2.6. Pi-Resupply-Mediated Phosphorylation of Arabidopsis Root and Cell Culture G6PD6 at Ser18 Is Correlated with a Significant Increase in G6PD Activity and Biomass
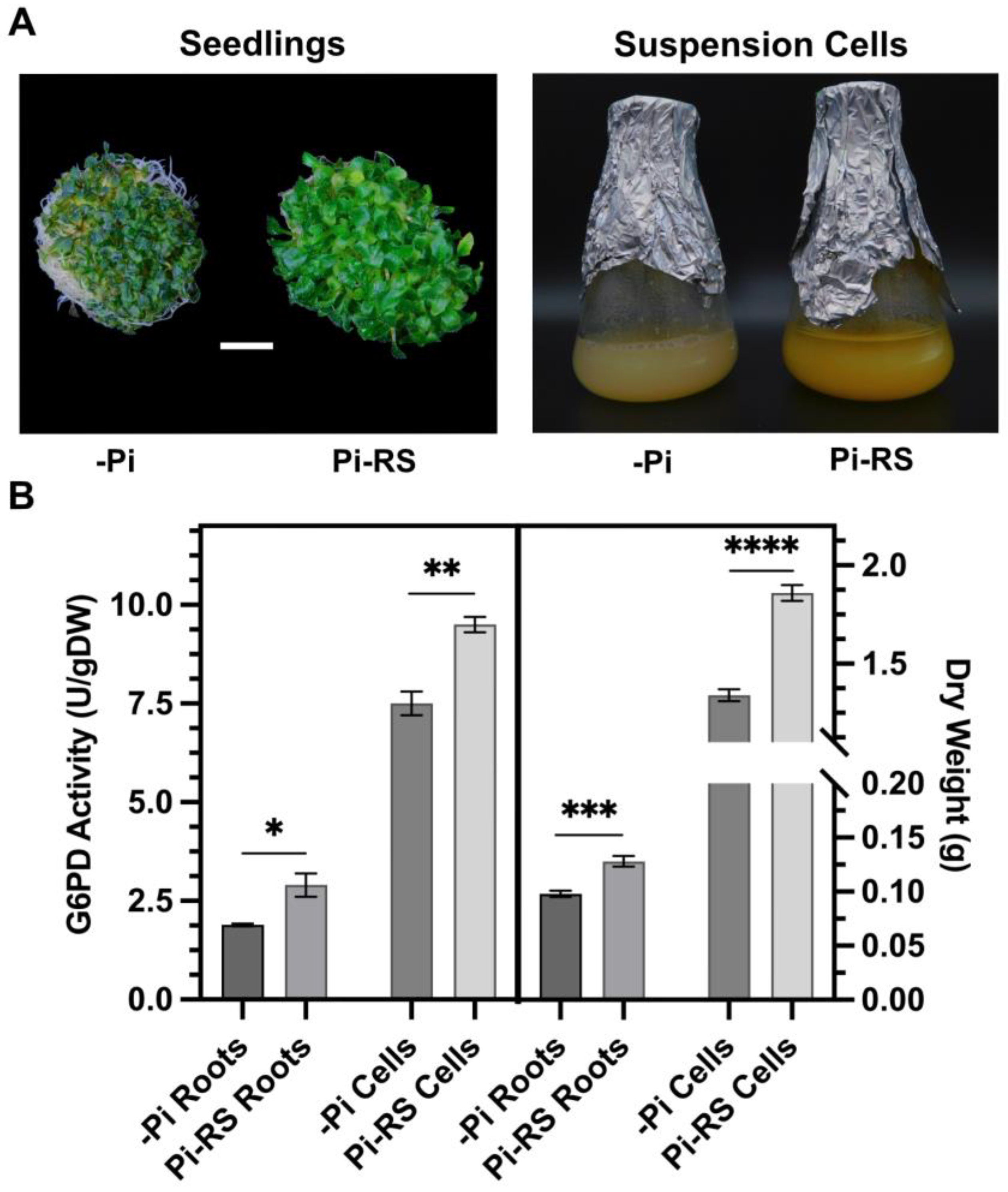
G6PD activity was assayed in desalted protein extracts from –Pi and Pi-resupplied Arabidopsis seedlings (roots) and suspension cells cultivated in liquid nutrient media (Figure 7A,B). No contaminating ‘NADP+ reductase’ activity was detected in G6PD reaction mixtures lacking G6P. Furthermore, the addition of 2 mM dithiothreitol, which reduces disulfide bonds to dithiols [45], had no effect on G6PD activities of the root or cell culture extracts. This corroborates evidence that G6PD activity of heterotrophic plant tissues is largely cytosolic since G6PDp isozymes are reversibly inactivated following dithiothreitol-mediated disulfide-to-dithiol interconversion [1,2]. Pi resupply to –Pi seedlings correlated with a significant (54%) increase in the G6PD activity of root extracts (Figure 7B). Additionally, a significant 31% increase in root biomass was observed in the Pi-resupplied seedlings (Figure 7B). Likewise, Pi resupply induced a significant 27% and 39% increase in G6PD activity and biomass, respectively, of the Arabidopsis suspension cells (Figure 7B).
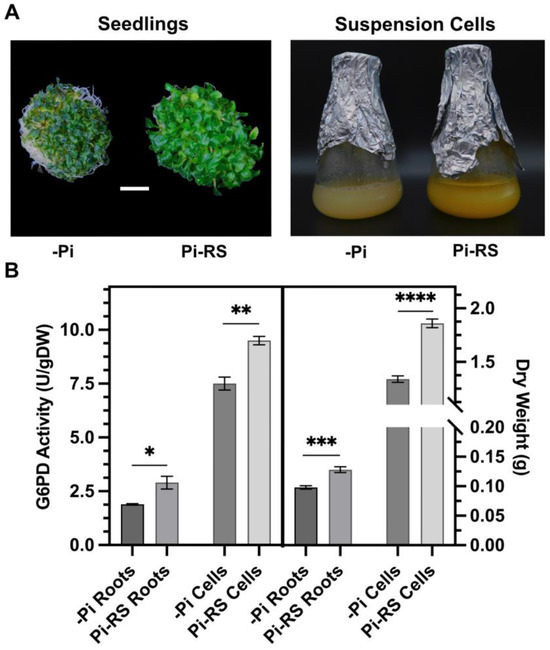
Figure 7.
Impact of Pi deprivation (–Pi) and 48 h Pi resupply (Pi-RS) on appearance, G6PD activity, and biomass accumulation of Arabidopsis seedling roots and suspension cells. (A) Images are representative of at least five replicates (bar = 1 cm). (B) Dry weight values represent average gDW per flask of liquid cultured seedlings (roots) or suspension cells. All values represent means (±SE) of n ≥ 3 biological replicates; statistical significance was evaluated via a two-tailed, unpaired Student’s t-test (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
2.7. G6PD6 Phosphorylation at Ser18 Appears to Be Catalyzed by a CDPK
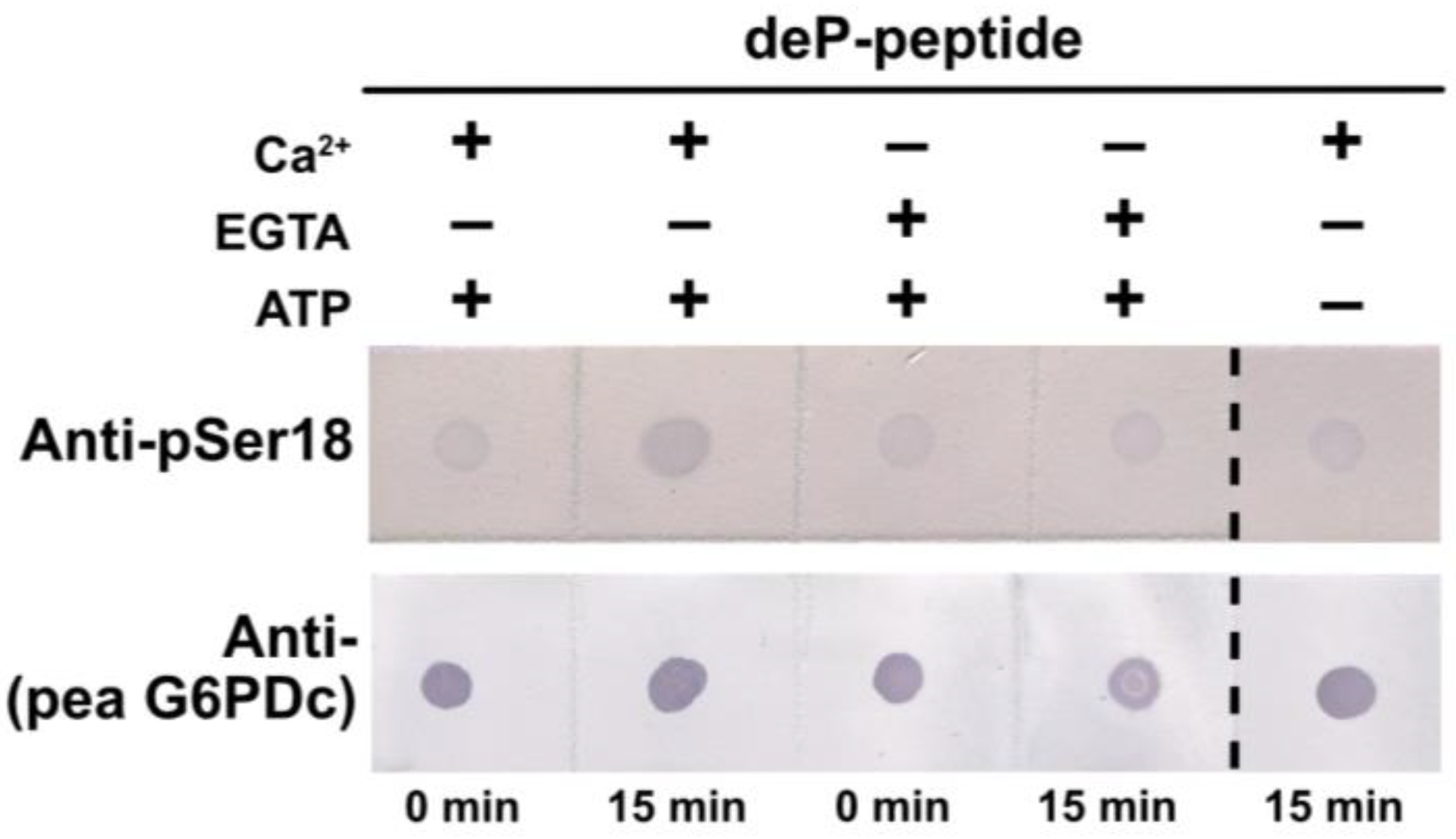
Semi-quantitative immunodot blot kinase assays using anti-pSer18 indicated that a CDPK present in clarified extracts of the Pi-resupplied Arabidopsis suspension cells catalyzes Ca2+-dependent phosphorylation of the synthetic G6PD6 deP-peptide shown in Figure 5A at Ser18 (Figure 8).
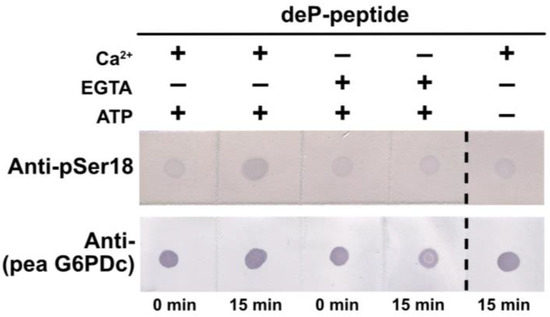
Figure 8.
Evidence that a CDPK catalyzes G6PD6 phosphorylation at Ser18. Reactions were incubated at 30 °C for 15 min with (+) and without (−) 0.5 mM ATP, and either 0.2 mM Ca2+ (+) or 5 mM EGTA (−) in a final volume of 10 µL. The reactions containing 0.4 mg/mL of synthetic dephospho- (deP-) peptide (corresponding to residues 10–24 of G6PD6; Figure 5A) were incubated with 4 µL of desalted clarified extract (equivalent to 36 µg of protein) from Pi-resupplied Arabidopsis cell cultures, and then dot-blotted with anti-pSer18 (+10 μg/mL deP-peptide) or anti-(pea G6PDc) (400 ng peptide/dot).
3. Discussion
G6PD is an integral enzyme of central C-metabolism that balances cellular redox status and maintains a critical control point for the diversion of G6P flux towards the OPPP for NADPH and pentose phosphate production (Figure 1). The present study presents evidence regarding the occurrence and possible function of multisite N-terminal phosphorylation of Arabidopsis G6PD6 in the context of Pi-nutrition. Sequence alignment and interrogation of phosphoproteomic datasets indicated that G6PD6’s N-terminal phosphosites are conserved in its paralogs and orthologs, and that G6PD6 and several of its orthologs from vascular plant and animal species are in vivo phosphorylated at similar sites in a range of tissues under various physiological conditions (Figure 2 and Table 1). It is notable that barley G6PDc (HvG6PD) and Arabidopsis G6PD6 were both phosphorylated at a comparable N-terminal serine residues following Pi resupply to –Pi plants (i.e., Ser14 and Ser13, respectively) (Figure 2 and Table 1), suggesting that this phosphorylation event may be a widespread G6PDc PTM of Pi-refed plants. By contrast, fungal and prokaryotic species have no conservation of Arabidopsis G6PD6’s N-terminal residues nor phosphosites (Figure 2). These results imply that N-terminal phosphorylation is a feature of most eukaryotic G6PDcs. Sequence analysis revealed that the flanking basophilic residues of G6PD6’s N-terminal phosphosites are reminiscent of motifs implicated in CDPK- and SnRK-mediated protein phosphorylation (Figure 3) [40,41,42]. Interestingly, peptide kinase assays using the synthetic G6PD6 deP-peptide (Figure 5A) as a substrate suggested that a CDPK phosphorylates G6PD6 at Ser18 in Pi-resupplied Arabidopsis (Figure 8). CDPKs catalyze site-specific phosphorylation of several regulatory enzymes of the central plant metabolism including nitrate reductase, sucrose synthase, sucrose phosphate synthase, and bacterial-type PEPC [41,46,47,48]. CDPKs also participate in nutrient signaling; for example, Liu et al. [49] found that Arabidopsis nitrate responses were regulated by CDPK-mediated phosphorylation, whereas Mehta et al. [18] reported that several CDPK isozymes and basophilic, CDPK-like motifs were enriched in the phosphoproteome of the Pi-resupplied Arabidopsis cell cultures.
Structural modeling and MobiDB interrogation indicated that the first 22 residues of G6PD6’s N-terminus correspond to an intrinsically disordered region. Annotation of the modeled G6PD6 structure (Figure 4) was based upon analysis of the human G6PD (HsG6PD) crystal structure, which revealed that the first 30 residues of its N-terminus are disordered [50]. Truncated HsG6PD mutants lacking these residues demonstrated normal catalytic activity, suggesting this region is neither essential for catalysis nor involved in tetramer/dimer formation [43,50]. Interestingly, Cys13 of HsG6PD forms a disulfide bridge with Cys446, which likely tethers down the mobile N-terminus to prevent interference with subunit association and catalysis. [50]. By contrast, Arabidopsis G6PD6 has no Cys residues within its N-terminal disordered domain (Figure 2) and would therefore lack the ability to form a stabilizing disulfide bridge. Disordered regions are frequently subject to site-specific phosphorylation and/or promote protein–protein interactions by providing a docking site for binding partners [51]. For example, castor plant bacterial-type PEPC contains an intrinsically disordered domain that (i) mediates its interaction with plant-type PEPC to form class-2 PEPC complexes that associate with the mitochondrial surface [52], and (ii) contains Ser425 and Ser451 phosphorylation sites that inhibit bacterial-type PEPC subunit activity within the class-2 PEPC complex [53,54]. Multisite N-terminal phosphorylation of G6PD6 following Pi resupply to –Pi Arabidopsis [18] could potentially stabilize its N-terminal disordered region and/or mediate the association with a binding partner, thus directly or indirectly influencing catalytic activity. However, the possibility that 14-3-3 proteins bind to and thereby regulate the activity of phospho-G6PD6 is unlikely since none of its N-terminal phosphosites are representative of a 14-3-3 binding motif (i.e., RSXpSXP or RXXpSXP where X is any amino acid) (Figure 2) [55]. Given the variability in the N-terminal sequences of orthologous plant G6PDc isozymes (Figure 2), perhaps the precise locations of G6PD6’s N-terminal phosphosites are not so important. Instead, the collective impact of multisite N-terminal phosphorylation would impart a pronounced, local negative charge that could potentially trigger downstream structural changes.
Phosphosite-specific antibodies provide a remarkably sensitive and specific immunochemical tool to visualize and quantify alterations in the phosphorylation status of a particular amino acid residue within a full-length protein sequence [56]. Immunoblotting using anti-pSer18 and anti-(pea G6PDc) antisera, respectively, revealed that Pi-resupply-mediated Ser18 phosphorylation of 59 kD G6PD6 polypeptides occurs in both Arabidopsis suspension cells and roots, with no obvious change in the amount of G6PD6 (Figure 6). These results corroborate those of Mehta et al. [18] who determined that although G6PD6 was in vivo phosphorylated at Ser18 (as well as Ser12 and Thr13) following Pi resupply to –Pi Arabidopsis suspension cells, its protein abundance was unaffected. The absence of G6PD6 Ser18 phosphorylation in the shoots of Pi-resupplied seedlings (Figure 6) implies that this PTM may play a greater role during Pi stress recovery in roots. It is important to note that the anti-pSer18 could potentially cross-react with pSer18 of G6PD5, as only three G6PD5 residues in positions 10–24 differ from the corresponding residues of G6PD6 (Figure 2). However, the occurrence of pSer18 in G6PD5 has rarely been documented (Table 1), and no change in G6PD5 phosphorylation nor protein abundance was detected following Pi resupply to the –Pi Arabidopsis cell cultures [18]. Furthermore, the cross-reaction of our anti-pSer18 antiserum with a Ser18 phosphorylated G6PD5 is unlikely since phosphosite-specific antibodies are capable of discriminating single-amino-acid substitutions occurring between the sequence of residues flanking phosphosites of paralogous or orthologous proteins [56].
Pi resupply significantly enhanced the G6PD activity of Arabidopsis roots and cell culture extracts (Figure 7B). This increase appears to occur post-translationally since G6PD6 protein abundance appeared to be unaffected between the two treatments (Figure 6) [18]. Similarly, Wakao et al. [12] concluded that the compensatory increase in G6PD6 activity of atg6pd5 T-DNA mutants was likely due to a post-translational mechanism since G6PD6 gene expression was unaffected in the atg6pd5 mutants. The Pi-resupply-mediated increase in root and cell culture G6PD activity and biomass paralleled in vivo phosphorylation of 59 kD G6PD6 polypeptides at Ser18 (Figure 6 and Figure 7), suggesting this PTM helps to activate G6PD6. This novel mechanism of post-translational G6PD6 control is hypothesized to contribute to the activation of cytosolic OPPP flux, and thus the production of NADPH and C-skeletons (e.g., ribose-5-phosphate) needed to support the rapid resumption of cell growth that ensues Pi resupply to –Pi plants [14]. In this regard, the steady-state metabolic flux analysis carried out by Masakapalli et al. [17] was employed to define the metabolic phenotype of heterotrophic Arabidopsis suspension cells cultured in liquid nutrient media containing 0, 1.25, or 5 mM Pi. Fluxes through the central metabolic network were deduced from the redistribution of 13C into metabolic intermediates and end-products when the cells were labeled with 13C-glucose. Their results indicated that G6P flux through the cytosolic OPPP increased in parallel with the levels of Pi in the media. Moreover, Pi starvation triggered a complete cessation of the cytosolic OPPP, thus redirecting metabolic flux away from biomass accumulation towards glycolytic conversion of hexose to organic acid anions for Pi-conserving and Pi-scavenging pathways (e.g., PEPC, malic enzyme, etc.) [14,17].
It is interesting to note that the involvement of N-terminal phosphorylation in fine-tuning catalytic activity is a feature of several other regulatory branchpoint enzymes of the central plant C-metabolism. For example, peach leaf aldose-6-phosphate reductase, a dehydrogenase that converts G6P into glucitol (thus serving as a critical control point of sugar–alcohol metabolism), undergoes inhibitory N-terminal phosphorylation, hypothesized to coordinate sucrose and glucitol production during the diel cycle [57]. Pi deprivation of Arabidopsis cell cultures and seedlings triggered in vivo phosphorylation of the plant-type PEPC isozyme AtPPC1 at Ser11, which enhanced PEPC activity [15]. Similarly, sucrose synthase is phosphorylated at Ser11 during castor oil seed development, and sucrose phosphate synthase activity is modulated by N-terminal phosphorylation under cold and osmotic stress, as well as during the light–dark transition [58,59].
During salinity stress, Arabidopsis G6PD6 was reported to be phosphorylated by ASKα (a glycogen synthase kinase3 ortholog) at a conserved C-terminal threonine residue (Thr467) that is proximal to the structural NADP+-binding domain (Figure 4) [8]. This phosphorylation event was suggested to activate G6PD6 to maintain cellular redox balance and regulate stress tolerance; askα mutants were hypersensitive to salt stress as demonstrated by their increased ROS levels and impaired G6PD activity, whereas plants overexpressing ASKα exhibited the reverse phenotype [8]. This same phosphorylation event also appears to occur during Arabidopsis immune signaling: g6pd6 mutants were more susceptible to Pseudomonas syringae infection than wild-type plants; this was reversed in g6pd6 mutants complemented with T467D phosphomimetic, but not phosphoablative T467A G6PD6 mutants [60]. However, to the best of our knowledge, phosphorylation of Arabidopsis G6PD6 (or any of its orthologs) at Thr467 has not yet been corroborated in any plant phosphoproteomic study, including the Arabidopsis Pi resupply analyses of Mehta et al. [18]. Nevertheless, the possible involvement of Thr467 phosphorylation in post-translational G6PD6 control [8] in response to Pi nutrition should also be considered.
Unfortunately, our attempts to purify native G6PD6 from Arabidopsis cell cultures for detailed assessment of the impact of reversible phosphorylation on its kinetic properties have been hampered by the enzyme’s exceptional instability, resulting in unacceptably poor yields. To circumvent this issue, the kinetic and structural properties of recombinant wild-type G6PD6 [4] with corresponding site-directed phosphomimetic G6PD6 mutants (i.e., S3D, S12D, T13D and S18D) could be compared. This would not only ensure high-yield preparations, but also guarantee the purity of the enzyme for more definitive conclusions to be drawn. Furthermore, targeted LC-MS/MS analyses would provide a valuable indication of tissue- and site-specific changes in the phosphorylation status of G6PD6 and other proteins following Pi resupply to –Pi Arabidopsis seedlings. It will also be of considerable interest to identify the protein kinases and upstream signaling pathways responsible for in vivo multisite phosphorylation of G6PD6 in response to Pi nutrition. Mehta et al. [18] reported that (i) a total of 70 different protein kinases were differentially phosphorylated following Pi resupply to the –Pi Arabidopsis cell cultures (including CDPKs, mitogen-activated protein kinases, receptor-like cytoplasmic kinases and leucine-rich repeat receptor-like kinases), and (ii) CDPK-like motifs were enriched in the phosphoproteome of the Pi-resupplied cells. Our motif analysis of G6PD6 N-terminal phosphosites (Figure 3), and Ser18 peptide kinase assays (Figure 8) are consistent with the results of Mehta et al. [18] indicating the likely contribution of CDPKs in phosphoproteome remodeling in response to Pi nutrition.
4. Materials and Methods
4.1. Plant Material
Heterotrophic Arabidopsis (cv. Landsburg erecta) suspension cells were maintained at 21 °C in the dark as previously described [61]. Routine subculturing was performed by inoculating 40 mL of sterile Murashige and Skoog media [62] containing 1.25 mM Pi with 10 mL of a 7 d old culture. Cells used for time-course studies were obtained by subculturing 10 mL of a 5 d old culture into 40 mL of media containing 0 mM Pi (–Pi). Five days later, the flasks were supplemented with 2 mM Pi or maintained under –Pi; then, 48 h following Pi resupply, the cells were harvested via vacuum filtration through Whatman 541 filter paper, rinsed with ultrapure water, frozen in liquid N2 and stored at −80 °C. For liquid seedling cultures, 5 mg of seeds was surface sterilized via rinsing in 70% EtOH for 2 min, 50% bleach for 10 min; then, the seeds were thoroughly washed and suspended in sterile ultrapure water. Sterile seeds were stratified at 4 °C in the dark for 2 d, then placed in 250 mL Magenta boxes containing 50 mL of sterile 0.5x Murashige and Skoog medium (pH 5.7) with 1% (w/v) sucrose and 0.2 mM Pi. Seedling cultures were then placed on a New Brunswick Platform Shaker (model C10; Edison, CT, USA) at 80 rpm and 23 °C under continuous illumination (100 μmol m−2 s−1) (Figure 7A). After 7 d, the seedlings were transferred into 50 mL of fresh –Pi media and cultivated for an additional 5 d, at which point they were either supplemented with 2 mM Pi or maintained under –Pi; 48 h later, the roots and shoots were rapidly separated, weighed, frozen in liquid N2, and lyophilized.
4.2. Preparation of Clarified Protein Extracts
Tissues were ground to a powder under liquid N2 and then homogenized (1:2 (w/v) for quick-frozen suspension cells; 1:40 (w/v) for lyophilized shoots or roots) using a mortar and pestle with a small scoop of sand in 50 mM HEPES-KOH (pH 7.5), containing 10 mM MgCl2, 1 mM EDTA, 10% (v/v) glycerol, phosphatase inhibitor cocktail (20 mM NaF, 1 mM NaMoO4, 1 mM Na3VO4), 1 mM phenylmethylsulfonyl fluoride, 2% (w/v) poly(vinylpolypyrrolidone), and 0.02 mM NADP+. Homogenates were centrifuged at 14,000× g and 4 °C for 10 min. Clarified extracts (0.5 mL) were rapidly desalted at 400× g on 3 mL G-Biosciences (Saint Louis, MO, USA) spin columns containing 2.5 mL of Sephadex G-25 pre-equilibrated in extraction buffer lacking poly(vinylpolypyrrolidone) [63], then prepared for SDS/PAGE and immunoblotting, or protein and G6PD activity assays.
4.3. G6PD Activity Assays and Protein Concentration Determination
G6PD activity was assayed at 23 °C by continuously monitoring the reduction of NADP+ to NADPH at 340 nm using a Spectramax Plus Microplate reader (Molecular Devices, San Jose, CA, USA). The optimized assay conditions were as follows: 50 mM Tris-HCl (pH 8.1) containing 20% (v/v) glycerol, 10 mM MgCl2, 0.2 mM NADP+ and either 0.5 mM 6PG plus 2.5 mM G6P, or 0.5 mM 6PG in a final reaction volume of 0.2 mL. To correct for contaminating 6PG-dehydrogenase activity, the activity measured in the presence of 6PG was subtracted from that measured with G6P and 6PG. The activity was proportional to the assay time and amount of enzyme added. One unit (U) of G6PD activity is the amount of enzyme resulting in the production of 1 μmol of NADPH per min.
Protein concentrations were determined using a bicinchoninic acid assay [64] with bovine γ-globulin as the protein standard.
4.4. Preparation of Anti-(phosphoSer18-Specific AtG6PD6) Antibody
Rabbit antiserum against pSer18 of AtG6PD6 was prepared using a synthetic phosphorylated peptide (LifeTein, Somerset, NJ, USA) corresponding to residues 10 through 24 of AtG6PD6’s N-terminus, with a phosphoryl group at the target Ser18 residue, and an additional N-terminal Cys residue to facilitate conjugation to keyhole limpet hemocyanin (KLH). KLH-conjugated P-peptide (1 mg) was reconstituted in Pi-buffered saline and emulsified 1:1 in Titermax Gold adjuvant (ThermoFisher Scientific, Mississauga, ON, Canada), then subcutaneously injected (500 µg) into a rabbit. Booster injections (250 µg each) were administered at 4 and 7 weeks; then, 7 d following the final injection, blood was collected into Vacutainers (Beckton Dickinson, Mississauga, ON, Canada) via cardiac puncture. Blood was centrifuged at 1000× g to remove coagulated blood cells, and the anti-pSer18 immune serum was frozen in liquid N2 and stored at −80 °C. The Animal Use Protocol for the production of rabbit antibodies was reviewed and approved by the Queen’s University Animal Care Committee, and the procedure was performed by Queen’s University Animal Care Services following the guidelines and policies of the Canadian Council on Animal Care.
4.5. Electrophoresis and Immunoblotting
SDS/PAGE was performed using a Mini-PROTEAN 3 gel electrophoresis mini-gel apparatus (Bio-Rad, Mississauga, ON, Canada). Stacking and resolving gels were made up of 4% and 10% (w/v) acrylamide, respectively, and were run at 200 V for 55 min. Following PAGE, the gels were electroblotted onto poly(vinylidene) difluoride membranes for immunoblotting. Immunoblots probed with anti-(pea G6PDc) were blocked in 3% (w/v) skim milk powder, whereas 3% (w/v) gelatin from cold water fish skin (Millipore-Sigma, Toronto, ON, Canada) was used to block anti-pSer18 immunoblots. Rabbit anti-(pea G6PDc) and anti-pSer18 immune sera were diluted 1:1000 and 1:500, respectively, in tris-buffered saline (20 mM Tris (pH 7.5), 150 mM NaCl) with 0.1% (v/v) Tween-20, containing bovine serum albumin or gelatin from cold water fish skin (1% w/v each) for anti-(pea G6PDc) and anti-pSer18, respectively, and 0.05% (w/v) NaN3. Antigenic polypeptides were visualized chromogenically using an alkaline-phosphatase linked secondary antibody as previously described [61]. All immunoblots were replicated a minimum of two times (with representative results shown), and the intensity of immunoreactive polypeptides was proportional to the amount of protein loaded.
4.6. G6PD Peptide Kinase Assays
Semiquantitative immunodot blot assays were performed using the synthetic G6PD6 deP-peptide (Figure 5A) as substrate to test for G6PD kinase activity in Arabidopsis suspension cell extracts. Clarified protein extracts prepared from liquid N2-frozen, 48 h Pi-resupplied cells were desalted as described above. The standard G6PD6 kinase assay mixture contained 50 mM HEPES-KOH (pH 7.3), 10 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, and 0.5 mM MgATP. To assess Ca2+-dependent kinase activity, the samples were incubated in the standard G6PD kinase assay mixture with the addition of 0.2 mM CaCl2 or 5 mM EGTA. Clarified extract (4 µL, equivalent to 36 µg protein) was incubated for 0 or 15 min at 30 °C in the respective G6PD kinase assay mixture, containing 0.4 mg/mL deP-peptide in a final volume of 10 µL. Aliquots (1 µL) were pipetted onto a nitrocellulose membrane and probed (after blocking as described above) using anti-(pea G6PDc) or anti-pSer18 (+10 μg/mL deP-peptide). This experiment was replicated 3 times, with representative results shown in Figure 8.
4.7. Bioinformatic Analyses
G6PD amino acid sequences were retrieved from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/protein/, accessed on 22 August 2023), and the protein BLAST tool was used to identify Arabidopsis G6PD6 orthologs. Multiple sequence alignments were executed using ClustalW version 2.1 (https://www.genome.jp/tools-bin/clustalw, accessed on 22 August 2023). The Eukaryotic Phosphorylation Site Database [65] (http://epsd.biocuckoo.cn/index.php, accessed on 22 August 2023) and Plant PTM Viewer version 2.0 [66] (https://bioinformatics.psb.ugent.be/webtools/ptm-viewer//, accessed on 22 August 2023) were used to identify experimental phosphorylation sites. The in silico predicted structure of AtG6PD6 was obtained from AlphaFold [67] (https://alphafold.ebi.ac.uk/, accessed on 15 August 2023) (model = AF-Q9FJI5-F1), and the residues were manually annotated in the SWISS-MODEL workspace [68] (https://swissmodel.expasy.org/, accessed on 15 August 2023) based on pairwise alignment with human G6PD using EMBOSS Matcher (https://www.ebi.ac.uk/Tools/psa/emboss_matcher/, accessed on 15 August 2023). The coenzyme and β+α domains (IPR036291 and IPR022675, respectively) were classified using InterPro [69] (https://www.ebi.ac.uk/interpro/, accessed on 15 August 2023). MobiDB (https://mobidb.bio.unipd.it/, accessed on 15 August 2023) was used to predict disordered regions [70].
4.8. Statistics
Unless stated otherwise, all statistical analyses were performed on a minimum of n = 3 biological replicates using Student’s t-test (two-tailed; unpaired) in GraphPad Prism (version 9.0). Data are reported as means ± SEM; statistical differences were deemed significant at p < 0.05.
5. Conclusions
Results of the current study indicate that Arabidopsis G6PD6 and its orthologs may be post-translationally regulated by multisite phosphorylation within their N-terminal disordered region. In particular, in vivo G6PD6 phosphorylation at Ser18 correlated with a significant increase in extractable G6PD activity and biomass accumulation in both Pi-resupplied Arabidopsis cell cultures and seedlings (i.e., roots). This mechanism of post-translational G6PDc control is hypothesized to help activate the enzyme to enhance cytosolic OPPP flux needed for anabolism and cell growth following Pi resupply. Understanding the mechanisms of post-translational G6PDc control, particularly those driven by abiotic stressors (or recovery from such), is important for broadening our understanding of the OPPP’s role in plant carbohydrate metabolism and redox homeostasis. Such research is foundational for the development of biotechnological tools and sustainable crop varieties, including those designed to optimize Pi utilization and acquisition, thus limiting our overuse of unsustainable and polluting Pi fertilizers in agricultural production.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/2223-7747/13/1/31/s1. Table S1: Protein accession numbers of G6PD orthologs aligned in Figure 2; Figure S1: Full, uncropped immunoblots used in Figure 6; Figure S2: Specificity of anti-pSer18.
Author Contributions
M.A.S., K.H.B. and W.C.P. planned and designed the research. K.H.B. conducted the G6PD6 peptide kinase assays and immunoblots shown in Figure S2, whereas M.A.S. performed all other experiments. M.A.S. and W.C.P. wrote and assembled the manuscript with input by K.H.B. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was provided by the Natural Sciences and Engineering Research Council of Canada (grant number RGPIN/04476-2018) and a Queen’s Research Chair Award (to W.C.P.).
Data Availability Statement
Publicly available datasets were analyzed in this study. All data generated and analyzed during this study are included in this article.
Acknowledgments
We are grateful to Jennifer Selinski (Kiel University) for providing anti-(pea G6PDc) immune serum. We also thank Maria Aristizabal, George diCenzo, and Wayne Snedden (Queen’s University) for their helpful comments and advice.
Conflicts of Interest
The authors declare no conflict of interest.
Correction Statement
This article has been republished with a minor correction to change the word "ensures" to "ensues". This change does not affect the scientific content of the article.
References
- Jiang, Z.; Wang, M.; Nicolas, M.; Ogé, L.; Pérez-Garcia, M.-D.; Crespel, L.; Li, G.; Ding, Y.; Le Gourrierec, J.; Grappin, P.; et al. Glucose-6-Phosphate Dehydrogenases: The Hidden Players of Plant Physiology. Int. J. Mol. Sci. 2022, 23, 16128. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Carfagna, S.; Massaro, G.; Vona, V.; Di Martino Rigano, V. Glucose-6-phosphate dehydrogenase in barley roots: Kinetic properties and localisation of the isoforms. Planta 2001, 212, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Massaro, G.; Vona, V.; Di Martino Rigano, V.; Carfagna, S.; Rigano, C. Ammonium induction of a novel isoform of glucose-6P dehydrogenase in barley roots. Physiol. Plant. 2001, 113, 469–476. [Google Scholar] [CrossRef]
- Wakao, S.; Benning, C. Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J. 2005, 41, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Castiglia, D.; Cardi, M.; Landi, S.; Cafasso, D.; Esposito, S. Expression and characterization of a cytosolic glucose 6 phosphate dehydrogenase isoform from barley (Hordeum vulgare) roots. Protein Expr. Purif. 2015, 112, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Linnenbrügger, L.; Doering, L.; Lansing, H.; Fischer, K.; Eirich, J.; Finkemeier, I.; von Schaewen, A. Alternative splicing of Arabidopsis G6PD5 recruits NADPH-producing OPPP reactions to the endoplasmic reticulum. Front. Plant Sci. 2022, 13, 909624. [Google Scholar] [CrossRef] [PubMed]
- Cardi, M.; Castiglia, D.; Ferrara, M.; Guerriero, G.; Chiurazzi, M.; Esposito, S. The effects of salt stress cause a diversion of basal metabolism in barley roots: Possible different roles for glucose-6-phosphate dehydrogenase isoforms. Plant Physiol. Biochem. 2015, 86, 44–54. [Google Scholar] [CrossRef]
- Dal Santo, S.; Stampfl, H.; Krasensky, J.; Kempa, S.; Gibon, Y.; Petutschnig, E.; Rozhon, W.; Heuck, A.; Clausen, T.; Jonak, C. Stress-Induced GSK3 Regulates the Redox Stress Response by Phosphorylating Glucose-6-Phosphate Dehydrogenase in Arabidopsis. Plant Cell 2012, 24, 3380–3392. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, Y.; Huang, S.; Yu, J.; Wang, X.; Xin, D.; Li, X.; Liu, Y.; Dai, Y.; Qi, Z.; et al. Genome-Wide Analysis of the Glucose-6-Phosphate Dehydrogenase Family in Soybean and Functional Identification of GmG6PDH2 Involvement in Salt Stress. Front. Plant Sci. 2020, 11, 214. [Google Scholar] [CrossRef]
- Nemoto, Y.; Sasakuma, T. Specific expression of glucose-6-phosphate dehydrogenase (G6PDH) gene by salt stress in wheat (Triticum aestivum L.). Plant Sci. 2000, 158, 53–60. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; Chang, N.; Nan, W.; Wang, S.; Ruan, M.; Sun, L.; Li, S.; Bi, Y. Cytosolic Glucose-6-Phosphate Dehydrogenase Is Involved in Seed Germination and Root Growth Under Salinity in Arabidopsis. Front. Plant Sci. 2019, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Wakao, S.; Andre, C.; Benning, C. Functional Analyses of Cytosolic Glucose-6-Phosphate Dehydrogenases and Their Contribution to Seed Oil Accumulation in Arabidopsis. Plant Physiol. 2007, 146, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Shane, M.W. The role of post-translational enzyme modifications in the metabolic adaptations of phosphorus-deprived plants. In Annual Plant Reviews; Roberts, J.A., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2015; Volume 48, pp. 99–124. [Google Scholar]
- Dissanayaka, D.M.S.B.; Ghahremani, M.; Siebers, M.; Wasaki, J.; Plaxton, W.C. Recent insights into the metabolic adaptations of phosphorus-deprived plants. J. Exp. Bot. 2021, 72, 199–223. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.L.; Hurley, B.A.; Tran, H.T.; Valentine, A.J.; She, Y.-M.; Knowles, V.L.; Plaxton, W.C. In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem. J. 2009, 420, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Shane, M.W.; Fedosejevs, E.T.; Plaxton, W.C. Reciprocal Control of Anaplerotic Phosphoenolpyruvate Carboxylase by in Vivo Monoubiquitination and Phosphorylation in Developing Proteoid Roots of Phosphate-Deficient Harsh Hakea. Plant Physiol. 2013, 161, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Masakapalli, S.K.; Bryant, F.M.; Kruger, N.J.; Ratcliffe, R.G. The metabolic flux phenotype of heterotrophic Arabidopsis cells reveals a flexible balance between the cytosolic and plastidic contributions to carbohydrate oxidation in response to phosphate limitation. Plant J. 2014, 78, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Ghahremani, M.; Pérez-Fernández, M.; Tan, M.; Schläpfer, P.; Plaxton, W.C.; Uhrig, R.G. Phosphate and phosphite have a differential impact on the proteome and phosphoproteome of Arabidopsis suspension cell cultures. Plant J. 2021, 105, 924–941. [Google Scholar] [CrossRef]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.-J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.-K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and Phosphoproteomics Reveal a Protein Phosphorylation Network in the Abscisic Acid Signaling Pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Z.; You, C.; Zhang, X.; Lu, J.; Ma, H. Abundant protein phosphorylation potentially regulates Arabidopsis anther development. J. Exp. Bot. 2016, 67, 4993–5008. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, H.; Berke, L.; Heck, A.J.R.; Mohammed, S.; Scheres, B.; Menke, F.L.H. Quantitative Phosphoproteomics after Auxin-stimulated Lateral Root Induction Identifies an SNX1 Protein Phosphorylation Site Required for Growth. Mol. Cell. Proteom. 2013, 12, 1158–1169. [Google Scholar] [CrossRef] [PubMed]
- Reiland, S.; Messerli, G.; Baerenfaller, K.; Gerrits, B.; Endler, A.; Grossmann, J.; Gruissem, W.; Baginsky, S. Large-Scale Arabidopsis Phosphoproteome Profiling Reveals Novel Chloroplast Kinase Substrates and Phosphorylation Networks. Plant Physiol. 2009, 150, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, G.B.; Wen, T.-N.; Nguyen, T.T.; Verslues, P.E. Protein Phosphatase 2Cs and Microtubule-Associated Stress Protein 1 Control Microtubule Stability, Plant Growth, and Drought Response. Plant Cell 2017, 29, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Roitinger, E.; Hofer, M.; Köcher, T.; Pichler, P.; Novatchkova, M.; Yang, J.; Schlögelhofer, P.; Mechtler, K. Quantitative Phosphoproteomics of the Ataxia Telangiectasia-Mutated (ATM) and Ataxia Telangiectasia-Mutated and Rad3-related (ATR) Dependent DNA Damage Response in Arabidopsis thaliana. Mol. Cell. Proteom. 2015, 14, 556–571. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Wang, P.; Wang, L.; Renzi, E.; Radivojac, P.; Tang, H.; Arnold, R.; Zhu, J.-K.; Tao, W.A. Quantitative Measurement of Phosphoproteome Response to Osmotic Stress in Arabidopsis Based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol. Cell. Proteom. 2013, 12, 2354–2369. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Brachova, L.; Nikolau, B.J.; Jones, A.M.; Walley, J.W. Heterotrimeric G-Protein-Dependent Proteome and Phosphoproteome in Unstimulated Arabidopsis Roots. Proteomics 2018, 18, e1800323. [Google Scholar] [CrossRef]
- Reiland, S.; Finazzi, G.; Endler, A.; Willig, A.; Baerenfaller, K.; Grossmann, J.; Gerrits, B.; Rutishauser, D.; Gruissem, W.; Rochaix, J.-D.; et al. Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc. Natl. Acad. Sci. USA 2011, 108, 12955–12960. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, B.; Née, G.; Kramer, K.; Finkemeier, I.; Soppe, W.J.J. Sequence Polymorphisms at the REDUCED DORMANCY5 Pseudophosphatase Underlie Natural Variation in Arabidopsis Dormancy. Plant Physiol. 2016, 171, 2659–2670. [Google Scholar] [CrossRef]
- Engelsberger, W.R.; Schulze, W.X. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 2012, 69, 978–995. [Google Scholar] [CrossRef]
- Wang, X.; Bian, Y.; Cheng, K.; Gu, L.-F.; Ye, M.; Zou, H.; Sun, S.S.-M.; He, J.-X. A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J. Proteom. 2013, 78, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Wei, Y.; Xie, Y.; Liu, L.; Jiang, C.; Yu, Y. Quantitative phosphoproteomic analysis provides insights into the aluminum-responsiveness of Tamba black soybean. PLoS ONE 2020, 15, e0237845. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.; Yao, L.; Ma, Z.; Ren, P.; Si, E.; Li, B.; Meng, Y.; Ma, X.; Yang, K.; et al. Global proteome analyses of phosphorylation and succinylation of barley root proteins in response to phosphate starvation and recovery. Front. Plant Sci. 2022, 13, 917652. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Qiu, J.; Wang, Y.; Li, Z.; Zhao, J.; Tong, X.; Lin, H.; Zhang, J. A Quantitative Proteomic Analysis of Brassinosteroid-induced Protein Phosphorylation in Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 514. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Hou, Y.; Tong, X.; Wang, Y.; Lin, H.; Liu, Q.; Zhang, W.; Li, Z.; Nallamilli, B.R.; Zhang, J. Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tong, X.; Qiu, J.; Li, Z.; Zhao, J.; Hou, Y.; Tang, L.; Zhang, J. A phosphoproteomic landscape of rice (Oryza sativa) tissues. Physiol. Plant. 2017, 160, 458–475. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-Scale Comparative Phosphoproteomics Identifies Conserved Phosphorylation Sites in Plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef]
- Qiu, J.; Hou, Y.; Wang, Y.; Li, Z.; Zhao, J.; Tong, X.; Lin, H.; Wei, X.; Ao, H.; Zhang, J. A Comprehensive Proteomic Survey of ABA-Induced Protein Phosphorylation in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2017, 18, 60. [Google Scholar] [CrossRef]
- Xi, L.; Zhang, Z.; Herold, S.; Kassem, S.; Wu, X.N.; Schulze, W.X. Phosphorylation site motifs in plant protein kinases and their substrates. In Plant Phosphoproteomics: Methods and Protocols; Wu, X.N., Ed.; Springer: New York, NY, USA, 2021; pp. 1–16. [Google Scholar]
- Huang, J.-Z.; Huber, S.C. Phosphorylation of Synthetic Peptides by a CDPK and Plant SNF1-Related Protein Kinase. Influence of Proline and Basic Amino Acid Residues at Selected Positions. Plant Cell Physiol. 2001, 42, 1079–1087. [Google Scholar] [CrossRef]
- Harper, J.F.; Harmon, A. Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 2005, 6, 555–566. [Google Scholar] [CrossRef]
- Kotaka, M.; Gover, S.; Vandeputte-Rutten, L.; Au, S.W.N.; Lam, V.M.S.; Adams, M.J. Structural studies of glucose-6-phosphate and NADP+binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr. Sect. D Struct. Biol. 2005, 61, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Fickenscher, K.; Scheibe, R. Purification and properties of the cytoplasmic glucose-6-phosphate dehydrogenase from pea leaves. Arch. Biochem. Biophys. 1986, 247, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Lendzian, K.J. Modulation of glucose-6-phosphate dehydrogenase by NADPH, NADP+ and dithiothreitol at variable NADPH/NADP+ ratios in an illuminated reconstituted Spinach (Spinacia oleracea L.) chloroplast system. Planta 1980, 148, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Douglas, P.; Moorhead, G.; Hong, Y.; Morrice, N.; MacKintosh, C. Purification of a nitrate reductase kinase from Spinacea oleracea leaves, and its identification as a calmodulin-domain protein kinase. Planta 1998, 206, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Fedosejevs, E.T.; Gerdis, S.A.; Ying, S.; Pyc, M.; Anderson, E.M.; Snedden, W.A.; Mullen, R.T.; She, Y.-M.; Plaxton, W.C. The calcium-dependent protein kinase RcCDPK2 phosphorylates sucrose synthase at Ser11 in developing castor oil seeds. Biochem. J. 2016, 473, 3667–3682. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Hill, A.T.; Pyc, M.; Anderson, E.M.; Snedden, W.A.; Mullen, R.T.; She, Y.-M.; Plaxton, W.C. Regulatory Phosphorylation of Bacterial-Type PEP Carboxylase by the Ca2+-Dependent Protein Kinase RcCDPK1 in Developing Castor Oil Seeds. Plant Physiol. 2017, 174, 1012–1027. [Google Scholar] [CrossRef]
- Liu, K.-H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Chung, H.S.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Au, S.W.; Gover, S.; Lam, V.M.; Adams, M.J. Human glucose-6-phosphate dehydrogenase: The crystal structure reveals a structural NADP + molecule and provides insights into enzyme deficiency. Structure 2000, 8, 293–303. [Google Scholar] [CrossRef]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef]
- Park, J.; Khuu, N.; Howard, A.S.M.; Mullen, R.T.; Plaxton, W.C. Bacterial- and plant-type phosphoenolpyruvate carboxylase isozymes from developing castor oil seeds interact in vivo and associate with the surface of mitochondria. Plant J. 2012, 71, 251–262. [Google Scholar] [CrossRef]
- O’Leary, B.; Rao, S.K.; Plaxton, W.C. Phosphorylation of bacterial-type phosphoenolpyruvate carboxylase at Ser425 provides a further tier of enzyme control in developing castor oil seeds. Biochem. J. 2011, 433, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Dalziel, K.J.; O’Leary, B.; Brikis, C.; Rao, S.K.; She, Y.-M.; Cyr, T.; Plaxton, W.C. The bacterial-type phosphoenolpyruvate carboxylase isozyme from developing castor oil seeds is subject to in vivo regulatory phosphorylation at serine-451. FEBS Lett. 2012, 586, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Moorhead, G.B.G.; Templeton, G.W.; Tran, H.T. Role of protein kinases, phosphatases and 14-3-3 proteins in the control of primary plant metabolism. In Annual Plant Reviews Volume 22: Control of Primary Metabolism in Plants; Plaxton, W.C., McManus, M.T., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 121–143. [Google Scholar]
- Brumbaugh, K.; Johnson, W.; Liao, W.-C.; Lin, M.-S.; Houchins, J.P.; Cooper, J.; Stoesz, S.; Campos-Gonzalez, R. Overview of the generation, validation, and application of phosphosite-specific antibodies. In Signal Transduction Immunohistochemistry: Methods and Protocols; Kalyuzhny, A.E., Ed.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2011; Volume 717, pp. 3–43. [Google Scholar]
- Hartman, M.D.; Rojas, B.E.; Ferrero, D.M.; Leyva, A.; Durán, R.; Iglesias, A.A.; Figueroa, C.M. Phosphorylation of aldose-6-phosphate reductase from Prunus persica leaves. Plant Physiol. Biochem. 2023, 194, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Fedosejevs, E.T.; Ying, S.; Park, J.; Anderson, E.M.; Mullen, R.T.; She, Y.-M.; Plaxton, W.C. Biochemical and Molecular Characterization of RcSUS1, a Cytosolic Sucrose Synthase Phosphorylated in Vivo at Serine 11 in Developing Castor Oil Seeds. J. Biol. Chem. 2014, 289, 33412–33424. [Google Scholar] [CrossRef]
- Huber, S.C.; Huber, J.L. Role and regulation of sucrose-phosphate synthase in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 431–444. [Google Scholar] [CrossRef]
- Stampfl, H.; Fritz, M.; Dal Santo, S.; Jonak, C. The GSK3/Shaggy-like kinase ASKα contributes to pattern-triggered immunity in Arabidopsis thaliana. Plant Physiol. 2016, 171, 1366–1377. [Google Scholar] [CrossRef]
- Veljanovski, V.; Vanderbeld, B.; Knowles, V.L.; Snedden, W.A.; Plaxton, W.C. Biochemical and Molecular Characterization of AtPAP26, a Vacuolar Purple Acid Phosphatase Up-Regulated in Phosphate-Deprived Arabidopsis Suspension Cells and Seedlings. Plant Physiol. 2006, 142, 1282–1293. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Penefsky, H.S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 1977, 252, 2891–2899. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Lin, S.; Wang, C.; Zhou, J.; Shi, Y.; Ruan, C.; Tu, Y.; Yao, L.; Peng, D.; Xue, Y. EPSD: A well-annotated data resource of protein phosphorylation sites in eukaryotes. Brief. Bioinform. 2020, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.; Horne, A.; Van Parys, T.; Goormachtig, S.; De Smet, I.; Botzki, A.; Van Breusegem, F.; Gevaert, K. The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J. 2019, 99, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, A.G.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2022, 51, D418–D427. [Google Scholar] [CrossRef]
- Piovesan, D.; Del Conte, A.; Clementel, D.; Monzon, A.M.; Bevilacqua, M.; Aspromonte, M.C.; Iserte, A.J.; Orti, E.F.; Marino-Buslje, C.; Tosatto, S.C.E. MobiDB: 10 years of intrinsically disordered proteins. Nucleic Acids Res. 2023, 51, D438–D444. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).