Analysis of the Effects of the Vrn-1 and Ppd-1 Alleles on Adaptive and Agronomic Traits in Common Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Results
2.1. Verification of Spring Genotypes of Studied NILs
2.2. Identification of the Ppd-1 Allelic Composition of Studied NILs
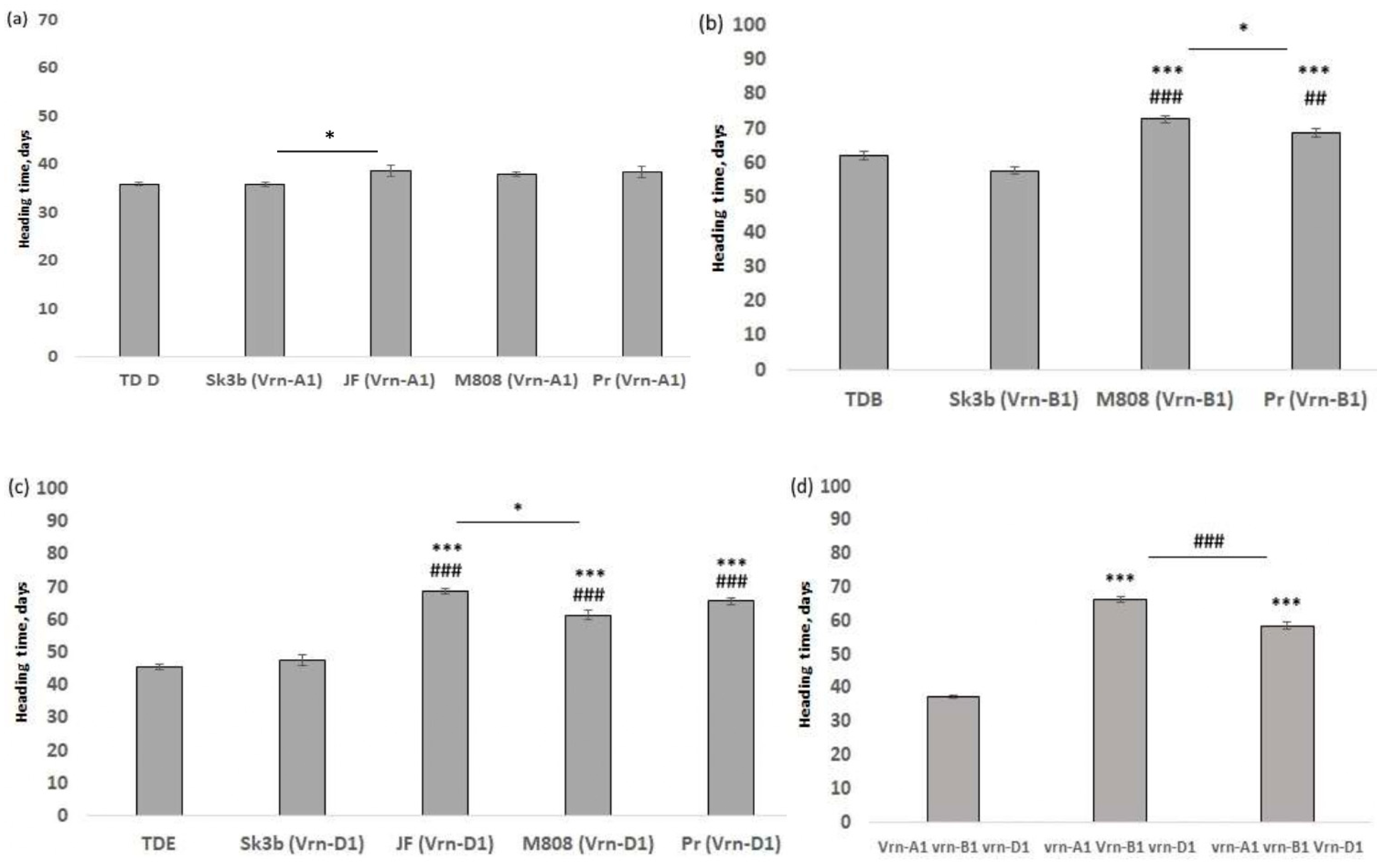
2.3. Influence of Ppd-1 and Vrn-1 Allelic Combinations on Wheat Earliness
2.4. PCA and Pearson’s Correlation Analysis to Study the Effect of Ppd-1 and Vrn-1 Allelic Combinations on Phenology and Agronomic Traits of NILs
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Plant Phenotyping
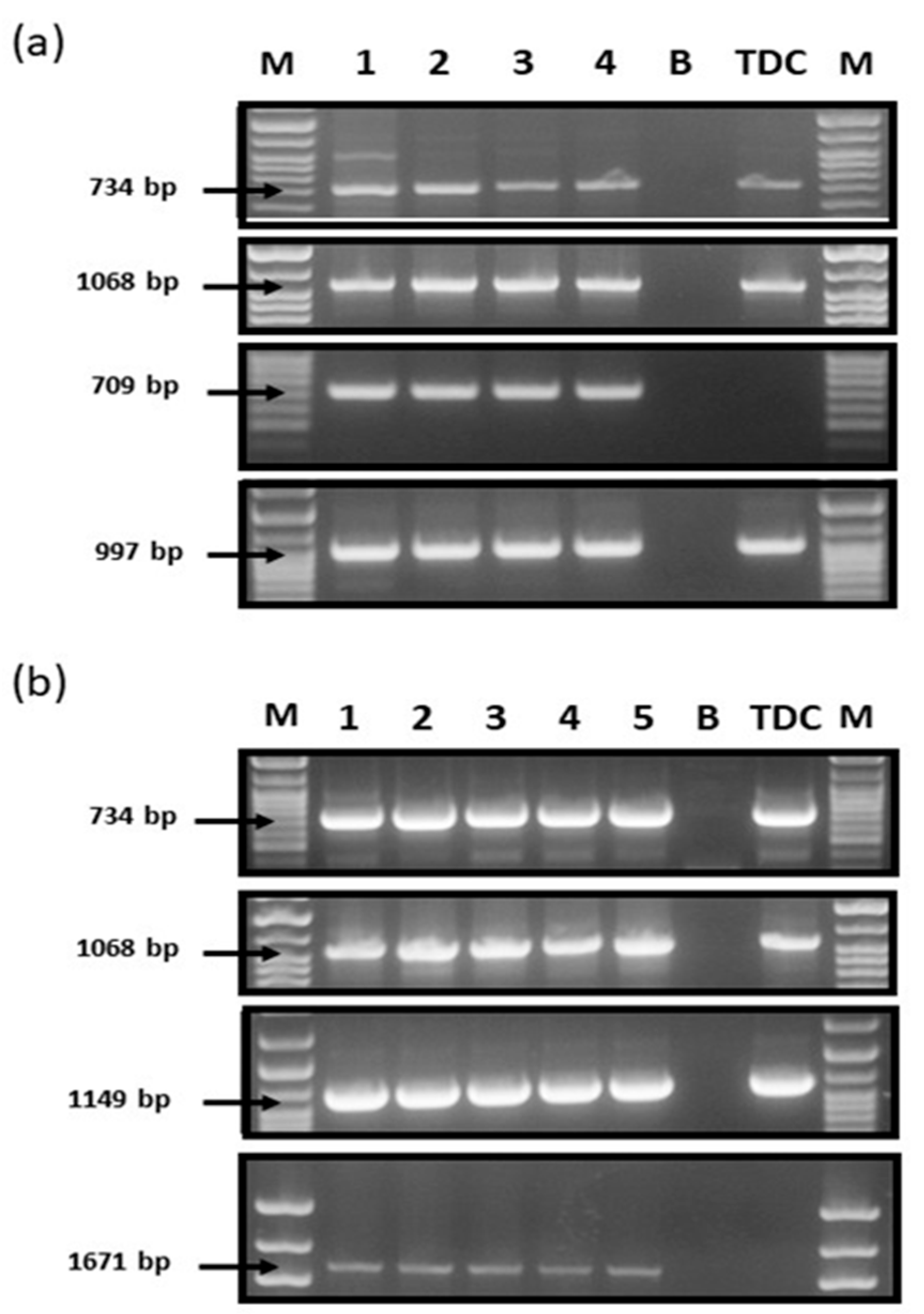
4.3. Genomic DNA Extraction and PCR Analysis
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Facts & Figures on Food and Biodiversity. Available online: https://idrc-crdi.ca/en/research-in-action/facts-figures-food-and-biodiversity (accessed on 14 December 2023).
- Grote, U.; Fasse, A.; Nguyen, T.T.; Erenstein, O. Food Security and the Dynamics of Wheat and Maize Value Chains in Africa and Asia. Front. Sustain. Food Syst. 2021, 4, 617009. [Google Scholar] [CrossRef]
- Kahiluoto, H.; Kaseva, J.; Balek, J.; Olesen, J.E.; Ruiz-Ramos, M.; Gobin, A.; Kersebaum, K.C.; Takáč, J.; Ruget, F.; Ferrise, R.; et al. Decline in Climate Resilience of European Wheat. Proc. Natl. Acad. Sci. USA 2019, 116, 123–128. [Google Scholar] [CrossRef]
- Hertel, T.W. The Global Supply and Demand for Agricultural Land in 2050: A Perfect Storm in the Making? Am. J. Agric. Econ. 2011, 93, 259–275. [Google Scholar] [CrossRef]
- Beddington, J.R.; Asaduzzaman, M.; Clark, M.E.; Fernández Bremauntz, A.; Guillou, M.D.; Howlett, D.J.B.; Jahn, M.M.; Lin, E.; Mamo, T.; Negra, C.; et al. What Next for Agriculture After Durban? Science 2012, 335, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate Variation Explains a Third of Global Crop Yield Variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef]
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising Temperatures Reduce Global Wheat Production. Nat. Clim. Change 2015, 5, 143–147. [Google Scholar] [CrossRef]
- Le Gouis, J.; Oury, F.-X.; Charmet, G. How Changes in Climate and Agricultural Practices Influenced Wheat Production in Western Europe. J. Cereal Sci. 2020, 93, 102960. [Google Scholar] [CrossRef]
- Juroszek, P.; Von Tiedemann, A. Climate Change and Potential Future Risks through Wheat Diseases: A Review. Eur. J. Plant Pathol. 2013, 136, 21–33. [Google Scholar] [CrossRef]
- Cockram, J.; Jones, H.; Leigh, F.J.; O’Sullivan, D.; Powell, W.; Laurie, D.A.; Greenland, A.J. Control of Flowering Time in Temperate Cereals: Genes, Domestication, and Sustainable Productivity. J. Exp. Bot. 2007, 58, 1231–1244. [Google Scholar] [CrossRef]
- Fjellheim, S.; Boden, S.; Trevaskis, B. The Role of Seasonal Flowering Responses in Adaptation of Grasses to Temperate Climates. Front. Plant Sci. 2014, 5, 431. [Google Scholar] [CrossRef]
- Horváth, Á.; Kiss, T.; Berki, Z.; Horváth, Á.D.; Balla, K.; Cseh, A.; Veisz, O.; Karsai, I. Effects of Genetic Components of Plant Development on Yield-Related Traits in Wheat (Triticum aestivum L.) under Stress-Free Conditions. Front. Plant Sci. 2023, 13, 1070410. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Navabi, A.; Salmon, D.F.; Yang, R.-C.; Murdoch, B.M.; Moore, S.S.; Spaner, D. Genetic Analysis of Flowering and Maturity Time in High Latitude Spring Wheat: Genetic Analysis of Earliness in Spring Wheat. Euphytica 2007, 154, 207–218. [Google Scholar] [CrossRef]
- Iqbal, M.; Navabi, A.; Yang, R.-C.; Salmon, D.F.; Spaner, D. The Effect of Vernalization Genes on Earliness and Related Agronomic Traits of Spring Wheat in Northern Growing Regions. Crop Sci. 2007, 47, 1031–1039. [Google Scholar] [CrossRef]
- Smolenskaya, S.E.; Efimov, V.M.; Kruchinina, Y.V.; Nemtsev, B.F.; Chepurnov, G.Y.; Ovchinnikova, E.S.; Belan, I.A.; Zuev, E.V.; Zhou, C.; Piskarev, V.V.; et al. Earliness and Morphotypes of Common Wheat Cultivars of Western and Eastern Siberia. Vavilov J. Genet. Breed. 2022, 26, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Worland, A.J.; Börner, A.; Korzun, V.; Li, W.M.; Petrovíc, S.; Sayers, E.J. The Influence of Photoperiod Genes on the Adaptability of European Winter Wheats. Euphytica 1998, 100, 385–394. [Google Scholar] [CrossRef]
- Würschum, T.; Leiser, W.L.; Langer, S.M.; Tucker, M.R.; Longin, C.F.H. Phenotypic and Genetic Analysis of Spike and Kernel Characteristics in Wheat Reveals Long-Term Genetic Trends of Grain Yield Components. Theor. Appl. Genet. 2018, 131, 2071–2084. [Google Scholar] [CrossRef] [PubMed]
- Semenov, M.A.; Shewry, P.R. Modelling Predicts That Heat Stress, Not Drought, Will Increase Vulnerability of Wheat in Europe. Sci. Rep. 2011, 1, 66. [Google Scholar] [CrossRef]
- Sheehan, H.; Bentley, A. Changing Times: Opportunities for Altering Winter Wheat Phenology. Plants People Planet 2021, 3, 113–123. [Google Scholar] [CrossRef]
- Snape, J.W.; Butterworth, K.; Whitechurch, E.; Worland, A.J. Waiting for fine times: Genetics of flowering time in wheat. Euphytica 2001, 119, 185–190. [Google Scholar] [CrossRef]
- Goncharov, N.P. Response to Vernalization in Wheat: Its Quantitative or Qualitative Nature. Cereal Res. Commun. 2004, 32, 323–330. [Google Scholar] [CrossRef]
- Distelfeld, A.; Li, C.; Dubcovsky, J. Regulation of Flowering in Temperate Cereals. Curr. Opin. Plant Biol. 2009, 12, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Slafer, G.A. Differences in Phasic Development Rate amongst Wheat Cultivars Independent of Responses to Photoperiod and Vernalization. A Viewpoint of the Intrinsic Earliness Hypothesis. J. Agric. Sci. 1996, 126, 403–419. [Google Scholar] [CrossRef]
- Appendino, M.L.; Slafer, G.A. Earliness per Se and Its Dependence upon Temperature in Diploid Wheat Lines Differing in the Major Gene Eps-A m1 Alleles. J. Agric. Sci. 2003, 141, 149–154. [Google Scholar] [CrossRef]
- Alvarez, M.A.; Tranquilli, G.; Lewis, S.; Kippes, N.; Dubcovsky, J. Genetic and Physical Mapping of the Earliness per Se Locus Eps-A m 1 in Triticum monococcum Identifies EARLY FLOWERING 3 (ELF3) as a Candidate Gene. Funct. Integr. Genom. 2016, 16, 365–382. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.; Faricelli, M.E.; Appendino, M.L.; Valarik, M.; Dubcovsky, J. The Chromosome Region Including the Earliness per Se Locus Eps-Am1 Affects the Duration of Early Developmental Phases and Spikelet Number in Diploid Wheat. J. Exp. Bot. 2008, 59, 3595–3607. [Google Scholar] [CrossRef] [PubMed]
- Law, C.N.; Worland, A.J.; Giorgi, B. The Genetic Control of Ear-Emergence Time by Chromosomes 5A and 5D of Wheat. Heredity 1976, 36, 49–58. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Lijavetzky, D.; Appendino, L.; Tranquilli, G. Comparative RFLP Mapping of Triticum monococcum Genes Controlling Vernalization Requirement. Theor. Appl. Genet. 1998, 97, 968–975. [Google Scholar] [CrossRef]
- Yan, L.; Loukoianov, A.; Tranquilli, G.; Helguera, M.; Fahima, T.; Dubcovsky, J. Positional Cloning of the Wheat Vernalization Gene VRN1. Proc. Natl. Acad. Sci. USA 2003, 100, 6263–6268. [Google Scholar] [CrossRef]
- Yan, L.; Helguera, M.; Kato, K.; Fukuyama, S.; Sherman, J.; Dubcovsky, J. Allelic Variation at the VRN-1 Promoter Region in Polyploid Wheat. Theor. Appl. Genet. 2004, 109, 1677–1686. [Google Scholar] [CrossRef]
- Stelmakh, A.F. Growth Habit in Common Wheat (Triticum aestivum L. Em. Thell.). Euphytica 1987, 36, 513–519. [Google Scholar] [CrossRef]
- Fu, D.; Szűcs, P.; Yan, L.; Helguera, M.; Skinner, J.S.; Von Zitzewitz, J.; Hayes, P.M.; Dubcovsky, J. Large Deletions within the First Intron in VRN-1 Are Associated with Spring Growth Habit in Barley and Wheat. Mol. Genet. Genom. 2005, 273, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Loukoianov, A.; Yan, L.; Blechl, A.; Sanchez, A.; Dubcovsky, J. Regulation of VRN-1 Vernalization Genes in Normal and Transgenic Polyploid Wheat. Plant Physiol. 2005, 138, 2364–2373. [Google Scholar] [CrossRef]
- Stelmakh, A.F. Genetic Effects of Vrn Genes on Heading Date and Agronomic Traits in Bread Wheat. Euphytica 1992, 65, 53–60. [Google Scholar] [CrossRef]
- Stelmakh, A.F. Genetic Systems Regulating Flowering Response in Wheat. Euphytica 1998, 100, 359–369. [Google Scholar] [CrossRef]
- Pánková, K.; Košner, J. Chromosome Substitutions with Dominant Loci Vrn-1 and Their Effect on Developmental Stages of Wheat. Czech J. Genet. Plant Breed. 2004, 40, 37–44. [Google Scholar] [CrossRef]
- Goncharov, N.P. Genetic Resources of Wheat Related Species: The Vrn Genes Controlling Growth Habit (Spring vs. Winter). Euphytica 1998, 100, 371–376. [Google Scholar] [CrossRef]
- Stelmakh, A.F. Geographic Distribution of Vrn-Genes in Landraces and Improved Varieties of Spring Bread Wheat. Euphytica 1990, 45, 113–118. [Google Scholar] [CrossRef]
- Santra, D.K.; Santra, M.; Allan, R.E.; Campbell, K.G.; Kidwell, K.K. Genetic and Molecular Characterization of Vernalization Genes Vrn-A1, Vrn-B1, and Vrn-D1 in Spring Wheat Germplasm from the Pacific Northwest Region of the U.S.A. Plant Breed. 2009, 128, 576–584. [Google Scholar] [CrossRef]
- Shcherban, A.B.; Börner, A.; Salina, E.A. Effect of VRN-1 and PPD-D1 Genes on Heading Time in European Bread Wheat Cultivars. Plant Breed. 2015, 134, 49–55. [Google Scholar] [CrossRef]
- Zhang, X.K.; Xiao, Y.G.; Zhang, Y.; Xia, X.C.; Dubcovsky, J.; He, Z.H. Allelic Variation at the Vernalization Genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese Wheat Cultivars and Their Association with Growth Habit. Crop Sci. 2008, 48, 458–470. [Google Scholar] [CrossRef]
- Iwaki, K.; Haruna, S.; Niwa, T.; Kato, K. Adaptation and Ecological Differentiation in Wheat with Special Reference to Geographical Variation of Growth Habit and Vrn Genotype. Plant Breed. 2001, 120, 107–114. [Google Scholar] [CrossRef]
- Eagles, H.A.; Cane, K.; Kuchel, H.; Hollamby, G.J.; Vallance, N.; Eastwood, R.F.; Gororo, N.N.; Martin, P.J. Photoperiod and Vernalization Gene Effects in Southern Australian Wheat. Crop Pasture Sci. 2010, 61, 721. [Google Scholar] [CrossRef]
- Andeden, E.; Yediay, F.; Baloch, F.; Shaaf, S.; Kilian, B.; Nachit, M.; Özkan, H. Distribution of Vernalization and Photoperiod Genes (Vrn-A1, Vrn-B1, Vrn-D1, Vrn-B3, Ppd-D1) in Turkish Bread Wheat Cultivars and Landraces. Cereal Res. Commun. 2011, 39, 352–364. [Google Scholar] [CrossRef]
- Whitechurch, E.M.; Slafer, G.A. Contrasting Ppd Alleles in Wheat: Effects on Sensitivity to Photoperiod in Different Phases. Field Crops Res. 2002, 73, 95–105. [Google Scholar] [CrossRef]
- Law, C.N.; Sutka, J.; Worland, A.J. A Genetic Study of Day-Length Response in Wheat. Heredity 1978, 41, 185–191. [Google Scholar] [CrossRef]
- Scarth, R.; Law, C.N. The Location of the Photoperiod Gene, Ppd2 and an Additional Genetic Factor for Ear-Emergence Time on Chromosome 2B of Wheat. Heredity 1983, 51, 607–619. [Google Scholar] [CrossRef]
- Scarth, R.; Law, C.N. The Control of the Day-Length Response in Wheat by the Group 2 Chromosomes. Z. Pfl. Zücht. 1984, 92, 140–150. [Google Scholar]
- McIntosh, R.A.; Yamazaki, Y.; Devos, K.M.; Dubcovsky, J.; Rogers, W.J.; Appels, R. Catalogue of Gene Symbols for Wheat. In Proceedings of 10th International Wheat Genetics Symposium; Istituto Sperimentale per la Cerealicoltura: Rome, Italy, 2003; pp. 1–47. [Google Scholar]
- Nishida, H.; Yoshida, T.; Kawakami, K.; Fujita, M.; Long, B.; Akashi, Y.; Laurie, D.A.; Kato, K. Structural Variation in the 5′ Upstream Region of Photoperiod-Insensitive Alleles Ppd-A1a and Ppd-B1a Identified in Hexaploid Wheat (Triticum aestivum L.), and Their Effect on Heading Time. Mol. Breed. 2013, 31, 27–37. [Google Scholar] [CrossRef]
- Beales, J.; Turner, A.; Griffiths, S.; Snape, J.W.; Laurie, D.A. A Pseudo-Response Regulator Is Misexpressed in the Photoperiod Insensitive Ppd-D1a Mutant of Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 115, 721–733. [Google Scholar] [CrossRef]
- Díaz, A.; Zikhali, M.; Turner, A.S.; Isaac, P.; Laurie, D.A. Copy Number Variation Affecting the Photoperiod-B1 and Vernalization-A1 Genes Is Associated with Altered Flowering Time in Wheat (Triticum aestivum). PLoS ONE 2012, 7, e33234. [Google Scholar] [CrossRef]
- Vanzetti, L.S.; Yerkovich, N.; Chialvo, E.; Lombardo, L.; Vaschetto, L.; Helguera, M. Genetic Structure of Argentinean Hexaploid Wheat Germplasm. Genet. Mol. Biol. 2013, 36, 391–399. [Google Scholar] [CrossRef]
- Ferrara, G.O.; Mosaad, M.G.; Mahalakshmi, V.; Rajaram, S. Photoperiod and vernalisation response of Mediterranean wheats, and implications for adaptation. Euphytica 1998, 100, 377–384. [Google Scholar] [CrossRef]
- Seki, M.; Chono, M.; Nishimura, T.; Sato, M.; Yoshimura, Y.; Matsunaka, H.; Fujita, M.; Oda, S.; Kubo, K.; Kiribuchi-Otobe, C.; et al. Distribution of Photoperiod-Insensitive Allele Ppd-A1a and Its Effect on Heading Time in Japanese Wheat Cultivars. Breed. Sci. 2013, 63, 309–316. [Google Scholar] [CrossRef]
- Yang, F.P.; Zhang, X.K.; Xia, X.C.; Laurie, D.A.; Yang, W.X.; He, Z.H. Distribution of the Photoperiod Insensitive Ppd-D1a Allele in Chinese Wheat Cultivars. Euphytica 2009, 165, 445–452. [Google Scholar] [CrossRef]
- Grogan, S.M.; Brown-Guedira, G.; Haley, S.D.; McMaster, G.S.; Reid, S.D.; Smith, J.; Byrne, P.F. Allelic Variation in Developmental Genes and Effects on Winter Wheat Heading Date in the U.S. Great Plains. PLoS ONE 2016, 11, e0152852. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Koval, S.F.; Koval, V.S. Wheat Near-Isogenic Lines; Sankeisha: Nagoya, Japan, 2003; ISBN 978-4-88361-131-7. [Google Scholar]
- Pugsley, A. A Genetic Analysis of the Spring-Winter Habit of Growth in Wheat. Aust. J. Agric. Res. 1971, 22, 21. [Google Scholar] [CrossRef]
- Pugsley, A.T. Additional Genes Inhibiting Winter Habit in Wheat. Euphytica 1972, 21, 547–552. [Google Scholar] [CrossRef]
- Stelmakh, A.F.; Avsenin, V.I. Development of Vrn1-3 near-Isogenic Lines. Nauch. Tech. Bull. VSGI 1983, 48, 24–28. (In Russian) [Google Scholar]
- Zeven, A.C.; Waninge, J.; Colon, L.T. The Extent of Similarity between Near-Isogenic Lines of the Australian Spring Wheat Variety Triple Dirk with Their Recurrent Parent. Euphytica 1986, 35, 381–393. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal Component Analysis. WIREs Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Gower, J.C. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika 1966, 53, 325. [Google Scholar] [CrossRef]
- Fiore, M.C.; Mercati, F.; Spina, A.; Blangiforti, S.; Venora, G.; Dell’Acqua, M.; Lupini, A.; Preiti, G.; Monti, M.; Pè, M.E.; et al. High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily. Plants 2019, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- Chernook, A.G.; Kroupin, P.Y.; Bespalova, L.A.; Panchenko, V.V.; Kovtunenko, V.Y.; Bazhenov, M.S.; Nazarova, L.A.; Karlov, G.I.; Kroupina, A.Y.; Divashuk, M.G. Phenotypic Effects of the Dwarfing Gene Rht-17 in Spring Durum Wheat under Two Climatic Conditions. Vavilov J. Genet. Breed. 2019, 23, 916–925. [Google Scholar] [CrossRef]
- Hill, C.B.; Li, C. Genetic Architecture of Flowering Phenology in Cereals and Opportunities for Crop Improvement. Front. Plant Sci. 2016, 7, 1906. [Google Scholar] [CrossRef] [PubMed]
- Steinfort, U.; Fukai, S.; Trevaskis, B.; Glassop, D.; Chan, A.; Dreccer, M.F. Vernalisation and Photoperiod Sensitivity in Wheat: The Response of Floret Fertility and Grain Number Is Affected by Vernalisation Status. Field Crops Res. 2017, 203, 243–255. [Google Scholar] [CrossRef]
- Zeven, A.C.; Zeven-Hissink, N.C. Genealogies of 14000 Wheat Varieties. In Genealogies of 14000 Wheat Varieties; The Netherlands Cereals Centre-NGC: Wageningen, The Netherlands; The International Maize and Wheat Improvement Center: Texcoco, Mexico, 1976. [Google Scholar]
- Sehgal, D.; Dixon, L.; Pequeno, D.; Hyles, J.; Lacey, I.; Crossa, J.; Bentley, A.; Dreisigacker, S. Genomic Insights on Global Journeys of Adaptive Wheat Genes That Brought Us to Modern Wheat. In The Wheat Genome; Appels, R., Eversole, K., Feuillet, C., Gallagher, D., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2024; pp. 213–239. ISBN 978-3-031-38292-5. [Google Scholar]
- González, F.G.; Slafer, G.A.; Miralles, D.J. Pre-Anthesis Development and Number of Fertile Florets in Wheat as Affected by Photoperiod Sensitivity Genes Ppd-D1and Ppd-B1. Euphytica 2005, 146, 253–269. [Google Scholar] [CrossRef]
- Acreche, M.M.; Slafer, G.A. Grain Weight Response to Increases in Number of Grains in Wheat in a Mediterranean Area. Field Crops Res. 2006, 98, 52–59. [Google Scholar] [CrossRef]
- Kato, K.; Miura, H.; Sawada, S. Mapping QTLs Controlling Grain Yield and Its Components on Chromosome 5A of Wheat. Theor. Appl. Genet. 2000, 101, 1114–1121. [Google Scholar] [CrossRef]
- Fait, V.I.; Balashova, I.A. Distribution of Photoperiod-Insensitive Alleles Ppd-D1a, Ppd-B1a, and Ppd-B1c in Winter Common Wheat Cultivars (Triticum aestivum L.) of Various Origin. Cytol. Genet. 2022, 56, 109–117. [Google Scholar] [CrossRef]
- Koval, S.F. Investigation of Spring Wheat Cultivars Model in Isogenic Lines and Analogous. In Problems and Prospects/Problems of Agricultural Plants Breeding; ICG SB AS USSR: Novosibirsk, Russia, 1983; pp. 56–68. (In Russian) [Google Scholar]
- Rebetzke, G.J.; Bonnett, D.G.; Reynolds, M.P. Awns Reduce Grain Number to Increase Grain Size and Harvestable Yield in Irrigated and Rainfed Spring Wheat. J. Exp. Bot. 2016, 67, 2573–2586. [Google Scholar] [CrossRef]
- Sanchez-Bragado, R.; Molero, G.; Araus, J.L.; Slafer, G.A. Awned versus Awnless Wheat Spikes: Does It Matter? Trends Plant Sci. 2023, 28, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Slafer, G.A.; Savin, R.; Sadras, V.O. Coarse and Fine Regulation of Wheat Yield Components in Response to Genotype and Environment. Field Crops Res. 2014, 157, 71–83. [Google Scholar] [CrossRef]
- Philipp, N.; Weichert, H.; Bohra, U.; Weschke, W.; Schulthess, A.W.; Weber, H. Grain Number and Grain Yield Distribution along the Spike Remain Stable despite Breeding for High Yield in Winter Wheat. PLoS ONE 2018, 13, e0205452. [Google Scholar] [CrossRef] [PubMed]
- Kamran, A.; Randhawa, H.S.; Yang, R.; Spaner, D. The Effect of VRN1 Genes on Important Agronomic Traits in High-yielding Canadian Soft White Spring Wheat. Plant Breed. 2014, 133, 321–326. [Google Scholar] [CrossRef]
- Wilhelm, E.P.; Boulton, M.I.; Al-Kaff, N.; Balfourier, F.; Bordes, J.; Greenland, A.J.; Powell, W.; Mackay, I.J. Rht-1 and Ppd-D1 Associations with Height, GA Sensitivity, and Days to Heading in a Worldwide Bread Wheat Collection. Theor. Appl. Genet. 2013, 126, 2233–2243. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Du, Y.; Lu, Q.; Chen, H.; Meng, R.; Cui, C.; Lu, S.; Yang, Y.; Chai, Y.; Li, J.; et al. The Photoperiod-Insensitive Allele Ppd-D1a Promotes Earlier Flowering in Rht12 Dwarf Plants of Bread Wheat. Front. Plant Sci. 2018, 9, 1312. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Gianmarco, T.I.; Slafer, G.A.; González, F.G. Photoperiod-Sensitivity Genes Shape Floret Development in Wheat. J. Exp. Bot. 2019, 70, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Miralles, D.J.; Richards, R.A.; Slafer, G.A. Duration of the Stem Elongation Period Influences the Number of Fertile Florets in Wheat and Barley. Funct. Plant Biol. 2000, 27, 931. [Google Scholar] [CrossRef]
- Ning, S.; Li, S.; Xu, K.; Liu, D.; Ma, L.; Ma, C.; Hao, M.; Zhang, L.; Chen, W.; Zhang, B.; et al. Development and Characterization of Near-Isogenic Lines Derived from Synthetic Wheat Revealing the 2 Kb Insertion in the PPD-D1 Gene Responsible for Heading Delay and Grain Number Improvement. Int. J. Mol. Sci. 2023, 24, 10834. [Google Scholar] [CrossRef]
- Arjona, J.M.; Royo, C.; Dreisigacker, S.; Ammar, K.; Villegas, D. Effect of Ppd-A1 and Ppd-B1 Allelic Variants on Grain Number and Thousand Kernel Weight of Durum Wheat and Their Impact on Final Grain Yield. Front. Plant Sci. 2018, 9, 888. [Google Scholar] [CrossRef]
- Iqbal, M.; Shahzad, A.; Ahmed, I. Allelic Variation at the Vrn-A1, Vrn-B1, Vrn-D1, Vrn-B3 and Ppd-D1a Loci of Pakistani Spring Wheat Cultivars. Electron. J. Biotechnol. 2001, 14, 1–2. [Google Scholar] [CrossRef]
- Iwaki, K.; Nakagawa, K.; Kuno, H.; Kato, K. Ecogeographical differentiation in east Asian wheat, revealed from the geographical variation of growth habit and Vrn genotype. Euphytica 2000, 111, 137–143. [Google Scholar] [CrossRef]
- Olmstead, A.L.; Rhode, P.W. Adapting North American Wheat Production to Climatic Challenges, 1839–2009. Proc. Natl. Acad. Sci. USA 2011, 108, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Efremova, T.T.; Chumanova, E.V.; Trubacheeva, N.V.; Arbuzova, V.S.; Belan, I.A.; Pershina, L.A. Prevalence of VRN1 Locus Alleles among Spring Common Wheat Cultivars Cultivated in Western Siberia. Russ. J. Genet. 2016, 52, 146–153. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Sylvester-Bradley, R.; Worland, A.J.; Snape, J.W. Effects of a Photoperiod-Response Gene Ppd-D1 on Yield Potential and Drought Resistance in UK Winter Wheat. Euphytica 2004, 135, 63–73. [Google Scholar] [CrossRef]
- Zhou, Y.; Conway, B.; Miller, D.; Marshall, D.; Cooper, A.; Murphy, P.; Chao, S.; Brown-Guedira, G.; Costa, J. Quantitative Trait Loci Mapping for Spike Characteristics in Hexaploid Wheat. Plant Genome 2017, 10, plantgenome2016.10.0101. [Google Scholar] [CrossRef] [PubMed]
- Miralles, D.J.; Katz, S.D.; Colloca, A.; Slafer, G.A. Floret Development in near Isogenic Wheat Lines Differing in Plant Height. Field Crops Res. 1998, 59, 21–30. [Google Scholar] [CrossRef]
- Kamran, A.; Randhawa, H.S.; Pozniak, C.; Spaner, D. Phenotypic Effects of the Flowering Gene Complex in Canadian Spring Wheat Germplasm. Crop Sci. 2013, 53, 84–94. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, B.; He, Y. Understanding the Effects of Growing Seasons, Genotypes, and Their Interactions on the Anthesis Date of Wheat Sown in North China. Biology 2021, 10, 955. [Google Scholar] [CrossRef]
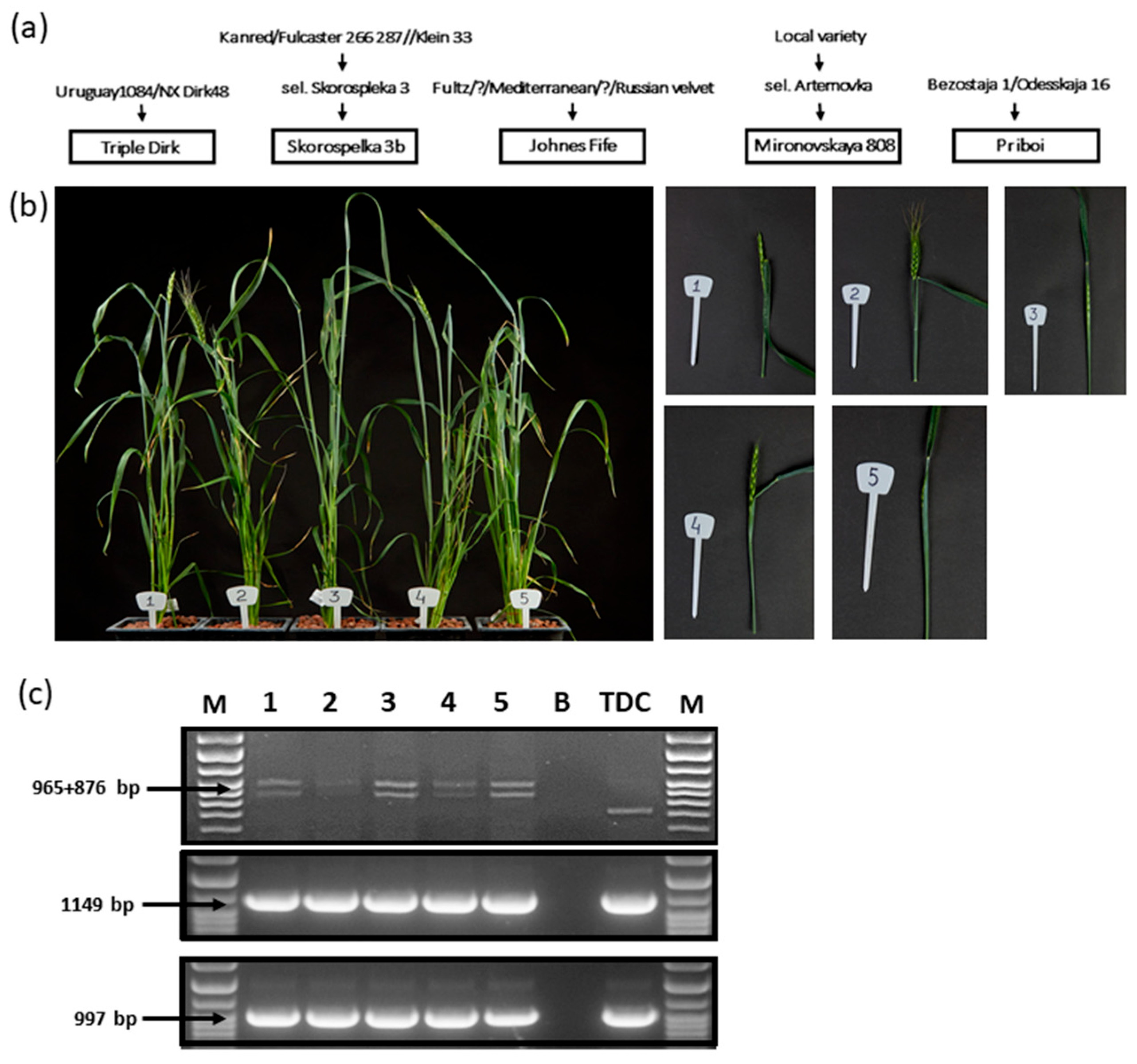
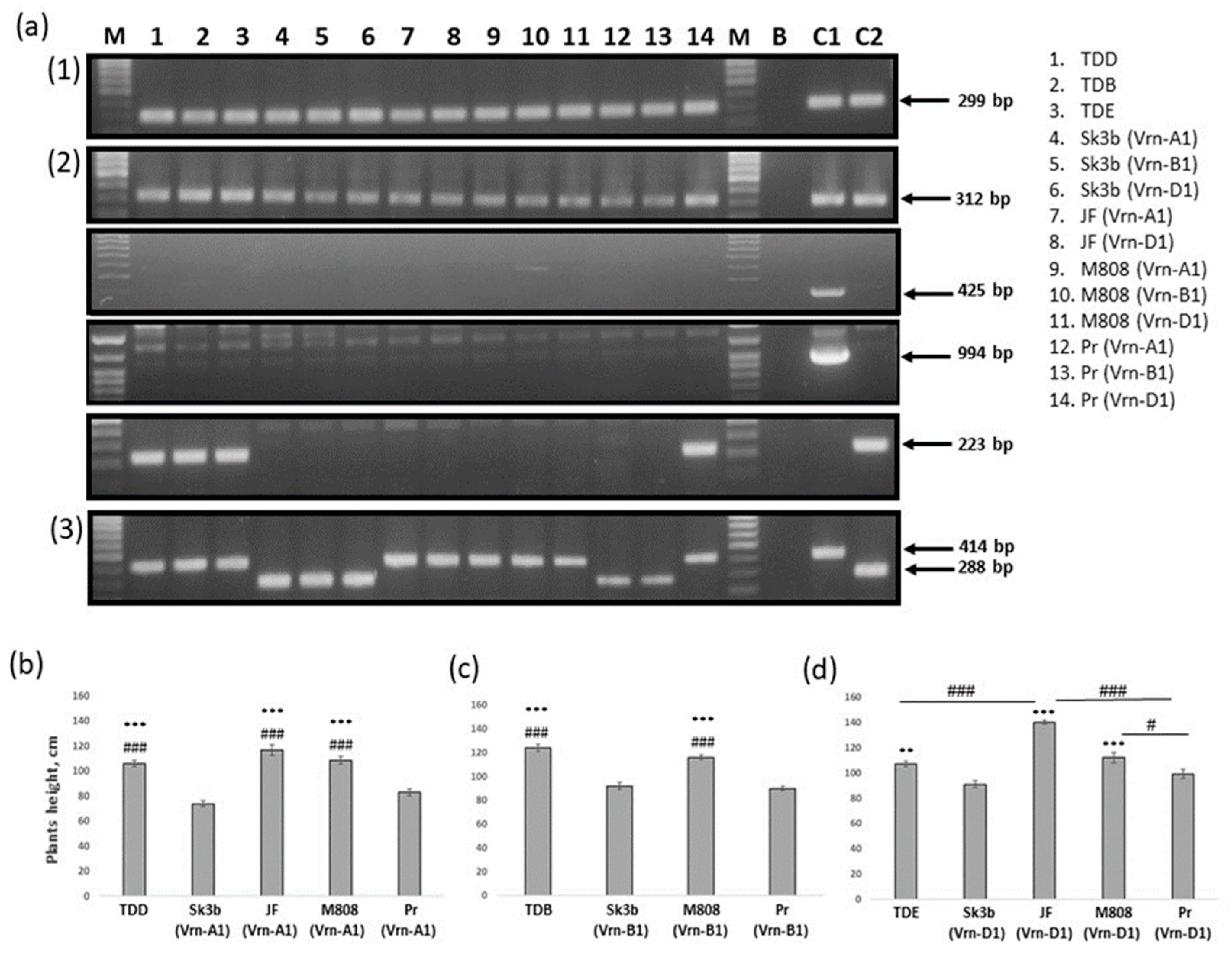

| NILs Original Name | Abbreviations Used in This Study | Presumed Vrn-1 Allelic Composition | Presumed Ppd-1 Allelic Composition | Breeding History (Donor//Recurrent cv) |
|---|---|---|---|---|
| Skorospelka 3b (Vrn1) | Sk3b (Vrn-A1) | Vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a | Triple Dirk D//9 * Skorospelka 3b |
| Skorospelka 3b (Vrn2) | Sk3b (Vrn-B1) | vrn-A1 Vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a | Triple Dirk B//9 * Skorospelka 3b |
| Skorospelka 3b (Vrn3) | Sk3b (Vrn-D1) | vrn-A1 vrn-B1 Vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a | Triple Dirk E//9 * Skorospelka 3b |
| Johnes Fife (Vrn1) | JF (Vrn-A1) | Vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1b | Triple Dirk D//9 * Johnes Fife |
| Johnes Fife (Vrn3) | JF (Vrn-D1) | vrn-A1 vrn-B1 Vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1b | Triple Dirk E//9 * Johnes Fife |
| Mironovskaya 808 (Vrn1) | M808 (Vrn-A1) | Vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1b | Triple Dirk D//9 * Mironovskaya 808 |
| Mironovskaya 808 (Vrn2) | M808 (Vrn-B1) | vrn-A1 Vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1b | Triple Dirk B//9 * Mironovskaya 808 |
| Mironovskaya 808 (Vrn3) | M808 (Vrn-D1) | vrn-A1 vrn-B1 Vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1b | Triple Dirk E//9 * Mironovskaya 808 |
| Priboi (Vrn1) | Pr (Vrn-A1) | Vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a | Triple Dirk D//9 * Priboi |
| Priboi (Vrn2) | Pr (Vrn-B1) | vrn-A1 Vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a | Triple Dirk B//9 * Priboi |
| Priboi (Vrn3) | Pr (Vrn-D1) | vrn-A1 vrn-B1 Vrn-D1 | Ppd-A1b Ppd-B1a Ppd-D1b | Triple Dirk E//9 * Priboi |
| Triple Dirk D | TDD | Vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1a Ppd-D1b | Winter Minflor/3–4 * TD |
| Triple Dirk B | TDB | vrn-A1 Vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1a Ppd-D1b | Winter Minflor/3–4 * TD |
| Triple Dirk E | TDE | vrn-A1 vrn-B1 Vrn-D1 | Ppd-A1b Ppd-B1a Ppd-D1b | Loro/3–4 * TD |
| NILs | HT, Days | PH, cm | SL, cm | SNS | Tillering | GNS | GWS, g | SF |
|---|---|---|---|---|---|---|---|---|
| TDD | 35.9 ± 0.4 | 106 ± 2.4 | 8.1 ± 0.2 | 14.3 ± 0.3 | 6.8 ± 0.5 | 24.8± 1 | 1.16 ± 0.05 | 1.8 ± 0.03 |
| TDB | 61.9 ± 1.2 | 124 ± 2.8 | 9± 0.3 | 20 ± 0.8 | 10.2 ± 1.1 | 24.7 ± 4.2 | 1.1 ± 0.18 | 1.5 ± 0.2 |
| TDE | 45.6 ± 0.9 | 107.6 ± 2 | 8.6 ± 0.1 | 15.5± 0.4 | 6 ±0.4 | 23.5 ± 1.4 | 1.23 ± 0.1 | 1.5 ± 0.1 |
| ANOVA | p < 0.001 | p < 0.001 | p < 0.05 | p < 0.001 | p < 0.001 | ns | ns | p < 0.05 |
| Sk3b (Vrn-A1) | 35.8 ± 0.4 | 73.8 ± 2.4 | 6.2 ± 0.2 | 13 ± 0.3 | 4.3 ± 0.3 | 24.1 ± 1.3 | 0.79 ± 0.1 | 1.8 ± 0.1 |
| Sk3b (Vrn-B1) | 57.7 ± 0.9 | 91.8 ± 3 | 6.7 ± 0.2 | 14.1 ± 0.6 | 5.9 ± 0.6 | 27.4 ± 2.3 | 1.1 ± 0.1 | 1.9 ± 0.1 |
| Sk3b (Vrn-D1) | 47.6 ± 1.7 | 91.3 ± 2.7 | 6.8 ± 0.2 | 14.3 ± 0.4 | 6.3 ± 0.7 | 29.9 ± 1.6 | 1.19 ± 0.1 | 2.1 ± 0.1 |
| ANOVA | p < 0.001 | p < 0.001 | p < 0.05 | ns | p < 0.05 | p < 0.05 | p < 0.001 | ns |
| JF (Vrn-A1) | 38.7 ± 1.2 | 117 ± 4.3 | 8.4 ± 0.3 | 14.8 ± 0.5 | 5.9 ± 0.9 | 35.6 ± 1.9 | 1.27 ± 0.1 | 2.3 ± 0.1 |
| JF (Vrn-D1) | 68.5 ± 0.8 | 140 ± 1.5 | 10.5 ± 0.2 | 23.2 ± 0.5 | 9.3 ± 1 | 43.4 ± 2.3 | 1.35 ± 0.1 | 2 ± 0.1 |
| ANOVA | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.05 | p < 0.05 | ns | p < 0.01 |
| M808 (Vrn-A1) | 38 ± 0.5 | 108.6 ± 3 | 9.4 ± 0.2 | 14.5 ± 0.2 | 7.5 ± 0.6 | 24.2 ± 1.5 | 1.13 ± 0.1 | 1.7 ± 0.1 |
| M808 (Vrn-B1) | 72.7 ± 1.1 | 116.4 ± 2.3 | 11 ± 0.3 | 19.5 ± 0.5 | 10.5 ± 1.2 | 34.3 ± 2.4 | 1.58 ± 0.2 | 2 ± 0.1 |
| M808 (Vrn-D1) | 61.3 ± 1.5 | 112.4 ± 3.9 | 10.3 ±0.4 | 18.2 ± 0.7 | 8.7 ± 1.5 | 31.9 ± 3.6 | 1.39 ± 0.2 | 1.8 ± 0.5 |
| ANOVA | p < 0.001 | ns | p < 0.01 | p < 0.001 | ns | p < 0.01 | ns | ns |
| Pr (Vrn-A1) | 38.5 ± 1.1 | 83.4± 2.5 | 7.5 ± 0.2 | 15.6 ± 0.2 | 7.8 ± 0.6 | 21.9 ± 1 | 0.78 ± 0.1 | 1.5 ± 0.1 |
| Pr (Vrn-B1) | 68.5 ± 1.2 | 90.2 ± 1.9 | 8.5 ± 0.3 | 20.2 ± 0.8 | 9.2 ± 0.5 | 26.7 ± 2.1 | 1.25 ± 0.1 | 1.5 ± 0.1 |
| Pr (Vrn-D1) | 65.7 ± 1 | 99.7 ± 3.6 | 9.3 ± 0.3 | 17.8 ± 0.6 | 9.5 ± 0.9 | 23.3 ± 2.9 | 0.94 ± 0.1 | 1.6 ± 0.1 |
| ANOVA | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | ns | ns | p < 0.05 | ns |
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 | PC10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Vrn-A1a | −0.112 | 0.166 | −0.280 | 0.205 | −0.253 | 0.052 | −0.151 | 0.083 | 0.211 | −0.032 |
| Vrn-B1 | 0.022 | −0.035 | 0.242 | 0.086 | −0.258 | −0.377 | 0.271 | −0.290 | 0.089 | −0.018 |
| Vrn-D1 | 0.090 | −0.131 | 0.038 | −0.291 | 0.511 | 0.325 | −0.120 | 0.207 | −0.300 | 0.050 |
| Ppd-B1a | 0.004 | −0.183 | −0.140 | 0.080 | 0.261 | 0.310 | 0.714 | −0.171 | 0.357 | 0.041 |
| Ppd-D1a | −0.203 | 0.258 | 0.364 | 0.043 | −0.103 | 0.054 | −0.158 | −0.162 | −0.093 | −0.093 |
| Awnless | 0.264 | −0.236 | −0.554 | −0.068 | −0.149 | −0.330 | −0.062 | 0.002 | −0.129 | 0.472 |
| HT | 0.220 | −0.379 | 0.479 | −0.287 | 0.091 | −0.416 | 0.051 | 0.061 | 0.016 | 0.050 |
| PH | 0.387 | −0.108 | −0.226 | −0.040 | 0.147 | 0.009 | −0.250 | −0.648 | −0.027 | −0.523 |
| SL | 0.410 | −0.121 | −0.040 | 0.110 | −0.239 | 0.047 | 0.055 | 0.584 | 0.075 | −0.519 |
| SNS | 0.372 | −0.149 | 0.282 | −0.029 | −0.283 | 0.395 | −0.200 | −0.053 | 0.448 | 0.267 |
| Tillering | 0.253 | −0.038 | 0.177 | 0.840 | 0.352 | −0.080 | −0.116 | 0.018 | −0.121 | 0.193 |
| GNS | 0.365 | 0.400 | 0.062 | −0.169 | −0.055 | 0.215 | −0.091 | −0.133 | 0.003 | 0.327 |
| GWS | 0.353 | 0.354 | 0.053 | −0.008 | −0.218 | 0.072 | 0.469 | −0.019 | −0.535 | −0.015 |
| SF | 0.204 | 0.565 | −0.058 | −0.148 | 0.408 | −0.391 | 0.020 | 0.159 | 0.443 | −0.044 |
| SD | 2.14 | 1.33 | 1.23 | 0.90 | 0.74 | 0.67 | 0.65 | 0.56 | 0.495 | 0.39 |
| Variability, % | 41.58 | 16.06 | 13.84 | 7.34 | 5.01 | 4.05 | 3.88 | 2.91 | 2.23 | 1.415 |
| Cumulative, % | 41.58 | 57.65 | 71.49 | 78.83 | 83.84 | 87.89 | 91.77 | 94.68 | 96.91 | 98.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plotnikov, K.O.; Klimenko, A.I.; Ovchinnikova, E.S.; Lashin, S.A.; Goncharov, N.P. Analysis of the Effects of the Vrn-1 and Ppd-1 Alleles on Adaptive and Agronomic Traits in Common Wheat (Triticum aestivum L.). Plants 2024, 13, 1453. https://doi.org/10.3390/plants13111453
Plotnikov KO, Klimenko AI, Ovchinnikova ES, Lashin SA, Goncharov NP. Analysis of the Effects of the Vrn-1 and Ppd-1 Alleles on Adaptive and Agronomic Traits in Common Wheat (Triticum aestivum L.). Plants. 2024; 13(11):1453. https://doi.org/10.3390/plants13111453
Chicago/Turabian StylePlotnikov, Kirill O., Alexandra I. Klimenko, Ekaterina S. Ovchinnikova, Sergey A. Lashin, and Nikolay P. Goncharov. 2024. "Analysis of the Effects of the Vrn-1 and Ppd-1 Alleles on Adaptive and Agronomic Traits in Common Wheat (Triticum aestivum L.)" Plants 13, no. 11: 1453. https://doi.org/10.3390/plants13111453
APA StylePlotnikov, K. O., Klimenko, A. I., Ovchinnikova, E. S., Lashin, S. A., & Goncharov, N. P. (2024). Analysis of the Effects of the Vrn-1 and Ppd-1 Alleles on Adaptive and Agronomic Traits in Common Wheat (Triticum aestivum L.). Plants, 13(11), 1453. https://doi.org/10.3390/plants13111453








