Exploring Genetics by Environment Interactions in Some Rice Genotypes across Varied Environmental Conditions
Abstract
:1. Introduction
2. Results
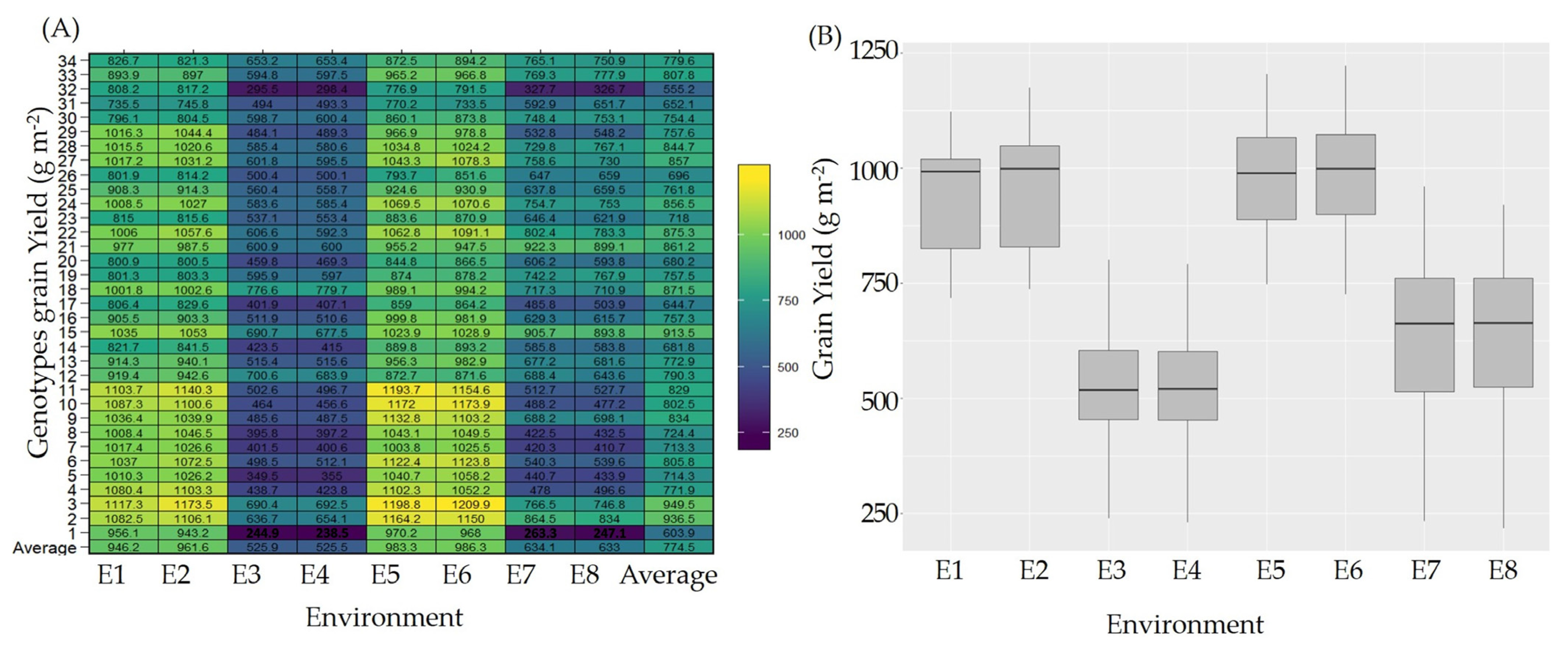
2.1. GY Combined AMMI Analysis of Variance and Genotypic Variability
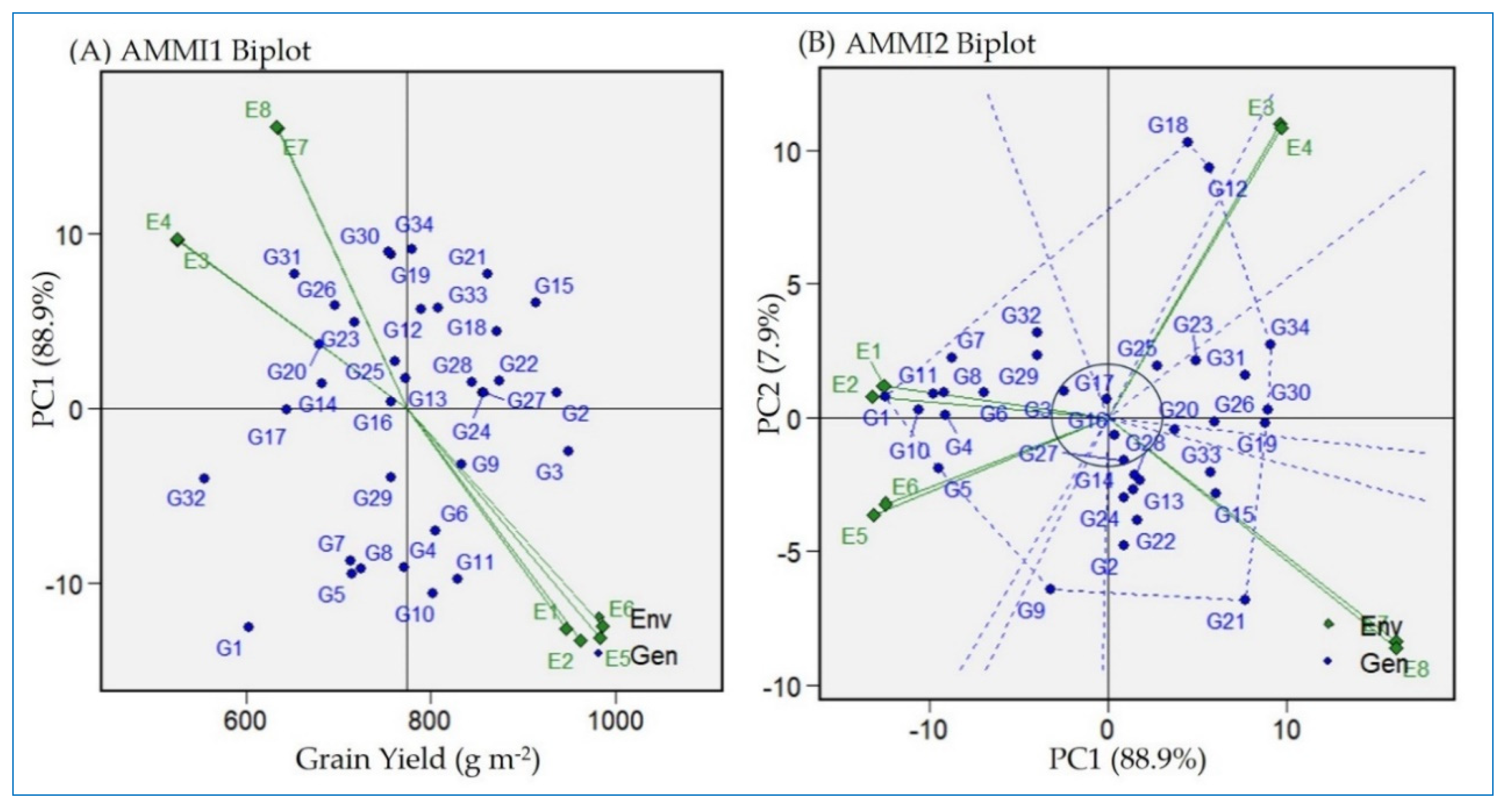
2.2. GEI-Structure-Based Additive Main Effect and Multiplicative Interaction Model
| Source | Df | Sum Sq | Mean Sq | F Value | Pr (>F) | Proportion | Accumulated |
|---|---|---|---|---|---|---|---|
| ENV | 7 | 32,277,639 | 4,611,091 | 5554.827 | 0 | ||
| REP(ENV) | 16 | 13,281.69 | 830.1054 | 1.703935 | 0.042241 | ||
| GEN | 33 | 6,685,134 | 202,579.8 | 415.83 | 0 | ||
| GEN:ENV | 231 | 6,311,279 | 27,321.56 | 56.08221 | 8.8 × 10−275 | ||
| PC1 | 39 | 5,612,970 | 143,922.3 | 295.43 | 0 | 88.9 | 88.9 |
| PC2 | 37 | 499,545.4 | 13,501.23 | 27.71 | 0 | 7.9 | 96.9 |
| PC3 | 35 | 141,380.5 | 4039.443 | 8.29 | 0 | 2.2 | 99.1 |
| PC4 | 33 | 28,625.39 | 867.4361 | 1.78 | 0.0054 | 0.5 | 99.5 |
| PC5 | 31 | 15,441.88 | 498.1253 | 1.02 | 0.439 | 0.2 | 99.8 |
| PC6 | 29 | 10,612.79 | 365.9583 | 0.75 | 0.8262 | 0.2 | 100 |
| PC7 | 27 | 2703 | 100.1111 | 0.21 | 1 | 0 | 100 |
| Residuals | 528 | 257,225.6 | 487.1697 | ||||
| Total | 1046 | 51,855,839 | 49,575.37 |
2.3. Detection of Stable and High-Yielding Genotypes via AMMIs Model Biplot
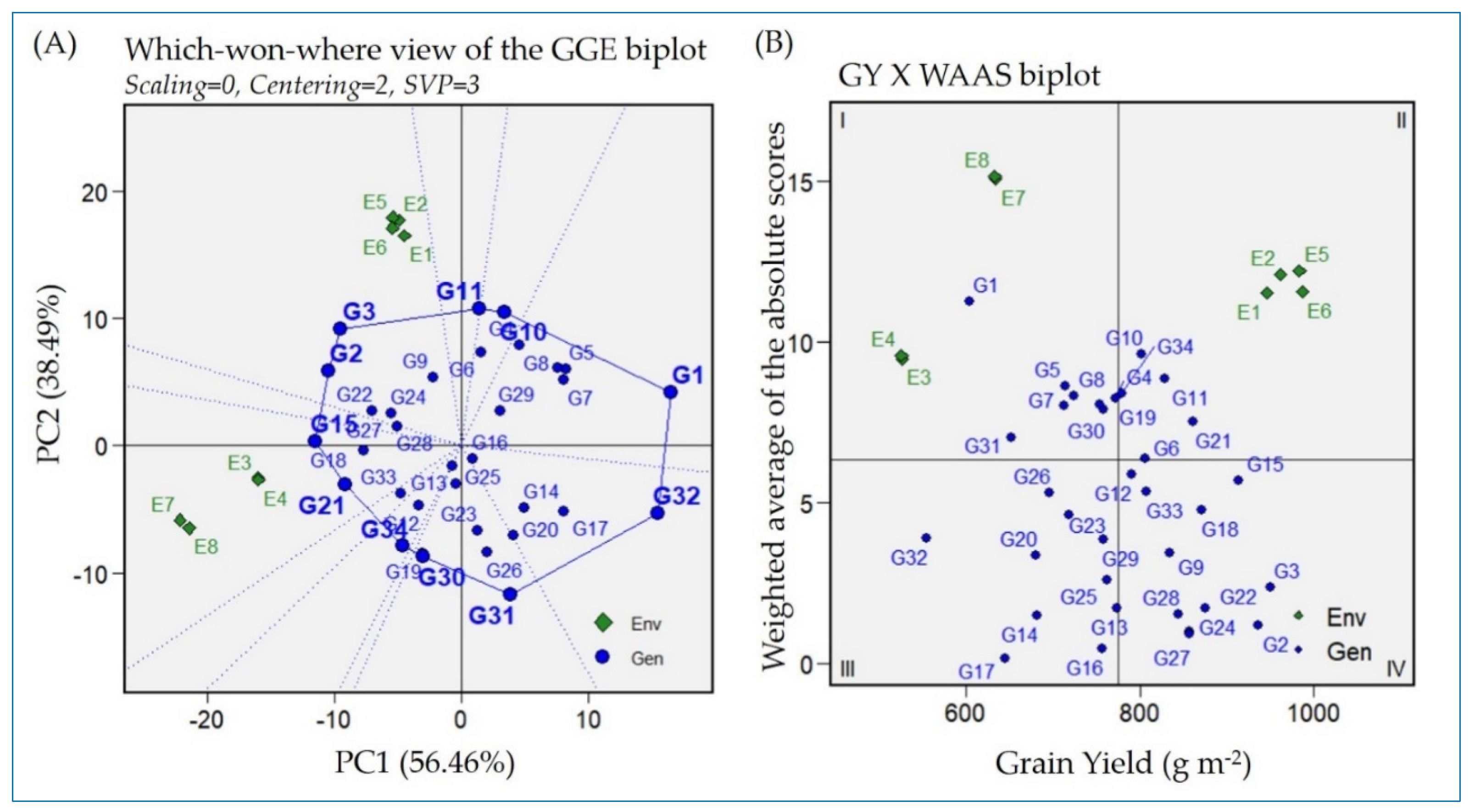
2.4. Which–Won–Where Approach Based on GGE Biplot for Detecting the Best-Performing Genotypes
2.5. Grain Yield Versus Weighted Average of Absolute Score Stability Index Biplot
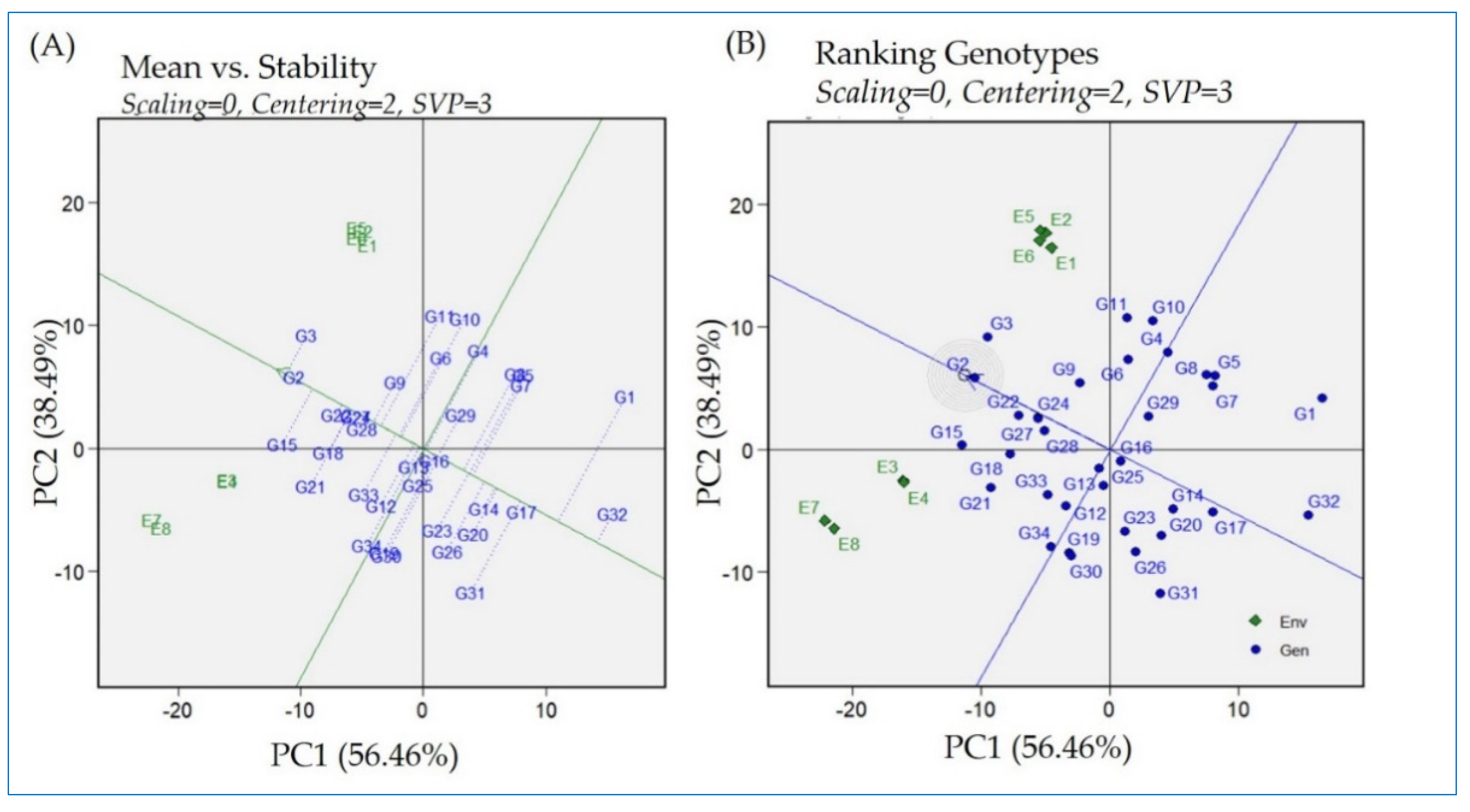
2.6. GGE Biplot—Means Versus Stability Model and Ranking of Rice Genotypes’ Performance
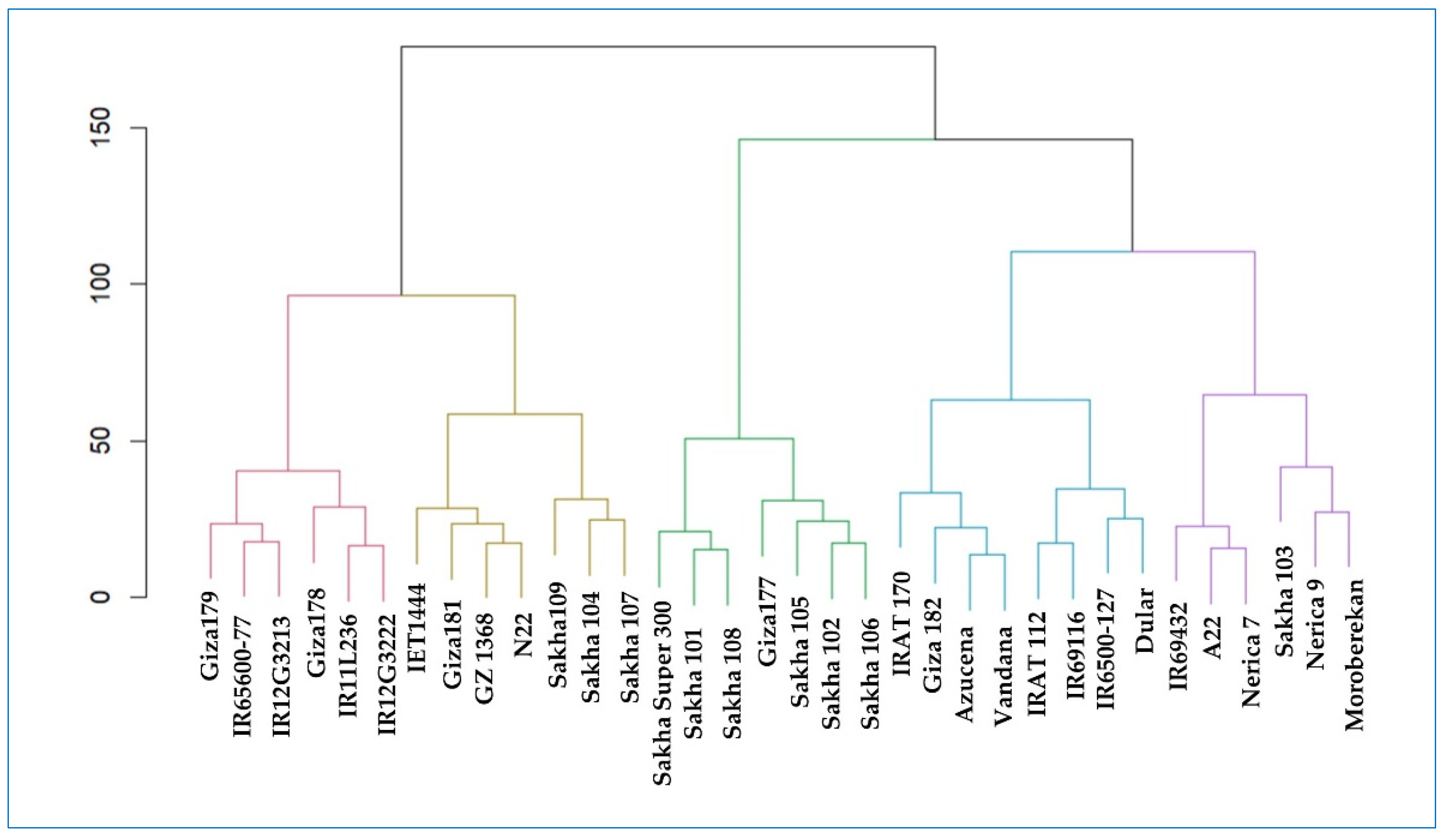
2.7. Cluster-Analysis-Based Stability Indices
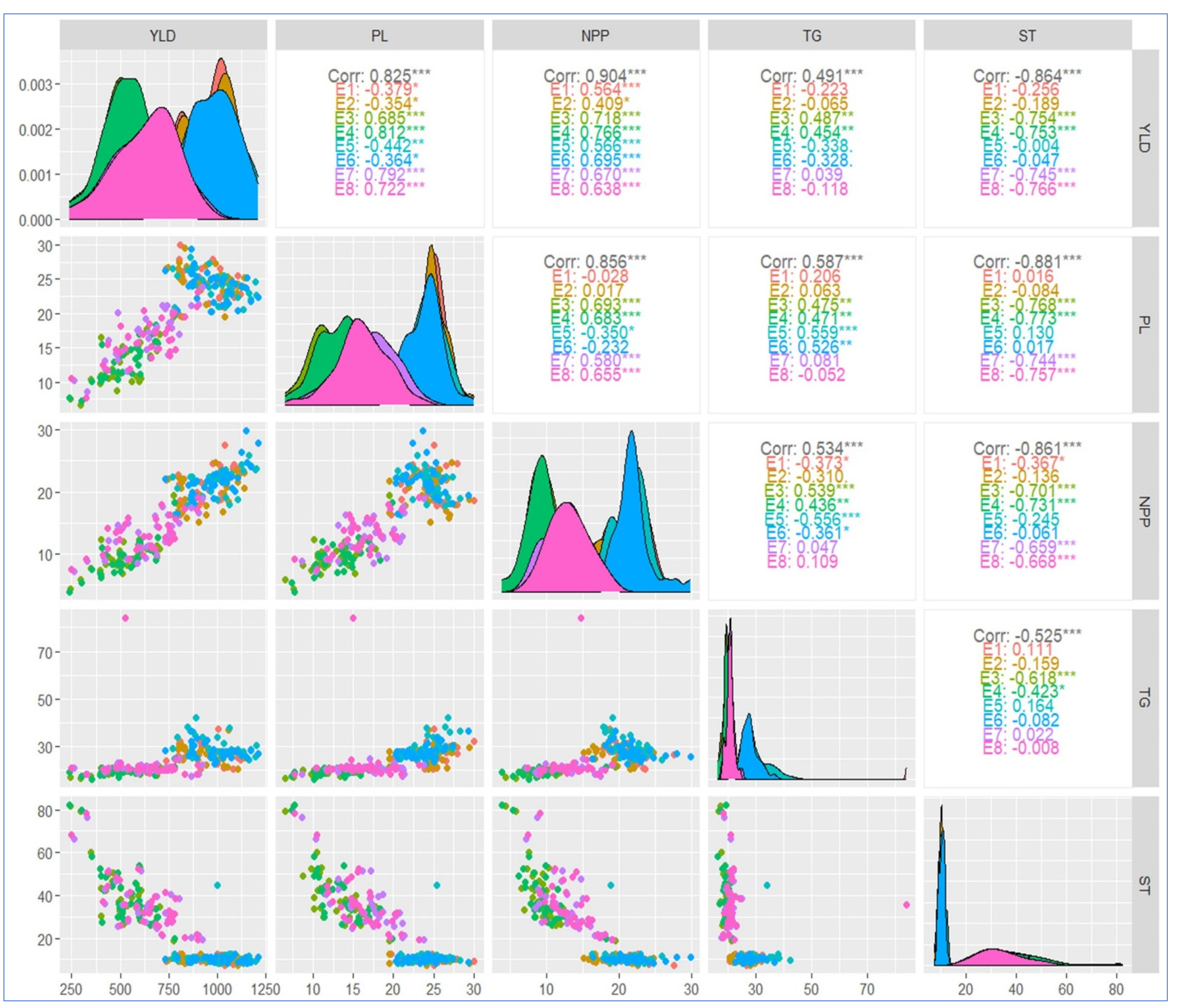
2.8. Correlation Coefficient Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
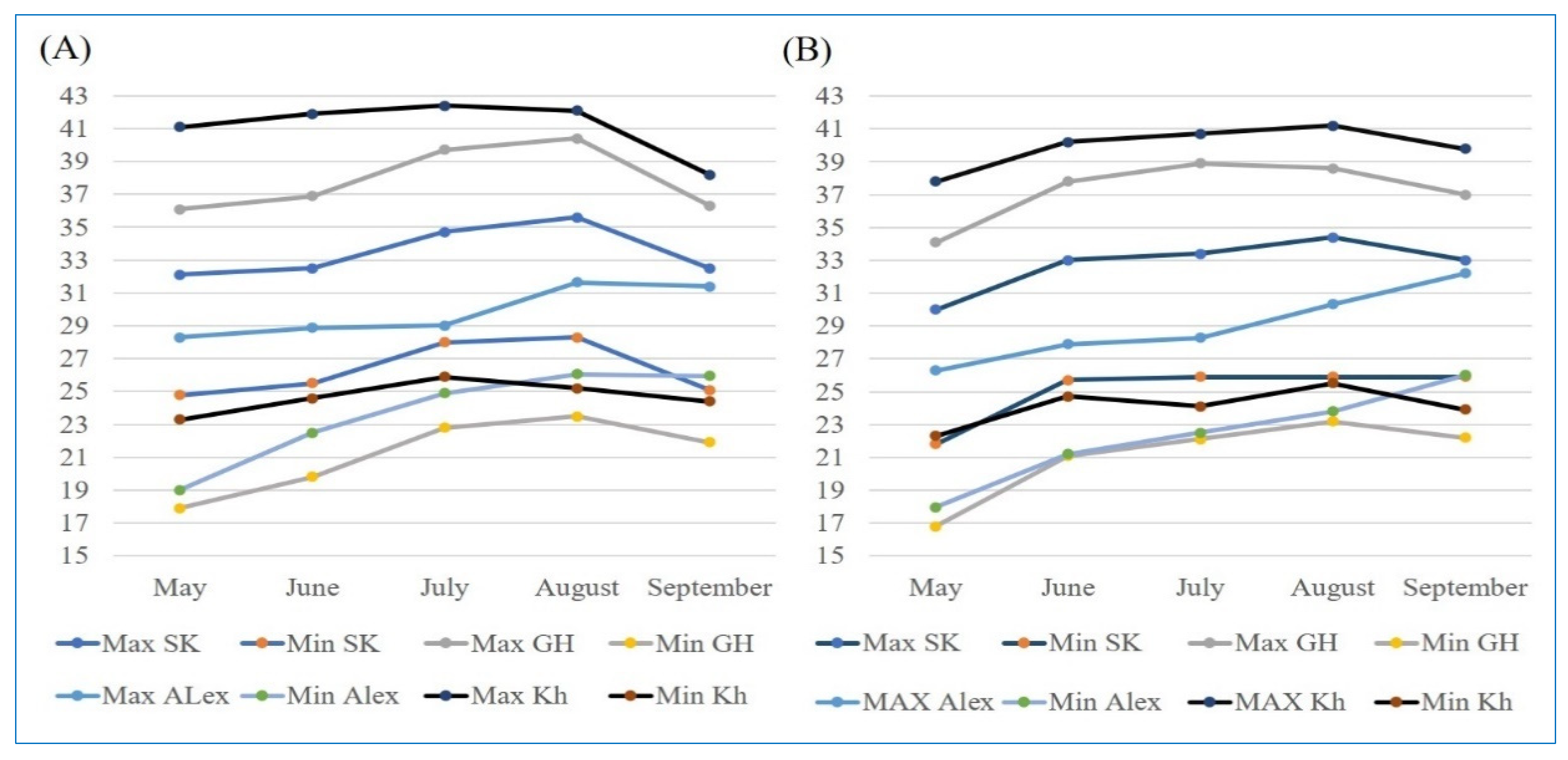
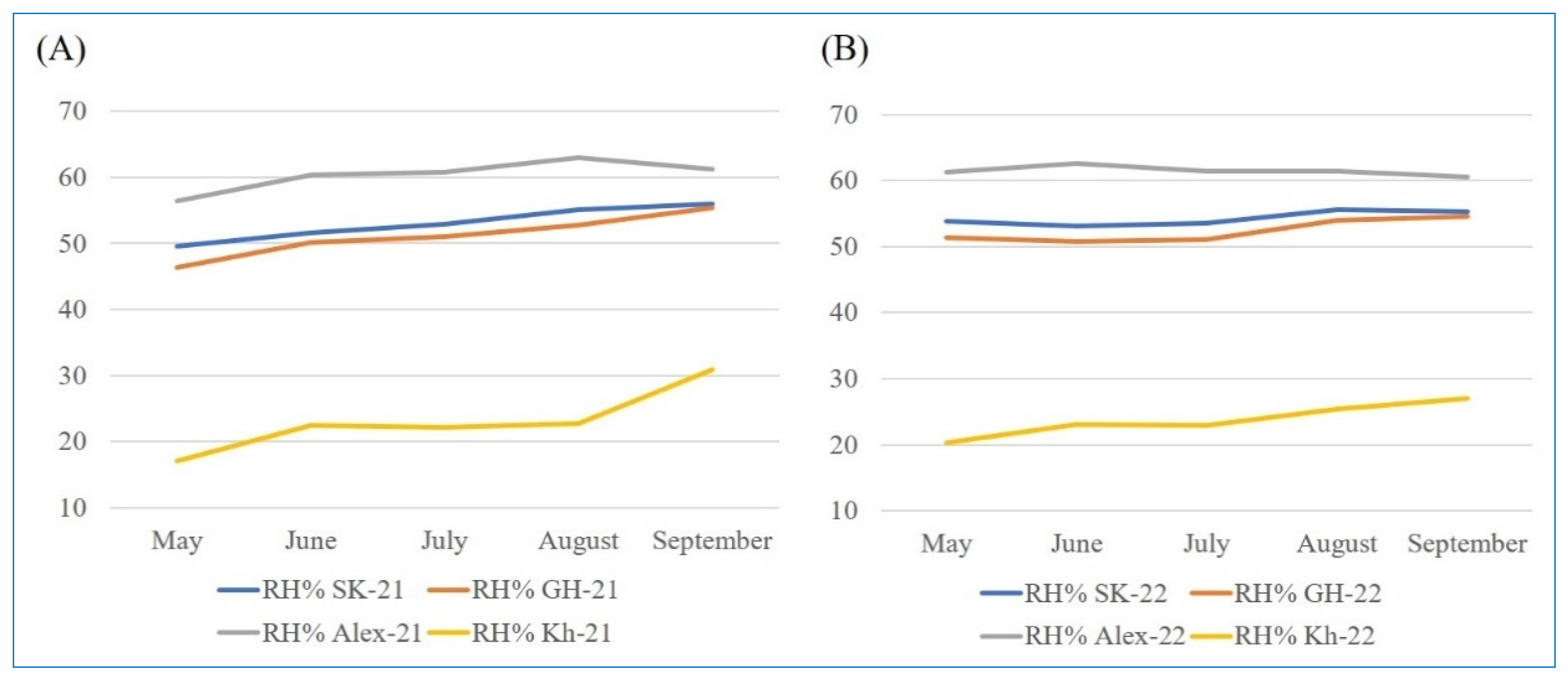
4.2. Experimental Locations and Climatic Conditions
4.3. Experimental Design and Data Recording
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chamie, J. Population Growth Diversity Continuing in the Twenty-first Century. In Population Levels, Trends, and Differentials: More Important Population Matters; Springer: Berlin/Heidelberg, Germany, 2023; pp. 179–182. [Google Scholar]
- Shaheen, S.M.; Antoniadis, V.; Shahid, M.; Yang, Y.; Abdelrahman, H.; Zhang, T.; Hassan, N.E.; Bibi, I.; Niazi, N.K.; Younis, S.A. Sustainable applications of rice feedstock in agro-environmental and construction sectors: A global perspective. Renew. Sustain. Energy Rev. 2022, 153, 111791. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, H.-S.; Lee, C.-M.; Ha, S.-K.; Park, H.-M.; Lee, S.-M.; Kwon, Y.; Jeung, J.-U.; Mo, Y. Multi-Environment Trials and Stability Analysis for Yield-Related Traits of Commercial Rice Cultivars. Agriculture 2023, 13, 256. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Zhao, K. Genome editing and rice grain quality. In The Future of Rice Demand: Quality Beyond Productivity; Springer: Cham, Switzerland, 2020; pp. 395–422. [Google Scholar]
- Chairi, F.; Aparicio, N.; Serret, M.D.; Araus, J.L. Breeding effects on the genotype× environment interaction for yield of durum wheat grown after the Green Revolution: The case of Spain. Crop J. 2020, 8, 623–634. [Google Scholar] [CrossRef]
- Khan, M.M.H.; Rafii, M.Y.; Ramlee, S.I.; Jusoh, M.; Al Mamun, M. AMMI and GGE biplot analysis for yield performance and stability assessment of selected Bambara groundnut (Vigna subterranea L. Verdc.) genotypes under the multi-environmental trials (METs). Sci. Rep. 2021, 11, 22791. [Google Scholar] [CrossRef] [PubMed]
- Omrani, A.; Omrani, S.; Khodarahmi, M.; Shojaei, S.H.; Illés, Á.; Bojtor, C.; Mousavi, S.M.N.; Nagy, J. Evaluation of grain yield stability in some selected wheat genotypes using AMMI and GGE biplot methods. Agronomy 2022, 12, 1130. [Google Scholar] [CrossRef]
- Yue, H.; Gauch, H.G.; Wei, J.; Xie, J.; Chen, S.; Peng, H.; Bu, J.; Jiang, X. Genotype by environment interaction analysis for grain yield and yield components of summer maize hybrids across the huanghuaihai region in China. Agriculture 2022, 12, 602. [Google Scholar] [CrossRef]
- Mehana, M.; Abdelrahman, M.; Emadeldin, Y.; Rohila, J.S.; Karthikeyan, R. Impact of genetic improvements of rice on its water use and effects of climate variability in Egypt. Agriculture 2021, 11, 865. [Google Scholar] [CrossRef]
- Anputhas, M.; Samita, S.; Abeysiriwardena, D. Stability and adaptability analysis of rice cultivars using environment-centered yield in two-way ANOVA model. Commun. Biometry Crop Sci. 2011, 6, 80–86. [Google Scholar]
- Sandhu, N.; Yadaw, R.B.; Chaudhary, B.; Prasai, H.; Iftekharuddaula, K.; Venkateshwarlu, C.; Annamalai, A.; Xangsayasane, P.; Battan, K.R.; Ram, M. Evaluating the performance of rice genotypes for improving yield and adaptability under direct seeded aerobic cultivation conditions. Front. Plant Sci. 2019, 10, 159. [Google Scholar] [CrossRef]
- Bocianowski, J.; Tratwal, A.; Nowosad, K. Genotype by environment interaction for main winter triticale varieties characteristics at two levels of technology using additive main effects and multiplicative interaction model. Euphytica 2021, 217, 26. [Google Scholar] [CrossRef]
- Banterng, P.; Patanothai, A.; Pannangpetch, K.; Jogloy, S.; Hoogenboom, G. Yield stability evaluation of peanut lines: A comparison of an experimental versus a simulation approach. Field Crops Res. 2006, 96, 168–175. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Alharbi, K.; El-Denary, M.E.; Abd El-Megeed, T.; Naeem, E.-S.; Monir, S.; Al-Shaye, N.A.; Ammar, M.H.; Attia, K.; Dora, S.A. Detection of Superior Rice Genotypes and Yield Stability under Different Nitrogen Levels Using AMMI Model and Stability Statistics. Plants 2022, 11, 2775. [Google Scholar] [CrossRef] [PubMed]
- Finlay, K.; Wilkinson, G. The analysis of adaptation in a plant-breeding programme. Aust. J. Agric. Res. 1963, 14, 742–754. [Google Scholar] [CrossRef]
- Eberhart, S.t.; Russell, W. Stability parameters for comparing varieties 1. Crop Sci. 1966, 6, 36–40. [Google Scholar] [CrossRef]
- Gauch, H.G.; Zobel, R.W. Predictive and postdictive success of statistical analyses of yield trials. Theor. Appl. Genet. 1988, 76, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yan, W. Singular-value partitioning in biplot analysis of multienvironment trial data. Agron. J. 2002, 94, 990–996. [Google Scholar]
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.C. metan: An R package for multi-environment trial analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Khalili, M.; Poczai, P.; Olivoto, T. Stability indices to deciphering the genotype-by-environment interaction (GEI) effect: An applicable review for use in plant breeding programs. Plants 2022, 11, 414. [Google Scholar] [CrossRef]
- Hossain, M.A.; Sarker, U.; Azam, M.G.; Kobir, M.S.; Roychowdhury, R.; Ercisli, S.; Ali, D.; Oba, S.; Golokhvast, K.S. Integrating BLUP, AMMI, and GGE Models to Explore GE Interactions for Adaptability and Stability of Winter Lentils (Lens culinaris Medik.). Plants 2023, 12, 2079. [Google Scholar] [CrossRef]
- Tanin, M.J.; Sharma, A.; Saini, D.K.; Singh, S.; Kashyap, L.; Srivastava, P.; Mavi, G.; Kaur, S.; Kumar, V.; Kumar, V. Ascertaining yield and grain protein content stability in wheat genotypes having the Gpc-B1 gene using univariate, multivariate, and correlation analysis. Front. Genet. 2022, 13, 1001904. [Google Scholar] [CrossRef] [PubMed]
- Artur, C.R.R.; Moiana, L.D.; Maleia, M.P.; Valentini, G.; Sumbuleiro, A.D.; Marcos, M.A. Evaluation of the grain yield performance of 5 soybean genotypes in Mozambique using the GGE Biplot method. Afr. J. Biotechnol. 2023, 22, 61–70. [Google Scholar]
- Rani, R.; Raza, G.; Ashfaq, H.; Rizwan, M.; Shimelis, H.; Tung, M.H.; Arif, M. Analysis of genotype× environment interactions for agronomic traits of soybean (Glycine max [L.] Merr.) using association mapping. Front. Genet. 2023, 13, 1090994. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Dhansu, P.; Kulshreshtha, N.; Meena, M.R.; Kumaraswamy, M.H.; Appunu, C.; Chhabra, M.L.; Pandey, S.K. Identification of Salinity Tolerant Stable Sugarcane Cultivars Using AMMI, GGE and Some Other Stability Parameters under Multi Environments of Salinity Stress. Sustainability 2023, 15, 1119. [Google Scholar] [CrossRef]
- Anisha, A.; Rajappa, P.; Parashuram, P.; Hemalatha, V.; Dhanyashree, R.; Tonapi, V.A.; Sujatha, K.; Girish, G.; Madhusudhana, R. Selection of post-rainy sorghum landraces combining multi-traits mean performance and stability. Euphytica 2022, 218, 176. [Google Scholar] [CrossRef]
- Rubiales Olmedo, D.; Moral, A.; Flores Gil, F. Agronomic Performance of Broomrape Resistant and Susceptible Faba Bean Accession. Agronomy 2022, 12, 1421. [Google Scholar] [CrossRef]
- Hussain, T.; Akram, Z.; Shabbir, G.; Manaf, A.; Ahmed, M. Identification of drought tolerant Chickpea genotypes through multi trait stability index. Saudi J. Biol. Sci. 2021, 28, 6818–6828. [Google Scholar] [CrossRef]
- Rossi, A.; Clemente, C.; Tavarini, S.; Angelini, L.G. Variety and Sowing Date Affect Seed Yield and Chemical Composition of Linseed Grown under Organic Production System in a Semiarid Mediterranean Environment. Agronomy 2022, 13, 45. [Google Scholar] [CrossRef]
- Krishna, K.S.; Chaudhary, R.K.; Kumar, M. Deciphering the Genotype× Environment Interaction for Identification of Superior Genotypes of Mango (Mangifera indica L.) using Ammi Stability Measures. Int. J. Environ. Clim. Chang. 2022, 12, 348–358. [Google Scholar] [CrossRef]
- Adil, N.; WANI, S.H.; RAFIQEE, S.; Mehrajuddin, S.; SOFI, N.R.; SHIKARI, A.B.; HUSSAIN, A.; MOHIDDIN, F.; JEHANGIR, I.A.; KHAN, G.H. Deciphering genotype× environment interaction by AMMI and GGE biplot analysis among elite wheat (Triticum aestivum L.) genotypes of himalayan region. Ekin J. Crop Breed. Genet. 2022, 8, 41–52. [Google Scholar]
- Erdemci, I. Investigation of genotype× environment interaction in chickpea genotypes using AMMI and GGE biplot analysis. Turk. J. Field Crops 2018, 23, 20–26. [Google Scholar] [CrossRef]
- Barona, M.A.; Díaz, R.S.; Suárez, R.R. Adaptability and grain yield stability of rice hybrids and varieties in Venezuela. Bioagro 2021, 33, 181–190. [Google Scholar] [CrossRef]
- Dewi, A.K.; Chozin, M.A.; Triwidodo, H.; Aswidinnoor, H. Genotype× environment interaction, and stability analysis in lowland rice promising genotypes. Int. J. Agron. Agric. Res. 2014, 5, 74–84. [Google Scholar]
- Tariku, S.; Lakew, T.; Bitew, M.; Asfaw, M. Genotype by environment interaction and grain yield stability analysis of rice (Oryza sativa L.) genotypes evaluated in north western Ethiopia. Net J. Agric. Sci. 2013, 1, 10–16. [Google Scholar]
- Zewdu, Z.; Abebe, T.; Mitiku, T.; Worede, F.; Dessie, A.; Berie, A.; Atnaf, M. Performance evaluation and yield stability of upland rice (Oryza sativa L.) varieties in Ethiopia. Cogent Food Agric. 2020, 6, 1842679. [Google Scholar] [CrossRef]
- Liang, S.; Ren, G.; Liu, J.; Zhao, X.; Zhou, M.; McNeil, D.; Ye, G. Genotype-by-environment interaction is important for grain yield in irrigated lowland rice. Field Crops Res. 2015, 180, 90–99. [Google Scholar] [CrossRef]
- Singh, G.; Khanna, R.; Kaur, R.; Kaur, K.; Kaur, R.; Sharma, N.; Mangat, G.S. Performance under multi-environment trial for quantitative traits of rice (Oryza sativa L.) genotypes in North-West India (Punjab). Ecol. Genet. Genom. 2023, 28, 100190. [Google Scholar] [CrossRef]
- Gauch Jr, H.G. A simple protocol for AMMI analysis of yield trials. Crop Sci. 2013, 53, 1860–1869. [Google Scholar] [CrossRef]
- Gauch Jr, H.G.; Piepho, H.P.; Annicchiarico, P. Statistical analysis of yield trials by AMMI and GGE: Further considerations. Crop Sci. 2008, 48, 866–889. [Google Scholar] [CrossRef]
- Daba, S.D.; Kiszonas, A.M.; McGee, R.J. Selecting High-Performing and Stable Pea Genotypes in Multi-Environmental Trial (MET): Applying AMMI, GGE-Biplot, and BLUP Procedures. Plants 2023, 12, 2343. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.; da Silva, J.A.; Marchioro, V.S.; de Souza, V.Q.; Jost, E. Mean performance and stability in multi-environment trials I: Combining features of AMMI and BLUP techniques. Agron. J. 2019, 111, 2949–2960. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Barati, A.; Gholipoor, A.; Zali, H.; Marzooghian, A.; Koohkan, S.A.; Shahbazi-Homonloo, K.; Houseinpour, A. Deciphering genotype-by-environment interaction in barley genotypes using different adaptability and stability methods. J. Crop Sci. Biotechnol. 2023, 26, 547–562. [Google Scholar] [CrossRef]
- Abo-Yousef, M.; Sedeek, S.; EL-Rafaee, I.; Hammoud, S.; EL-Abd, A.; El-Malkey, M.; EL-Namaky, R.; Ammar, M.; Abdelkhalik, A.; Zayed, B. Giza 179 Egyptian rice variety: As a new, early, high-yielding, tolerant to saline, and climate change challenge. Egypt. J. Agric. Res. 2023, 101, 567–588. [Google Scholar]
- Shrestha, J.; Subedi, S.; Acharya, R.; Sharma, S.; Subedi, M. Grain yield stability of maize (Zea mays L.) hybrids using ammi model and GGE biplot analysis. SAARC J. Agric. 2021, 19, 107–121. [Google Scholar] [CrossRef]
- Abdelaal, K.; Mazrou, Y.; Mohamed, A.; Ghazy, M.; Barakat, M.; Hafez, Y.; Gaballah, M. The different responses of rice genotypes to heat stress associated with morphological, chlorophyll and yield characteristics. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12550. [Google Scholar] [CrossRef]
- Gaballah, M.M.; Ghoneim, A.M.; Rehman, H.U.; Shehab, M.M.; Ghazy, M.I.; El-Iraqi, A.S.; Mohamed, A.E.; Waqas, M.; Shamsudin, N.A.A.; Chen, Y. Evaluation of morpho-physiological traits in rice genotypes for adaptation under irrigated and water-limited environments. Agronomy 2022, 12, 1868. [Google Scholar] [CrossRef]
- Jagadish, S.; Muthurajan, R.; Rang, Z.W.; Malo, R.; Heuer, S.; Bennett, J.; Craufurd, P.Q. Spikelet proteomic response to combined water deficit and heat stress in rice (Oryza sativa cv. N22). Rice 2011, 4, 1–11. [Google Scholar] [CrossRef]
- Shehab, M.; Iovene, M.; Ciancio, A.; Colagiero, M.; Finetti-Sialer, M. Transcriptome analysis provides novel insights into salt stress response in two egyptian rice varieties with different tolerance levels. Rice Sci. 2022, 29, 499–502. [Google Scholar] [CrossRef]
- Grondin, A.; Dixit, S.; Torres, R.; Venkateshwarlu, C.; Rogers, E.; Mitchell-Olds, T.; Benfey, P.N.; Kumar, A.; Henry, A. Physiological mechanisms contributing to the QTL qDTY 3.2 effects on improved performance of rice Moroberekan x Swarna BC 2 F 3: 4 lines under drought. Rice 2018, 11, 1–17. [Google Scholar] [CrossRef]
- Laza, M.R.C.; Peng, S.; Akita, S.; Saka, H. Effect of panicle size on grain yield of IRRI-released indica rice cultivars in the wet season. Plant Prod. Sci. 2004, 7, 271–276. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2010. [Google Scholar]
- Yan, W.; Kang, M.S. GGE biplot analysis: A graphical tool for breeders, geneticists, and agronomists; CRC press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. NbClust: An R package for determining the relevant number of clusters in a data set. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
| No. | Genotype | Origin | Type |
|---|---|---|---|
| 1 | Giza 177 | Egypt | Japonica |
| 2 | Giza 178 | Egypt | Indica/Japonica |
| 3 | Giza 179 | Egypt | Indica/Japonica |
| 4 | Sakha 101 | Egypt | Japonica |
| 5 | Sakha 102 | Egypt | Japonica |
| 6 | Sakha 104 | Egypt | Japonica |
| 7 | Sakha 105 | Egypt | Japonica |
| 8 | Sakha 106 | Egypt | Japonica |
| 9 | Sakha 107 | Egypt | Japonica |
| 10 | Sakha 108 | Egypt | Japonica |
| 11 | Sakha Super 300 | Egypt | Japonica |
| 12 | IRAT 170 | Ivory Cost | Tropical japonica |
| 13 | A22 | Srilanka | Indica |
| 14 | Nerica 9 | Ivory cost | Indica |
| 15 | IET 1444 | Inbdia | Indica |
| 16 | Nerica 7 | Ivory Cost | Indica |
| 17 | Moroberekan | Guinea | Japonica |
| 18 | GZ 1368-S-5-4 | Egypt | Indica |
| 19 | Azucena | Philippine | Japonica |
| 20 | IRAT 112 | Ivory Cost | Indica |
| 21 | N22 | India | Aus |
| 22 | IR65600-77 | Philippines | Indica |
| 23 | IR69116 | Philippines | Indica |
| 24 | IR12G3213 | Philippines | Indica |
| 25 | IR69432 | Philippines | Indica |
| 26 | IR6500-127 | Philippines | Indica |
| 27 | IR11L236 | Philippines | Indica |
| 28 | IR12G3222 | Philippines | Indica |
| 29 | Sakha 109 | Egypt | Japonica |
| 30 | Vandana | India | Indica |
| 31 | Dular | India | Indica |
| 32 | Sakha 103 | Egypt | Japonica |
| 33 | Giza 181 | Egypt | Indica |
| 34 | Giza 182 | Egypt | Indica |
| No. | Location (Governorate) | Altitude–Latitude |
|---|---|---|
| 1 | Sakha (Kafr El-Sheikh) | 31.09° N and 30.9° E |
| 2 | Gemmiza (ElGharbya) | 30.88° N and 31.05° E |
| 3 | Sobahya (Alexandria) | 31.2° N and 29.9° E |
| 4 | Kharga oasis (New Valley) | 25.4° N and 30.5° E |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghazy, M.I.; Abdelrahman, M.; El-Agoury, R.Y.; El-hefnawy, T.M.; EL-Naem, S.A.; Daher, E.M.; Rehan, M. Exploring Genetics by Environment Interactions in Some Rice Genotypes across Varied Environmental Conditions. Plants 2024, 13, 74. https://doi.org/10.3390/plants13010074
Ghazy MI, Abdelrahman M, El-Agoury RY, El-hefnawy TM, EL-Naem SA, Daher EM, Rehan M. Exploring Genetics by Environment Interactions in Some Rice Genotypes across Varied Environmental Conditions. Plants. 2024; 13(1):74. https://doi.org/10.3390/plants13010074
Chicago/Turabian StyleGhazy, Mohamed I., Mohamed Abdelrahman, Roshdy Y. El-Agoury, Tamer M. El-hefnawy, Sabry A. EL-Naem, Elhousini M. Daher, and Medhat Rehan. 2024. "Exploring Genetics by Environment Interactions in Some Rice Genotypes across Varied Environmental Conditions" Plants 13, no. 1: 74. https://doi.org/10.3390/plants13010074







