OsSHMT4 Is Required for Synthesis of Rice Storage Protein and Storage Organelle Formation in Endosperm Cells
Abstract
:1. Introduction
2. Results
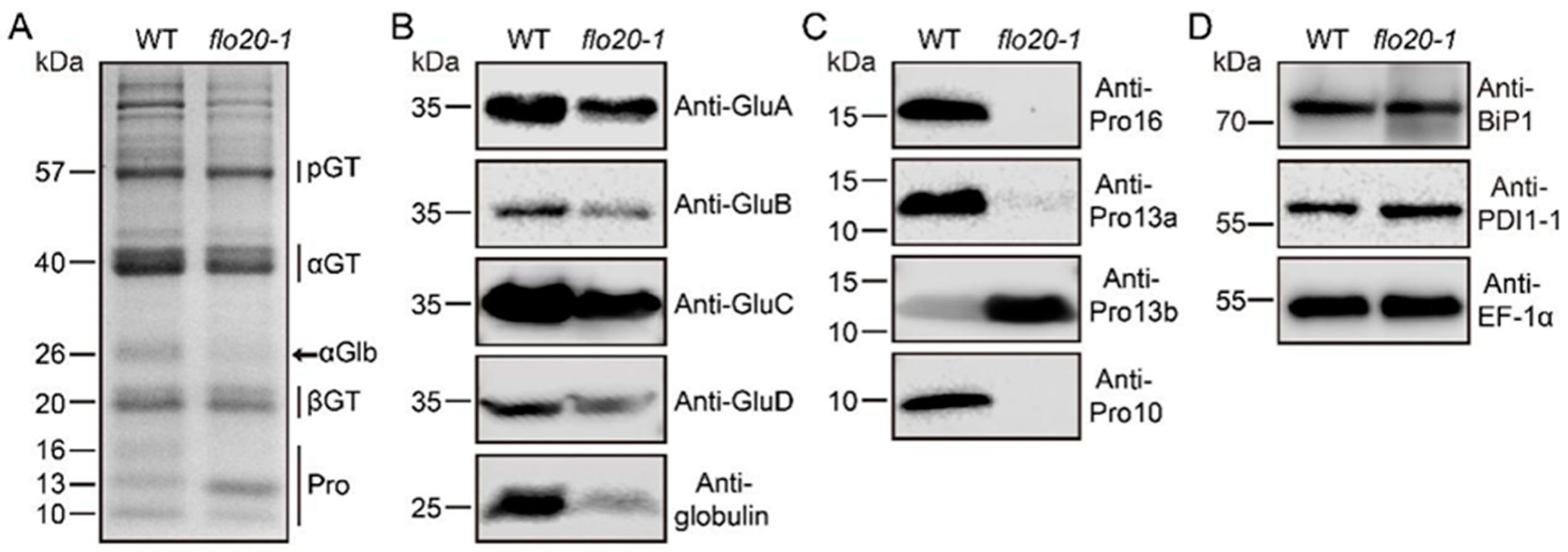
2.1. Storage Protein Composition Changed in the flo20-1 Mutant
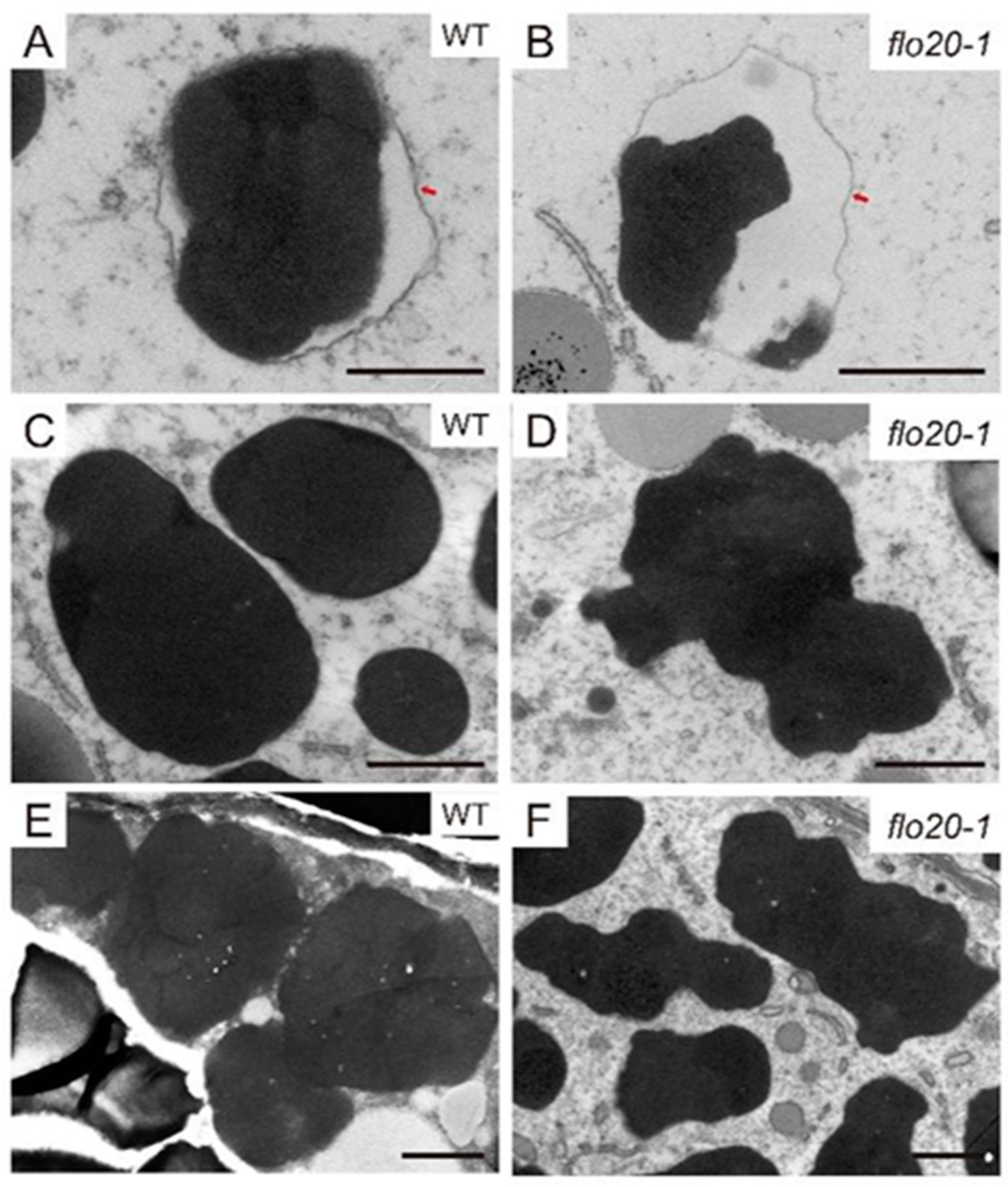
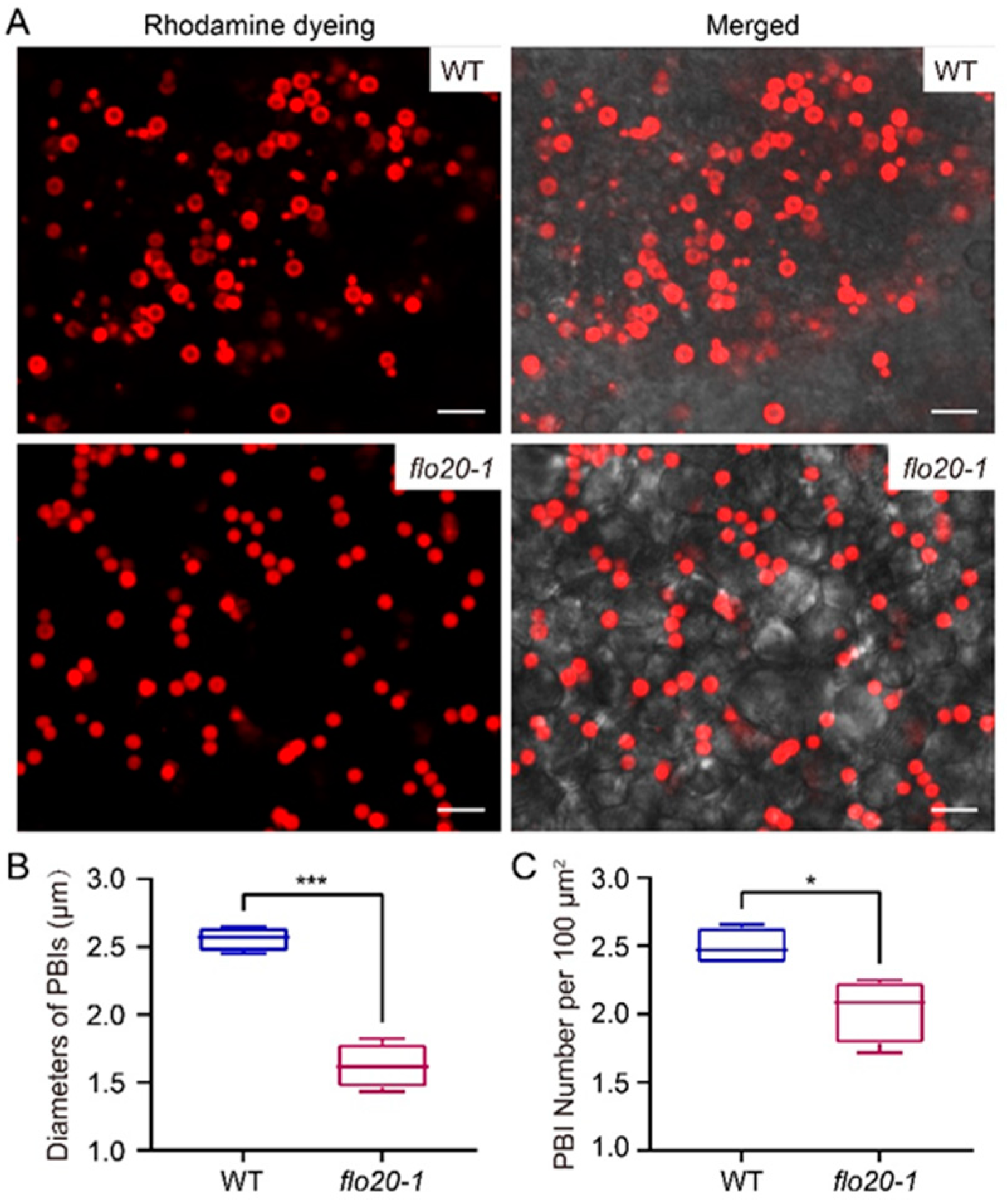
2.2. Amorphous Structure of PBII in the flo20-1 Mutant
2.3. Abnormal Morphological Structure of PBI in the flo20-1 Mutant
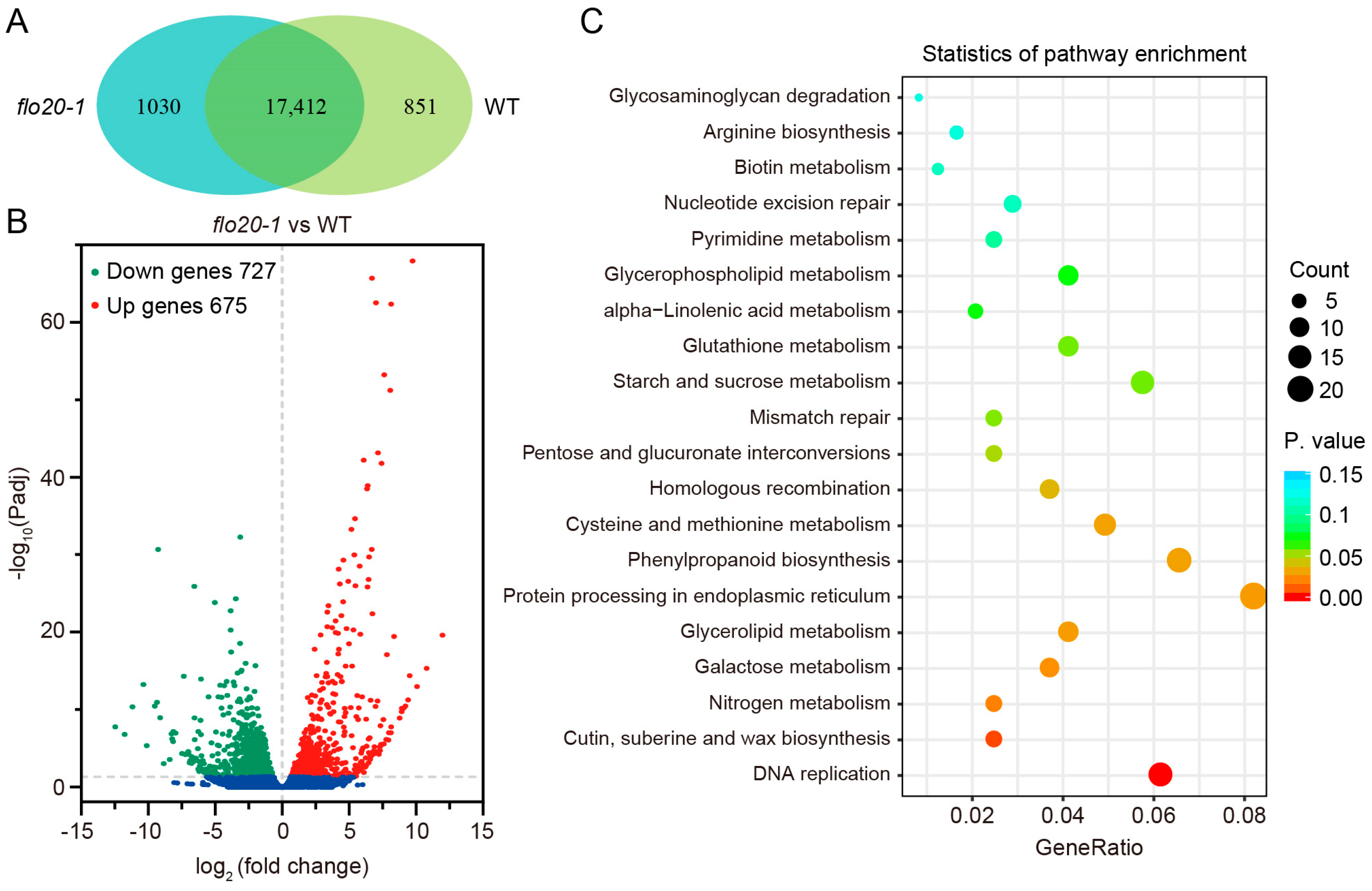
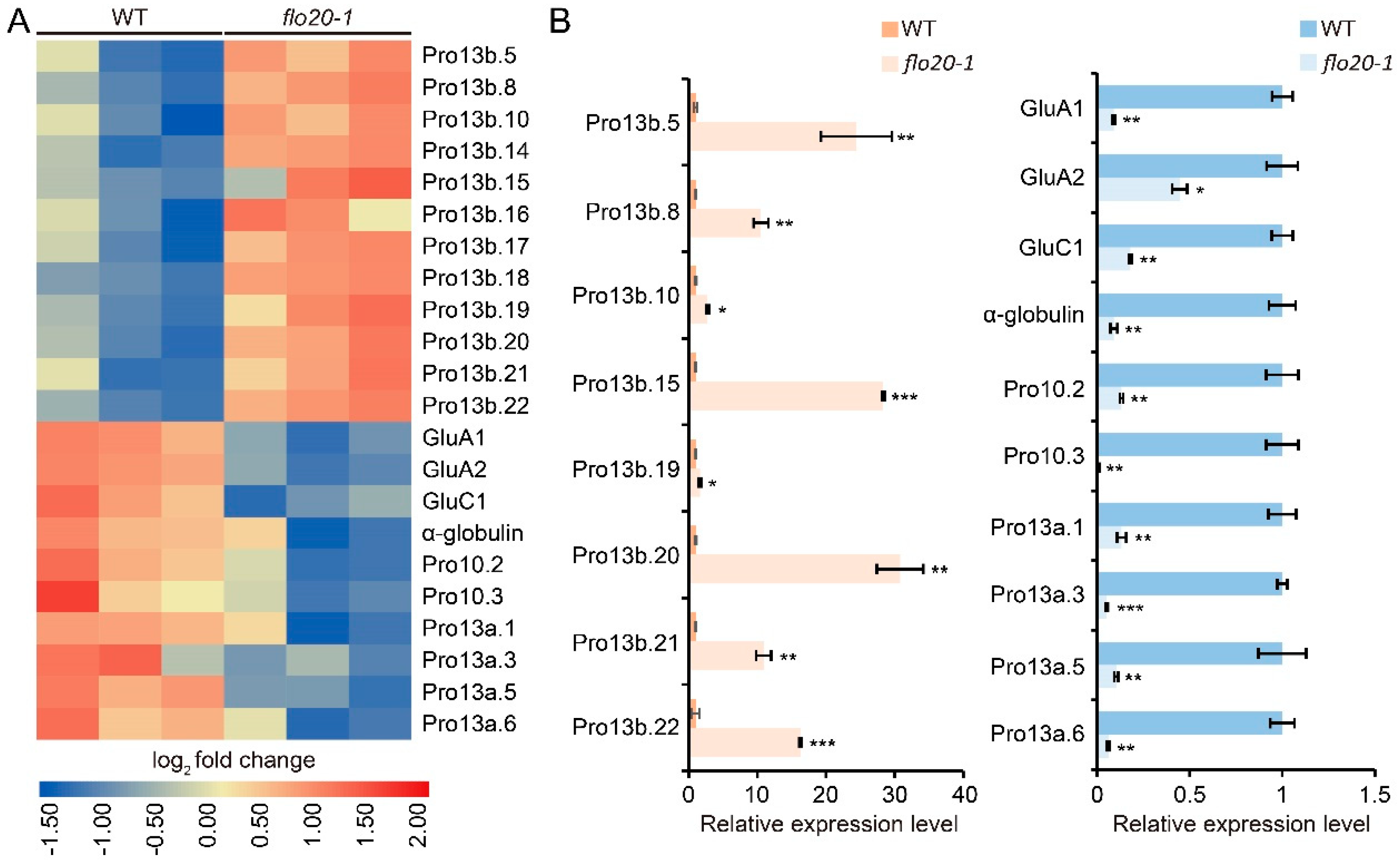
2.4. Transcriptome Sequencing Analysis of the flo20-1 Endosperm
2.5. Analysis of DEGs Related to the SSP Synthesis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials
5.2. SSP Extraction
5.3. Immunoblot Analyses
5.4. Antibodies
5.5. Microscopy Observation
5.6. RNA Extraction and RNA-seq Analysis
5.7. Quantitative Real-Time PCR (qRT-PCR) Validation of DEGs
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ufaz, S.; Galili, G. Improving the content of essential amino acids in crop plants: Goals and opportunities. Plant Physiol. 2008, 147, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Sugimoto, T.; Ogawa, M.; Kasai, Z. Isolation and characterization of two types of protein bodies in the rice endosperm. Agric. Biol. Chem. 1980, 44, 1633–1639. [Google Scholar]
- Yamagata, H.; Sugimoto, T.; Tanaka, K.; Kasai, Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982, 70, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Arcalis, E.; Ibl, V.; Peters, J.; Melnik, S.; Stoger, E. The dynamic behavior of storage organelles in developing cereal seeds and its impact on the production of recombinant proteins. Front. Plant Sci. 2014, 5, 439. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, L.; Lin, Q.; Yu, F. Rice seed storage proteins: Biosynthetic pathways and the effects of environmental factors. J. Integr. Plant Biol. 2021, 63, 1999–2019. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, Y.; Coughlan, S.J.; Okita, T.W.; Satoh, H.; Ogawa, M.; Kumamaru, T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002, 128, 1212–1222. [Google Scholar] [CrossRef]
- Liu, F.; Ren, Y.; Wang, Y.; Peng, C.; Zhou, K.; Lv, J.; Guo, X.; Zhang, X.; Zhong, M.; Zhao, S.; et al. OsVPS9A functions cooperatively with OsRAB5A to regulate post-Golgi dense vesicle-mediated storage protein trafficking to the protein storage vacuole in rice endosperm cells. Mol. Plant 2013, 6, 1918–1932. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, Y.; Liu, F.; Zhou, K.; Ding, Y.; Zhou, F.; Wang, Y.; Liu, K.; Gan, L.; Ma, W.; et al. GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 2014, 26, 410–425. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, F.; Ren, Y.; Wang, Y.; Liu, X.; Long, W.; Wang, D.; Zhu, J.; Zhu, X.; Jing, R.; et al. GOLGI TRANSPORT 1B regulates protein export from the endoplasmic reticulum in rice endosperm cells. Plant Cell 2016, 28, 2850–2865. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, Y.; Pan, T.; Wang, Y.; Wang, Y.; Gan, L.; Wei, Z.; Wang, F.; Wu, M.; Jing, R.; et al. GPA5 encodes a Rab5a effector required for post-Golgi trafficking of rice storage Proteins. Plant Cell 2020, 32, 758–777. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Takaiwa, F. Cereal seed storage protein synthesis: Fundamental processes for recombinant protein production in cereal grains. Plant Biotechnol. J. 2010, 8, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Hirano, H. Rice seed globulin: A protein similar to wheat seed glutenin. Phytochemistry 1992, 31, 3455–3459. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, H.; Tanaka, K. The Site of Synthesis and Accumulation of Rice Storage Proteins. Plant Cell Physiol. 1986, 27, 135–145. [Google Scholar]
- Li, X.; Wu, Y.; Zhang, D.; Gillikin, J.W.; Boston, R.S.; Franceschi, V.R.; Okita, T.W. Rice Prolamine Protein Body Biogenesis: A BiP-mediated process. Science 1993, 282, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Shigemitsu, T.; Tanaka, K.; Morita, S.; Satoh, S.; Masumura, T. Ultrastructure of mature protein body in the starchy endosperm of dry cereal grain. Biosci. Biotechnol. Biochem. 2010, 74, 1485–1487. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kumamaru, T.; Satoh, H.; Iwata, N.; Omura, T.; Kasai, Z.; Tanaka, K. Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 1987, 28, 1517–1527. [Google Scholar]
- Nagamine, A.; Matsusaka, H.; Ushijima, T.; Kawagoe, Y.; Ogawa, M.; Okita, T.W.; Kumamaru, T. A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol. 2011, 52, 1003–1016. [Google Scholar] [CrossRef]
- Kim, W.T.; Li, X.; Okita, T.W. Expression of storage protein multigene families in developing rice endosperm. Plant Cell Physiol. 1993, 34, 595–603. [Google Scholar]
- Saito, Y.; Shigemitsu, T.; Yamasaki, R.; Sasou, A.; Goto, F.; Kishida, K.; Kuroda, M.; Tanaka, K.; Morita, S.; Satoh, S.; et al. Formation mechanism of the internal structure of type I protein bodies in rice endosperm: Relationship between the localization of prolamin species and the expression of individual genes. Plant J. 2012, 70, 1043–1055. [Google Scholar] [CrossRef]
- Kusaba, M.; Miyahara, K.; Iida, S.; Fukuoka, H.; Takano, T.; Sassa, H.; Nishimura, M.; Nishio, T. Low glutelin content1: A dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 2003, 15, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Pan, T.; Zhu, Y.; Jiang, X.; Yu, M.; Wang, R.; Zhang, F.; Luo, S.; Bao, X.; Chen, Y.; et al. FLOURY ENDOSPERM20 encoding SHMT4 is required for rice endosperm development. Plant Biotechnol. J. 2022, 20, 1438–1440. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Satoh-Cruz, M.; Matsusaka, H.; Takemoto-Kuno, Y.; Fukuda, M.; Okita, T.W.; Ogawa, M.; Satoh, H.; Kumamaru, T. Gene-gene interactions between mutants that accumulate abnormally high amounts of proglutelin in rice seed. Breeding Sci. 2010, 60, 568–574. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Hirose, S.; Yasuda, H.; Takaiwa, F. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 2010, 154, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Ashida, K.; Saito, Y.; Masumura, T.; Iida, S. Ultrastructure of protein bodies in mutant rice (Oryza sativa L.) with altered storage protein composition. Breeding Sci. 2011, 61, 201–207. [Google Scholar] [CrossRef]
- Lee, H.J.; Jo, Y.M.; Lee, J.Y.; Lim, S.H.; Kim, Y.M. Lack of globulin synthesis during seed development alters accumulation of seed storage proteins in rice. Int. J. Mol. Sci. 2015, 16, 14717–14736. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, S.; Liu, S.; Jiang, L.; Chen, L.; Ren, Y.; Han, X.; Liu, F.; Ji, S.; Liu, X.; et al. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 2009, 58, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Kumamaru, T.; Uemura, Y.; Inoue, Y.; Takemoto, Y.; Siddiqui, S.U.; Ogawa, M.; Hara-Nishimura, I.; Satoh, H. Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 2010, 51, 38–46. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, Y.; Liu, X.; Jiang, L.; Chen, L.; Han, X.; Jin, M.; Liu, S.; Liu, F.; Lv, J.; et al. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J. 2010, 64, 812–824. [Google Scholar] [CrossRef]
- Zhu, J.; Ren, Y.; Wang, Y.; Liu, F.; Teng, X.; Zhang, Y.; Duan, E.; Wu, M.; Zhong, M.; Hao, Y.; et al. OsNHX5-mediated pH homeostasis is required for post-Golgi trafficking of seed storage proteins in rice endosperm cells. BMC Plant Biol. 2019, 19, 295. [Google Scholar] [CrossRef]
- Pan, T.; Wang, Y.; Jing, R.; Wang, Y.; Wei, Z.; Zhang, B.; Lei, C.; Qi, Y.; Wang, F.; Bao, X.; et al. Post-Golgi trafficking of rice storage proteins requires the small GTPase Rab7 activation complex MON1-CCZ1. Plant Physiol. 2021, 187, 2174–2191. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Ren, Y.; Zhang, Y.; Yang, J.; Duan, E.; Wang, Y.; Liu, F.; Wu, M.; Pan, T.; Wang, Y.; et al. Subunit E isoform 1 of vacuolar H+-ATPase OsVHA enables post-Golgi trafficking of rice seed storage proteins. Plant Physiol. 2021, 187, 2192–2208. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kumamaru, T.; Satoh, H.; Omura, T.; Park, T.; Shintaku, K.; Baba, K. Mutants for rice storage proteins 2. Isolation and characterization of protein bodies from rice mutants. Theor. Appl. Genet. 1989, 78, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Hanson, A.D.; Roje, S. One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 119–137. [Google Scholar] [CrossRef]
- Anderson, D.D.; Stover, P.J. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS ONE 2009, 4, e5839. [Google Scholar] [CrossRef]
- Anderson, D.D.; Woeller, C.F.; Chiang, E.P.; Shane, B.; Stover, P.J. Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J. Biol. Chem. 2012, 287, 7051–7062. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, M.; Zhou, Z.; Feng, J.; Bao, X.; Jiang, Z.; Dong, Z.; Chai, M.; Tan, M.; Li, L.; Cao, Y.; et al. OsSHMT4 Is Required for Synthesis of Rice Storage Protein and Storage Organelle Formation in Endosperm Cells. Plants 2024, 13, 81. https://doi.org/10.3390/plants13010081
Yan M, Zhou Z, Feng J, Bao X, Jiang Z, Dong Z, Chai M, Tan M, Li L, Cao Y, et al. OsSHMT4 Is Required for Synthesis of Rice Storage Protein and Storage Organelle Formation in Endosperm Cells. Plants. 2024; 13(1):81. https://doi.org/10.3390/plants13010081
Chicago/Turabian StyleYan, Mengyuan, Ziyue Zhou, Juling Feng, Xiuhao Bao, Zhengrong Jiang, Zhiwei Dong, Meijie Chai, Ming Tan, Libei Li, Yaoliang Cao, and et al. 2024. "OsSHMT4 Is Required for Synthesis of Rice Storage Protein and Storage Organelle Formation in Endosperm Cells" Plants 13, no. 1: 81. https://doi.org/10.3390/plants13010081
APA StyleYan, M., Zhou, Z., Feng, J., Bao, X., Jiang, Z., Dong, Z., Chai, M., Tan, M., Li, L., Cao, Y., Ke, Z., Wu, J., Feng, Z., & Pan, T. (2024). OsSHMT4 Is Required for Synthesis of Rice Storage Protein and Storage Organelle Formation in Endosperm Cells. Plants, 13(1), 81. https://doi.org/10.3390/plants13010081





