First Evidence of Thalassochory in the Ficus Genus: Seed Dispersal Using the Kuroshio Oceanic Current
Abstract
:1. Introduction
2. Material and Methods
2.1. Female Syconium flotation and Seed Germination Experiments
2.2. Sampling and DNA Extraction
2.3. ddRAD Library Preparation and Sequencing
2.4. ddRAD Data Processing
3. Results
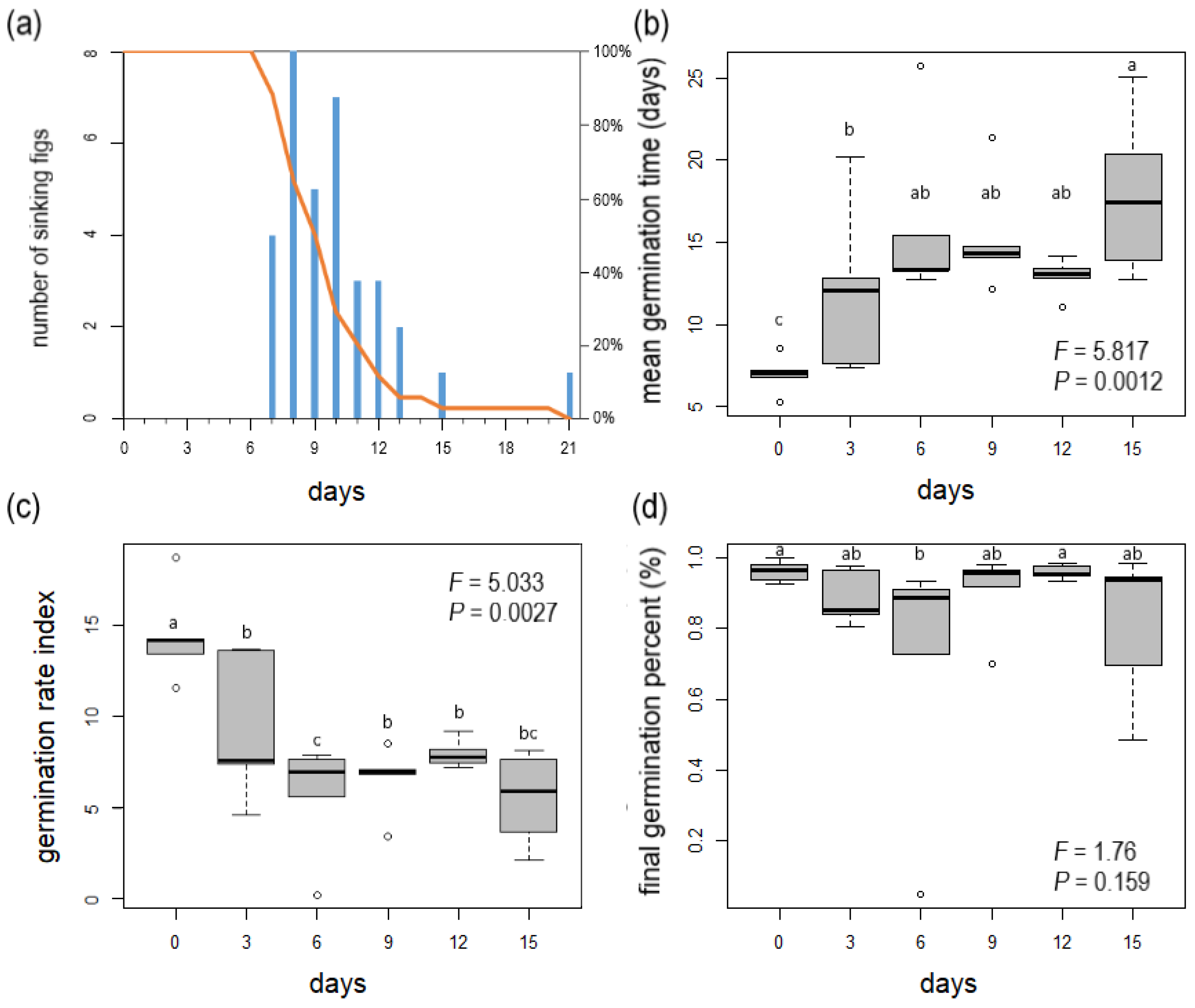
3.1. Female Syconium Flotation and Seed Germination Experiments
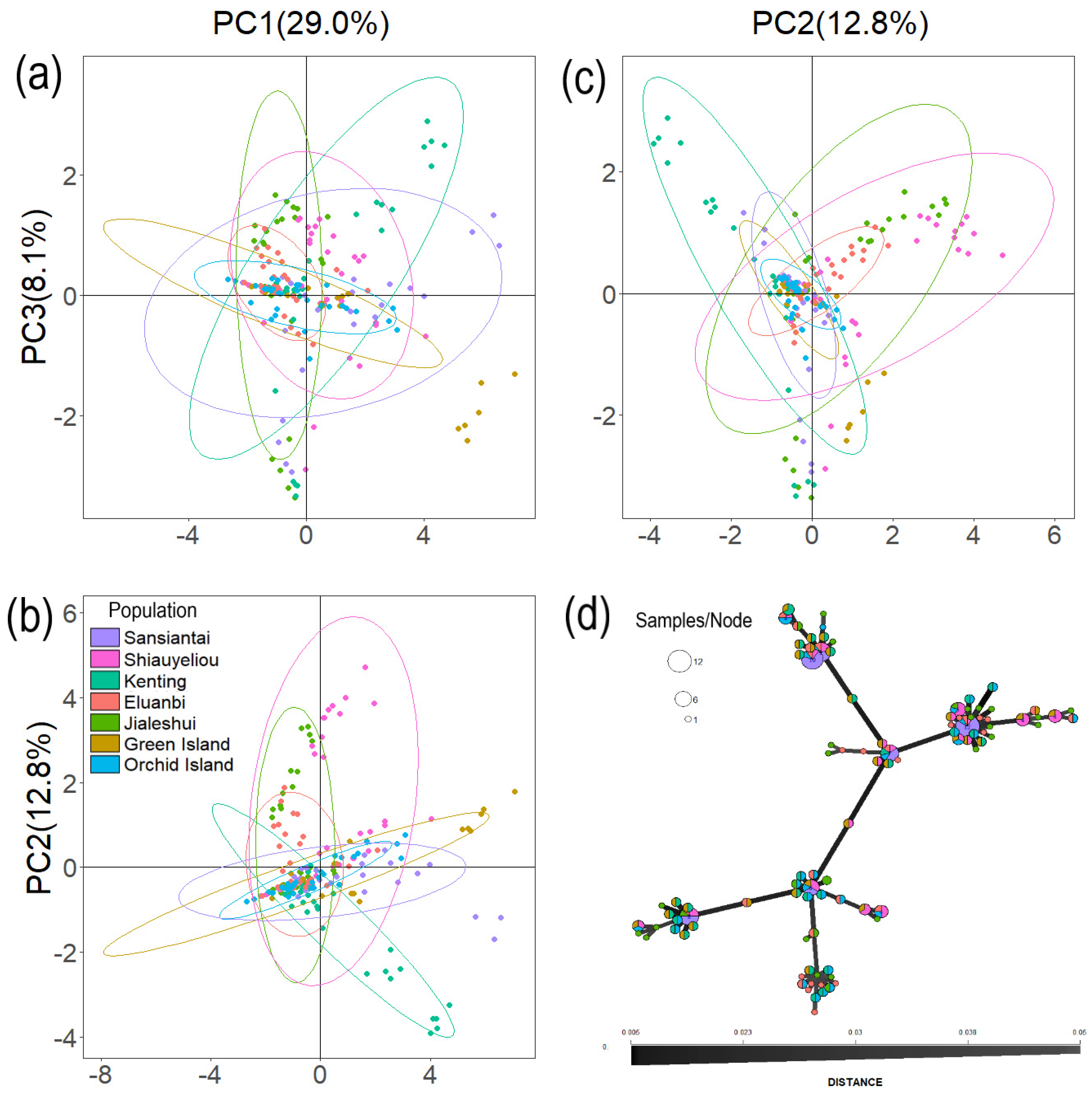
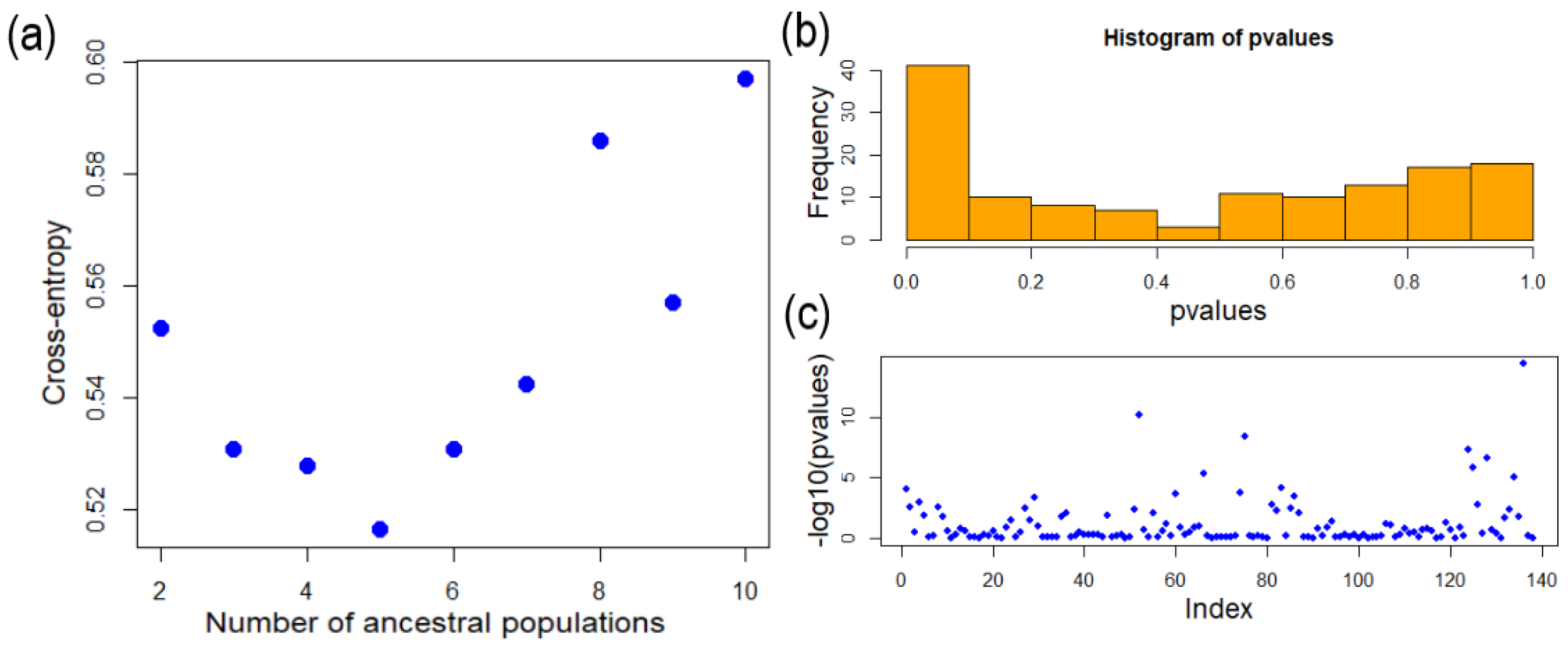
3.2. Mearns Fig Genetic Structure
4. Discussion
4.1. Buoyancy and Seed Dispersal
4.2. Population Genetic Structure and the Kuroshio Current
4.3. Overall Genetic Structure
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacArthur, R.H.; Wilson, E.O. The Theory of Island Biogeography; Princeton University Press: Princeton, NJ, USA, 1967. [Google Scholar]
- Heleno, R.; Vargas, P. How do islands become green? Glob. Ecol. Biogeogr. 2015, 24, 518–526. [Google Scholar] [CrossRef]
- Vargas, P.; Heleno, R.; Traveset, A.; Nogales, M. Colonization of the Galápagos Islands by plants with no specific syndromes for long-distance dispersal: A new perspective. Ecography 2012, 35, 33–43. [Google Scholar] [CrossRef]
- Banack, S.A.; Horn, M.H.; Gawlicka, A. Disperser- vs. establishment-limited distribution of a riparian fig tree (Ficus insipida) in a Costa Rican tropical rain forest. Biotropica 2006, 34, 232–243. [Google Scholar] [CrossRef]
- Berg, C.C.; Corner, E.J.H. Moraceae (Ficus). In Flora Malesiana; Nooteboom, H.P., Ed.; National Herbarium Nederland: Leiden, The Netherlands, 2005; Volume 17. [Google Scholar]
- Pothasin, P.; Compton, S.G.; Wangpakapattanawong, P. Seasonality of Leaf and Fig Production in Ficus squamosa, a Fig Tree with Seeds Dispersed by Water. PLoS ONE 2016, 11, e0152380. [Google Scholar] [CrossRef]
- Lambert, F.R.; Marshall, A.G. Keystone Characteristics of Bird-Dispersed Ficus in a Malaysian Lowland Rain Forest. J. Ecol. 1991, 79, 793. [Google Scholar] [CrossRef]
- Shanahan, M.; So, S.; Gompton, S.G.; Gorlett, R. Fig-eating by vertebrate frugivores: A global review. Biol. Rev. 2001, 76, 529–572. [Google Scholar] [CrossRef] [PubMed]
- Weiblen, G.D.; Lomascolo, S.B.; Oono, R.; Dumont, E.R. Nutritional Dimorphism in New Guinea Dioecious Figs. Biotropica 2010, 42, 656–663. [Google Scholar] [CrossRef]
- Berg, C.C.; Wiebes, J.T. African fig trees and fig wasps. K. Ned. Akad. Van Wet. 1992, 89, 1–298. [Google Scholar]
- Fan, K.-Y.; Bain, A.; Tzeng, H.-Y.; Chiang, Y.-P.; Chou, L.-S.; Kuo-Huang, L.-L. Comparative anatomy of the fig wall (Ficus, Moraceae). Botany 2019, 97, 417–426. [Google Scholar] [CrossRef]
- Correa, S.B.; Winemiller, K.O.; López-Fernández, H.; Galetti, M. Evolutionary Perspectives on Seed Consumption and Dispersal by Fishes. BioScience 2007, 57, 748–756. [Google Scholar] [CrossRef]
- Horn, M.H. Evidence for dispersal of fig seeds by the fruit-eating characid fish Brycon guatemalensis Regan in a Costa Rican tropical rain forest. Oecologia 1997, 109, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Kjellberg, F.; Jousselin, E.; Hossaert-McKey, M.; Rasplus, J.-Y. Biology, ecology, and evolution of fig-pollinating wasps (Chalcidoidea, Agaonidae). In Biology, Ecology and Evolution of Gall-Inducing Arthropods; Raman, A., Schaefer, W., Withers, T.M., Eds.; Science Publishers, Inc.: Enfield, NH, USA; Plymouth, UK, 2005; pp. 539–572. [Google Scholar]
- Cook, J.M.; Segar, S.T. Speciation in fig wasps. Ecol. Entomol. 2010, 35, 54–66. [Google Scholar] [CrossRef]
- Marussich, W.A.; Machado, C.A. Host-specificity and coevolution among pollinating and nonpollinating New World fig wasps. Mol. Ecol. 2007, 16, 1925–1946. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, H.-Y.; Lu, F.-Y.; Ou, C.-H.; Lu, K.-C.; Tseng, L.-J. Pollinational-mutualism strategy of Ficus erecta var. beecheyana and Blastophaga nipponica in seasonal Guandaushi Forest Ecosystem, Taiwan. Bot. Stud. 2006, 47, 307–318. [Google Scholar]
- Weiblen, G.D. How to be a Fig Wasp. Annu. Rev. Entomol. 2002, 47, 299–330. [Google Scholar] [CrossRef] [PubMed]
- Ghara, M.; Borges, R.M. Comparative life-history traits in a fig wasp community: Implications for community structure. Ecol. Entomol. 2010, 35, 139–148. [Google Scholar] [CrossRef]
- Dev, S.A.; Kjellberg, F.; Hossaert-McKey, M.; Borges, R.M. Fine-scale Population Genetic Structure of Two Dioecious Indian Keystone Species, Ficus hispida and Ficus exasperata (Moraceae). Biotropica 2011, 43, 309–316. [Google Scholar] [CrossRef]
- Ahmed, S.; Compton, S.G.; Butlin, R.K.; Gilmartin, P.M. Wind-borne insects mediate directional pollen transfer between desert fig trees 160 kilometers apart. Proc. Natl. Acad. Sci. USA 2009, 106, 20342–20347. [Google Scholar] [CrossRef] [PubMed]
- Kobmoo, N.; Hossaert-Mckey, M.; Rasplus, J.Y.; Kjellberg, F. Ficus racemosa is pollinated by a single population of a single agaonid wasp species in continental South-East Asia. Mol. Ecol. 2010, 19, 2700–2712. [Google Scholar] [CrossRef]
- Corner, E.J.H. Check-List of Ficus in Asia and Australasia with Keys to Identification; The Garden’s Bulletin: Singapore, 1965; Volume 21.
- Hatusima, S. An Enumeration of the Plants of Batan Island, N. Philippines. Mem. Fac. Agric. Kagoshima Univ. 1966, 5, 13–70. [Google Scholar]
- Kuo, C.-C.; Bain, A.; Chiu, Y.-T.; Ho, Y.-C.; Chen, W.-H.; Chou, L.-S.; Tzeng, H.-Y. Topographic effect on the phenology of Ficus pedunculosa var. mearnsii (Mearns fig) in its northern boundary distribution, Taiwan. Sci. Rep. 2017, 7, 14699. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-T.; Bain, A.; Deng, S.-L.; Ho, Y.-C.; Chen, W.-H.; Tzeng, H.-Y. Effects of climate change on a mutualistic coastal species: Recovery from typhoon damages and risks of population erosion. PLoS ONE 2017, 12, e0186763. [Google Scholar] [CrossRef] [PubMed]
- Taiwan Botanic Red List Editorial Committee. The Red List of Vascular Plants of Taiwan, 2017; Endemic Species Research Institute, Council of Agriculture, Executive Yuan, R.O.C (Taiwan): Nantou, Taiwan, 2017.
- Tzeng, H.-Y. Taxonomic Study of the Genus Ficus in Taiwan. Ph.D. Dissertation, Department of Forestry, National Chung Hsing University, Taichung, Taiwan, 2004; 396p. (In Chinese with English Summary). [Google Scholar]
- Chen, C.-H.; Chou, L.-Y. The Blastophagini of Taiwan (Hymenoptera: Agaonidae: Agaoninae). J. Taiwan Mus. 1997, 50, 113–154. [Google Scholar]
- Chou, P.-A.; Bain, A.; Chantarasuwan, B.; Tzeng, H.-Y. Parasitism features of a fig wasp of genus Apocrypta (Pteromalidae: Pteromalinae) associated with a host belonging to Ficus subgenus Ficus. Insects 2023, 14, 437. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.F.; Mayer, D.G. A critical analysis of Maguire’s germination rate index. J. Seed Technol. 1986, 10, 101–110. [Google Scholar]
- Ellis, R.A. The quantification of agent and survival in orthodox seeds. Seed Sci. Technol. 1981, 9, 373–409. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 9 May 2024).
- Shirasawa, K.; Hirakawa, H.; Isobe, S. Analytical workflow of double-digest restriction site-associated DNA sequencing based on empirical and in silico optimization in tomato. DNA Res. 2016, 23, 145–153. [Google Scholar] [CrossRef]
- Shirasawa, K.; Yakushiji, H.; Nishimura, R.; Morita, T.; Jikumaru, S.; Ikegami, H.; Toyoda, A.; Hirakawa, H.; Isobe, S. The Ficus erecta genome aids Ceratocystis canker resistance breeding in common fig (F. carica). Plant J. 2020, 102, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 9 May 2024).
- Paris, J.R.; Stevens, J.R.; Catchen, J.M. Lost in parameter space: A road map for stacks. Methods Ecol. Evol. 2017, 8, 1360–1373. [Google Scholar] [CrossRef]
- Eaton, D.A.R.; Overcast, I. ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 2020, 36, 2592–2594. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 3, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2014, e281. [Google Scholar] [CrossRef] [PubMed]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics, 4th ed.; Sinauer Associates, Inc. Publishers: Sunderland, MA, USA, 1997. [Google Scholar]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395. [Google Scholar] [CrossRef]
- Barton, N.H.; Slatkin, M. A Quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 1986, 56, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F.; Lopez, J.; Belkhir, K. Package ‘genepop’. R Package Version 1.1.7. 2020. Available online: https://cran.r-project.org/web/packages/genepop/genepop.pdf (accessed on 9 May 2024).
- Frichot, E.; François, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Frichot, E.; Mathieu, F.; Trouillon, T.; Bouchard, G.; François, O. Fast and Efficient Estimation of Individual Ancestry Coefficients. Genetics 2014, 196, 973–983. [Google Scholar] [CrossRef]
- Gain, C.; François, O. LEA 3: Factor models in population genetics and ecological genomics with R. Mol. Ecol. Resour. 2021, 21, 2738–2748. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. ldne: A program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 2008, 8, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; England, P.R. Estimating Contemporary Effective Population Size on the Basis of Linkage Disequilibrium in the Face of Migration. Genetics 2011, 189, 633–644. [Google Scholar] [CrossRef]
- Nazareno, A.G.; de Carvalho, D. What the reasons for no inbreeding and high genetic diversity of the neotropical fig tree Ficus arpazusa? Conserv. Genet. 2009, 10, 1789–1793. [Google Scholar] [CrossRef]
- Conceição de Souza-Stevaux, M.; Negrelle, R.R.B.; Citadini-Zanette, V. Seed dispersal by the fish Pterodoras granulosus in the Paraná River Basin, Brazil. J. Trop. Ecol. 1994, 10, 621–626. [Google Scholar] [CrossRef]
- Horn, M.H.; Correa, S.B.; Parolin, P.; Pollux, B.; Anderson, J.T.; Lucas, C.; Widmann, P.; Tjiu, A.; Galetti, M.; Goulding, M. Seed dispersal by fishes in tropical and temperate fresh waters: The growing evidence. Acta Oecologica 2011, 37, 561–577. [Google Scholar] [CrossRef]
- Renner, S. Plant Dispersal across the Tropical Atlantic by Wind and Sea Currents. Int. J. Plant Sci. 2004, 165, S23–S33. [Google Scholar] [CrossRef]
- Gandour, M.; Hessini, K.; Abdelly, C. Understanding the population genetic structure of coastal species (Cakile maritima): Seed dispersal and the role of sea currents in determining population structure. Genet. Res. 2008, 90, 167–178. [Google Scholar] [CrossRef]
- Walther, B.A.; Geier, J.; Chou, L.-S.; Bain, A. The figs of winter: Seasonal importance of fruiting fig trees (Ficus: Moraceae) for urban birds. Acta Oecologica 2018, 90, 28–34. [Google Scholar] [CrossRef]
- Liang, W.; Yang, Y.J.; Tang, T.Y.; Chuang, W. Kuroshio in the Luzon Strait. J. Geophys. Res. Oceans 2008, 113, C08048. [Google Scholar] [CrossRef]
- Bain, A.; Borges, R.M.; Chevallier, M.H.; Vignes, H.; Kobmoo, N.; Peng, Y.Q.; Cruaud, A.; Rasplus, J.Y.; Kjellberg, F.; Hossaert-Mckey, M. Geographic structuring into vicariant species-pairs in a wide-ranging, high-dispersal plant–insect mutualism: The case of Ficus racemosa and its pollinating wasps. Evol. Ecol. 2016, 30, 663–684. [Google Scholar] [CrossRef]
- Heer, K.; Kalko, E.K.V.; Albrecht, L.; García-Villacorta, R.; Staeps, F.C.; Herre, E.A.; Dick, C.W. Spatial Scales of Genetic Structure in Free-Standing and Strangler Figs (Ficus, Moraceae) Inhabiting Neotropical Forests. PLoS ONE 2015, 10, e0133581. [Google Scholar] [CrossRef] [PubMed]
- Anstett, M.-C.; Hossaert-McKey, M.; McKey, D. Modeling the persistence of small populations of strongly interdependent species: Figs and fig wasps. Conserv. Biol. 1997, 11, 204–213. [Google Scholar] [CrossRef]
- Borges, R.M. Figs, Malabar Giant Squirrels, and Fruit Shortages Within Two Tropical Indian Forests. Biotropica 1993, 25, 183. [Google Scholar] [CrossRef]
- Michaloud, G.; Michaloud-Pelletier, S. Ficus Hemi-Epiphytes (Moraceae) et Arbres Supports. Biotropica 1987, 19, 125. [Google Scholar] [CrossRef]
- Wang, R.; Ai, B.; Gao, B.; Yu, S.; Li, Y.; Chen, X. Spatial genetic structure and restricted gene flow in a functionally dioecious fig, Ficus pumila L. var. pumila (Moraceae). Popul. Ecol. 2008, 51, 307–315. [Google Scholar] [CrossRef]
- Baraket, G.; Chatti, K.; Saddoud, O.; Ben Abdelkarim, A.; Mars, M.; Trifi, M.; Hannachi, A.S. Comparative Assessment of SSR and AFLP Markers for Evaluation of Genetic Diversity and Conservation of Fig, Ficus carica L., Genetic Resources in Tunisia. Plant Mol. Biol. Rep. 2010, 29, 171–184. [Google Scholar] [CrossRef]
- Cabrita, L.F.; Aksoy, U.; Hepaksoy, S.; Leitão, J.M. Suitability of isozyme, RAPD and AFLP markers to assess genetic differences and relatedness among fig (Ficus carica L.) clones. Sci. Hortic. 2001, 87, 261–273. [Google Scholar] [CrossRef]
- Khadari, B.; Grout, C.; Santoni, S.; Kjellberg, F. Contrasted genetic diversity and differentiation among Mediterranean populations of Ficus carica L.: A study using mtDNA RFLP. Genet. Resour. Crop. Evol. 2005, 52, 97–109. [Google Scholar] [CrossRef]
- Harrison, R.D. Fig wasp dispersal and the stability of a keystone plant resource in Borneo. Proc. R. Soc. B 2003, 270, S76–S79. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.D.; Rasplus, J.-Y. Dispersal of fig pollinators in Asian tropical rain forests. J. Trop. Ecol. 2006, 22, 631–639. [Google Scholar] [CrossRef]
- Nazareno, A.G.; Alzate-Marin, A.L.; Pereira, R.A.S. Dioecy, more than monoecy, affects plant spatial genetic structure: The case study of Ficus. Ecol. Evol. 2013, 3, 3495–3508. [Google Scholar] [CrossRef] [PubMed]
- Bain, A. Colonization and Adaptations of Ficus in Taiwan. Ph.D. Dissertation, Institute of Ecology and Evolutionary Biology, National Taiwan University, Taipei, Taiwan, Université Montpellier 2, Montpellier, France, 2012. [Google Scholar]
- Rodriguez, L.J.; Bain, A.; Chou, L.-S.; Conchou, L.; Cruaud, A.; Gonzales, R.; Hossaert-McKey, M.; Rasplus, J.-Y.; Tzeng, H.-Y.; Kjellberg, F. Diversification and spatial structuring in the mutualism between Ficus septica and its pollinating wasps in insular South East Asia. BMC Evol. Biol. 2017, 17, 207. [Google Scholar] [CrossRef] [PubMed]
- Slarkin, M. Gene Flow in Natural Populations. Annu. Rev. Ecol. Syst. 1985, 16, 393–430. [Google Scholar] [CrossRef]
- Fischer, M.; Husi, R.; Prati, D.; Peintinger, M.; van Kleunen, M.; Schmid, B. RAPD variation among and within small and large populations of the rare clonal plant Ranunculus reptans (Ranunculaceae). Am. J. Bot. 2000, 87, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Lande, R. Natural Selection and Random Genetic Drift in Phenotypic Evolution. Evolution 1976, 30, 314. [Google Scholar] [CrossRef] [PubMed]
- Templeton, A.R. The theory of speciation via the founder principle. Genetics 1980, 94, 1011–1038. [Google Scholar] [CrossRef] [PubMed]
- Templeton, A.R. Experimental Tests of Genetic Transilience. Evolution 1999, 53, 1628. [Google Scholar] [CrossRef]
- Wright, S. Evolution in Mendelian populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. In Proceedings of the Sixth International Congress on Genetics, Ithaca, NY, USA, 29 October 1932; Volume 1, pp. 356–366. [Google Scholar]
- Barton, N.; Turelli, M. Effects of genetic drift on variance components under a general model of epistasis. Evolution 2004, 58, 2111–2132. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Barton, N.H.; Turelli, M. Prediction of effects of genetic drift on variance components under a general model of epistasis. Theor. Popul. Biol. 2006, 70, 56–62. [Google Scholar] [CrossRef]
- Robertson, A. The effect of inbreeding on the variation due to recessive genes. Genetics 1952, 37, 189–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Caballero, A.; Keightley, P.D.; Hill, W.G. Bottleneck Effect on Genetic Variance: A Theoretical Investigation of the Role of Dominance. Genetics 1998, 150, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Willi, Y.; VAN Buskirk, J.; Schmid, B.; Fischer, M. Genetic isolation of fragmented populations is exacerbated by drift and selection. J. Evol. Biol. 2007, 20, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.H.; Orr, H.A. Increased Heritable Variation Following Population Bottlenecks: The Role of Dominance. Evolution 1993, 47, 949. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-T.; Chao, C.-T.; Nakamura, K.; Liu, H.-L.; Luo, M.-X.; Liao, P.-C. Divergence with Gene Flow and Contrasting Population Size Blur the Species Boundary in Cycas Sect. Asiorientales, as Inferred from Morphology and RAD-Seq Data. Front. Plant Sci. 2022, 13, 824158. [Google Scholar] [CrossRef] [PubMed]
- Futai, K.; Isagi, Y.; Watanabe, H. The distribution pattern of Heritiera littoralis Dryand. on the Ryukyu Islands as affected by seed dispersal via ocean currents. Tropics 2010, 19, 21–27. [Google Scholar] [CrossRef]
- Hanaoka, S.; Chien, C.-T.; Chen, S.-Y.; Watanabe, A.; Setsuko, S.; Kato, K. Genetic structures of Calophyllum inophyllum L., a tree employing sea-drift seed dispersal in the northern extreme of its distribution. Ann. For. Sci. 2014, 71, 575–584. [Google Scholar] [CrossRef]
- Nakajima, Y.; Matsuki, Y.; Lian, C.; Fortes, M.D.; Uy, W.H.; Campos, W.L.; Nakaoka, M.; Nadaoka, K. The Kuroshio Current influences genetic diversity and population genetic structure of a tropical seagrass, Enhalus acoroides. Mol. Ecol. 2014, 23, 6029–6044. [Google Scholar] [CrossRef]
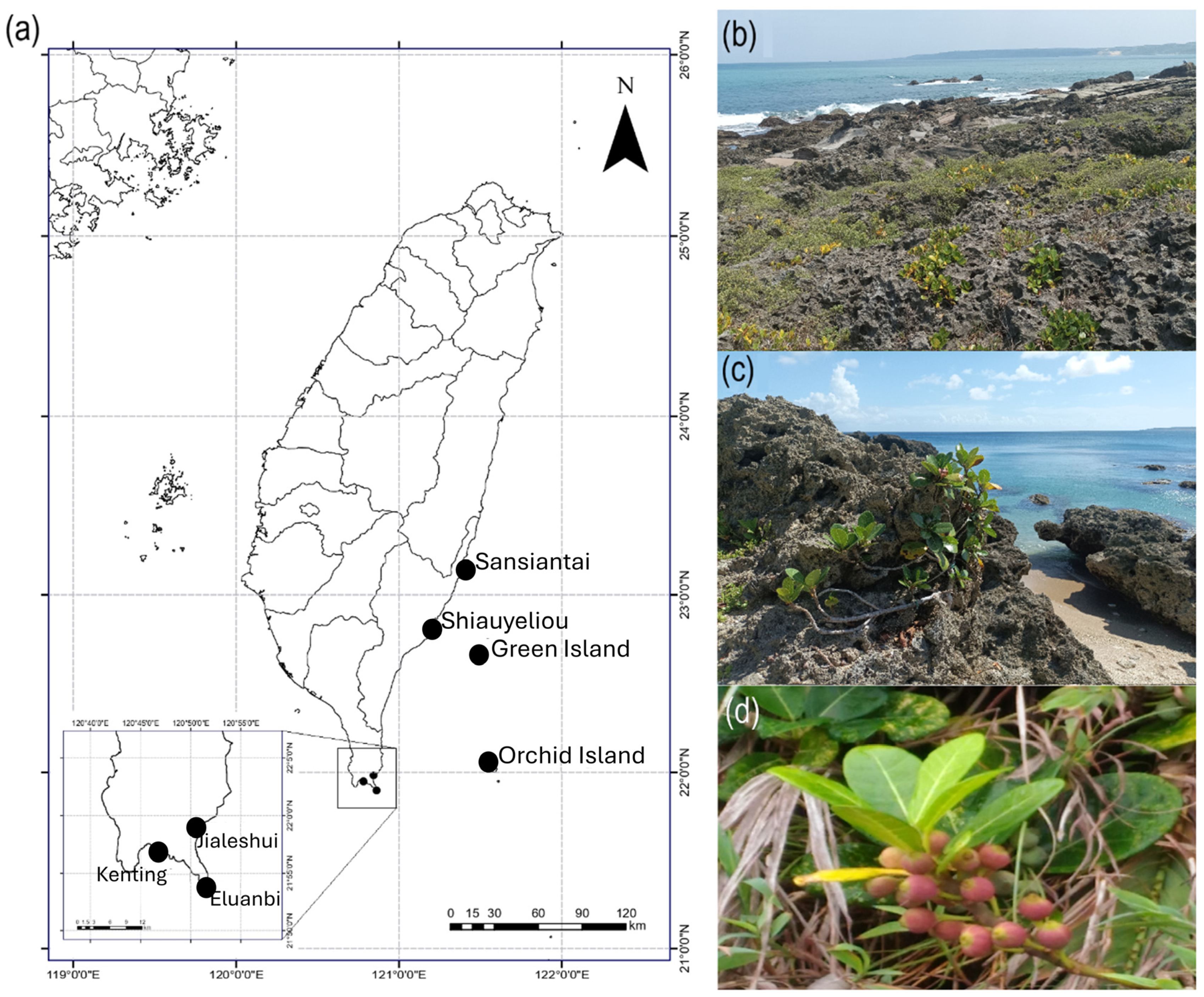

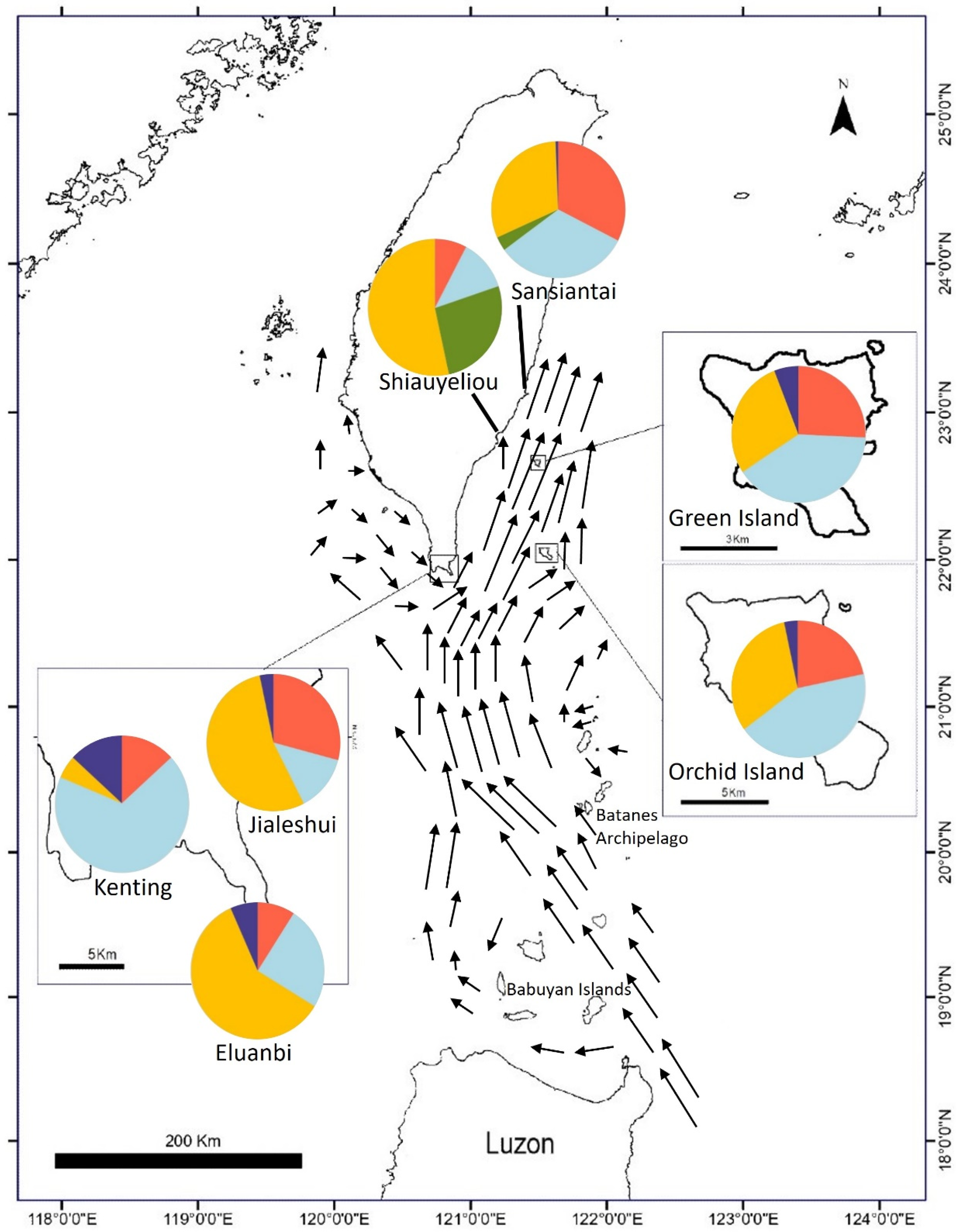
| Distribution | N | GPS Coordinates |
|---|---|---|
| Ficus pedunculosa var. mearnsii | 210 | |
| East Taiwan: | ||
| Sansiantai | 30 | 23°12′34″ N; 121°41′23″ E |
| Shiauyeliou | 30 | 22°79′49″ N; 121°19′76″ E |
| South Taiwan: | ||
| Kenting | 30 | 21°93′94 N; 120″79°79″′ E |
| Eluanbi | 30 | 21°89′93″ N; 120°85′7″ E |
| Jialeshui | 30 | 21°99′41″ N; 120°86′30″ E |
| Outlying Islands: | ||
| Green Island | 30 | 22°66′18″ N; 121°47′83″ E |
| Orchid Island | 30 | 22°2′67″ N; 121°56′79″ E |
| Sansiantai | Shiauyeliou | Kenting | Eluanbi | Jialeshui | Green Island | |
|---|---|---|---|---|---|---|
| Shiauyeliou | 42.6 | |||||
| Kenting | 146 | 104 | ||||
| Eluanbi | 148 | 106 | 7.03 | |||
| Jialeshui | 138 | 95.5 | 9.06 | 10.6 | ||
| Green Island | 51.8 | 32.7 | 107 | 107 | 97.5 | |
| Orchid Island | 123 | 93.5 | 80 | 75.3 | 72.7 | 71.2 |
| Mean Square | ||||
|---|---|---|---|---|
| Source | df | MGT | GRI | FGP |
| treatment | 5 | 0.018 ** | 0.019 ** | 0.104 |
| Residuals | 24 | 0.003 | 0.004 | 0.059 |
| Treatment (Days) | Percentage of Figs Still Floating at the End of the Treatments (%) | MGT * (Days) | GRI * | FGP * (%) |
|---|---|---|---|---|
| 0 | 100% | 6.98 ± 1.18 c | 14.47 ± 2.63 a | 96.23 ± 2.99 a |
| 3 | 100% | 12.00 ± 5.19 b | 9.38 ± 4.10 b | 88.83 ± 7.75 ab |
| 6 | 100% | 16.10 ± 5.49 ab | 5.64 ± 3.18 b | 70.23 ± 37.47 b |
| 9 | 50% | 15.32 ± 3.54 ab | 6.56 ± 1.90 c | 90.47 ± 11.69 ab |
| 12 | 12% | 12.89 ± 1.15 ab | 7.97 ± 0.79 b | 96.04 ± 2.10 a |
| 15 | 3% | 17.88 ± 5.01 a | 5.50 ± 2.60 bc | 81.02 ± 21.34 ab |
| Population | SNPs | FIS | Tajima’s D | π |
|---|---|---|---|---|
| Ficus pedunculosa var. mearnsii | 227,495 | 0.443 | −7.472 | 0.077 |
| East Taiwan | 190,227 | 0.420 | −2.429 | 0.071 |
| Sansiantai | 113,299 | 0.181 | −1.303 | 0.028 |
| Shiauyeliou | 129,315 | 0.432 | −2.736 | 0.027 |
| South Taiwan | 219,548 | 0.441 | −5.189 | 0.065 |
| Kenting | 155,846 | 0.251 | −3.254 | 0.096 |
| Eluanbi | 109,460 | 0.258 | −3.918 | 0.082 |
| Jialeshui | 136,694 | 0.425 | −2.479 | 0.060 |
| Outlying Islands | ||||
| Green Island | 52,733 | 0.239 | −4.476 | 0.029 |
| Orchid Island | 105,355 | 0.277 | −2.139 | 0.087 |
| Sansiantai | Shiauyeliou | Kenting | Eluanbi | Jialeshui | Green Island | ||
|---|---|---|---|---|---|---|---|
| Shiauyeliou | DXY | 0.022 | |||||
| FST | 0 | ||||||
| Nm | 0.284 | ||||||
| Kenting | DXY | 0.031 | 0.034 | ||||
| FST | 0.086 | 0.035 | |||||
| Nm | 0.372 | 0.252 | |||||
| Eluanbi | DXY | 0.060 | 0.068 | 0.081 | |||
| FST | 0.223 | 0.111 | 0 | ||||
| Nm | 0.253 | 0.254 | 0.235 | ||||
| Jialeshui | DXY | 0.065 | 0.057 | 0.054 | 0.071 | ||
| FST | 0.161 | 0.103 | 0.032 | 0.031 | |||
| Nm | 0.178 | 0.221 | 0.167 | 0.269 | |||
| Green Island | DXY | 0.052 | 0.056 | 0.122 | 0.109 | 0.166 | |
| FST | 0.836 | 0.724 | 0.578 | 0.369 | 0.623 | ||
| Nm | 0.376 | 0.376 | 0.357 | 0.471 | 0.298 | ||
| Orchid Island | DXY | 0.035 | 0.034 | 0.069 | 0.085 | 0.051 | 0.109 |
| FST | 0.040 | 0 | 0.033 | 0.069 | 0.056 | 0.362 | |
| Nm | 0.445 | 0.278 | 0.368 | 0.219 | 0.184 | 0.334 |
| Estimated Number of Individuals in the Wild * | Independent Comparisons | Overall r2 | Expected r2 | Ne | 95% CIs | ||
|---|---|---|---|---|---|---|---|
| Parametric | Jackknife Loci | ||||||
| All | 36,688 | 0.1496 | 0.0435 | 1.5 | 1.4–1.5 | 1.4–1.5 | |
| East Taiwan | 2269 | 0.5895 | 0.0896 | 0.3 | 0.3–0.3 | 0.3–0.3 | |
| Sansiantai | 100 | 171 | 0.4339 | 0.1825 | 0.6 | 0.4–0.9 | 0.4–0.9 |
| Shiauyeliou | 100 | 1229 | 0.4810 | 0.1083 | 0.4 | 0.4–0.5 | 0.4–0.4 |
| South Taiwan | 2000 | 65,251 | 0.0450 | 0.0300 | 20.0 | 19.3–20.7 | 18.8–21.3 |
| Kenting | 4496 | 0.1947 | 0.1381 | 2.7 | 2.4–3.2 | 2.3–3.3 | |
| Eluanbi | 51,404 | 0.0988 | 0.0959 | 104.6 | 73.1–179.6 | 71.7–188.1 | |
| Jialeshui | 89,667 | 0.1658 | 0.1238 | 4.7 | 4.3–5.1 | 4.1–5.2 | |
| Outlying Islands | |||||||
| Green Island | 100 | 405 | 0.2844 | 0.2110 | 2.1 | 1.3–5.7 | 1.3–6.4 |
| Orchid Island | 100 | 1115 | 0.2823 | 0.2416 | 5.0 | 2.3–14.6 | 2.3–13.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, S.-H.; Sun, Y.-H.; Tzeng, H.-Y.; Rodriguez, L.J.; Bain, A. First Evidence of Thalassochory in the Ficus Genus: Seed Dispersal Using the Kuroshio Oceanic Current. Plants 2024, 13, 1398. https://doi.org/10.3390/plants13101398
Pan S-H, Sun Y-H, Tzeng H-Y, Rodriguez LJ, Bain A. First Evidence of Thalassochory in the Ficus Genus: Seed Dispersal Using the Kuroshio Oceanic Current. Plants. 2024; 13(10):1398. https://doi.org/10.3390/plants13101398
Chicago/Turabian StylePan, Shin-Hung, Ying-Hsuan Sun, Hsy-Yu Tzeng, Lillian Jennifer Rodriguez, and Anthony Bain. 2024. "First Evidence of Thalassochory in the Ficus Genus: Seed Dispersal Using the Kuroshio Oceanic Current" Plants 13, no. 10: 1398. https://doi.org/10.3390/plants13101398
APA StylePan, S.-H., Sun, Y.-H., Tzeng, H.-Y., Rodriguez, L. J., & Bain, A. (2024). First Evidence of Thalassochory in the Ficus Genus: Seed Dispersal Using the Kuroshio Oceanic Current. Plants, 13(10), 1398. https://doi.org/10.3390/plants13101398






