Phospholipid Signaling in Crop Plants: A Field to Explore
Abstract
1. Introduction
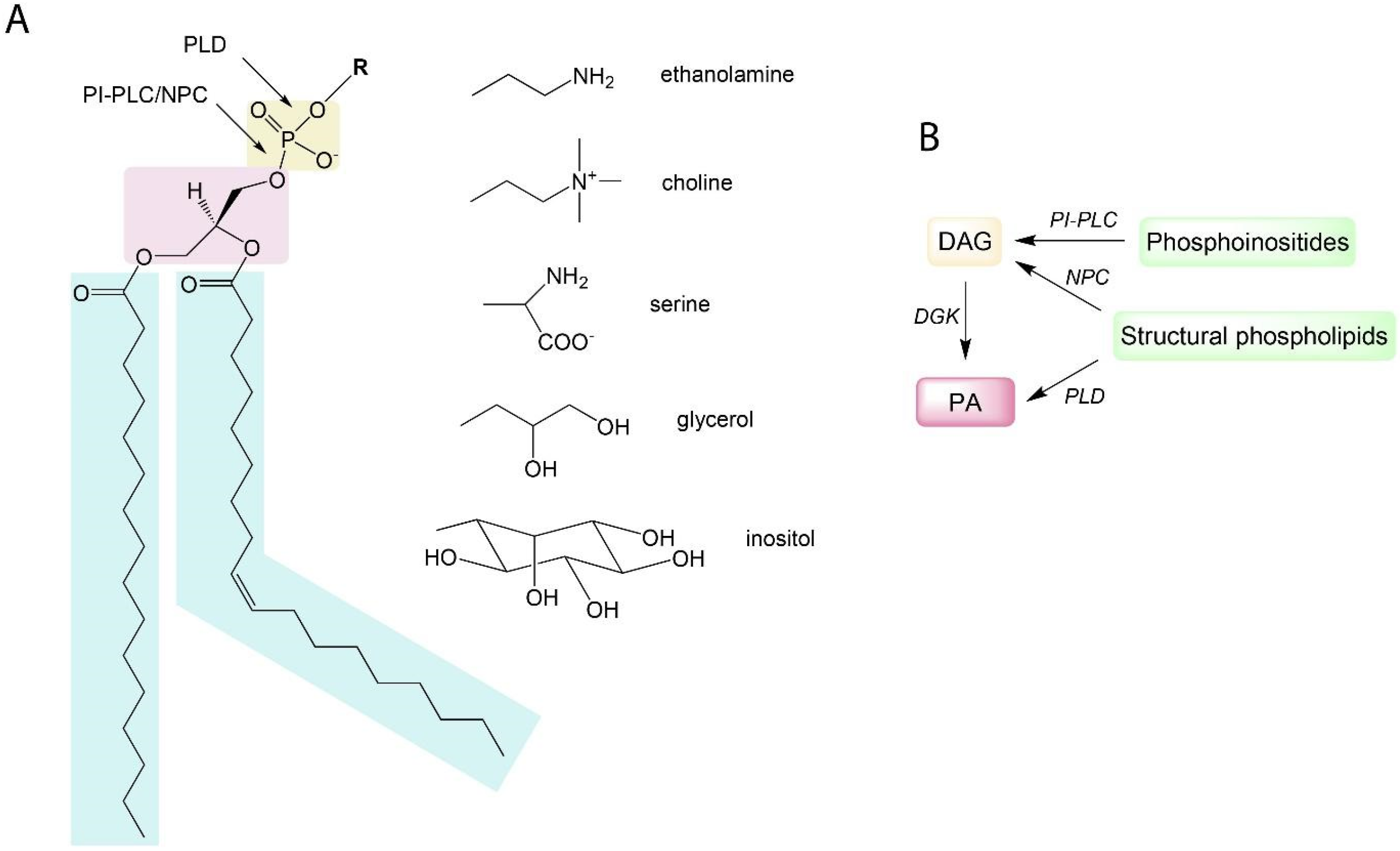
2. Results
2.1. Enzymes of the Lipid Signaling Pathways in Crop Plants
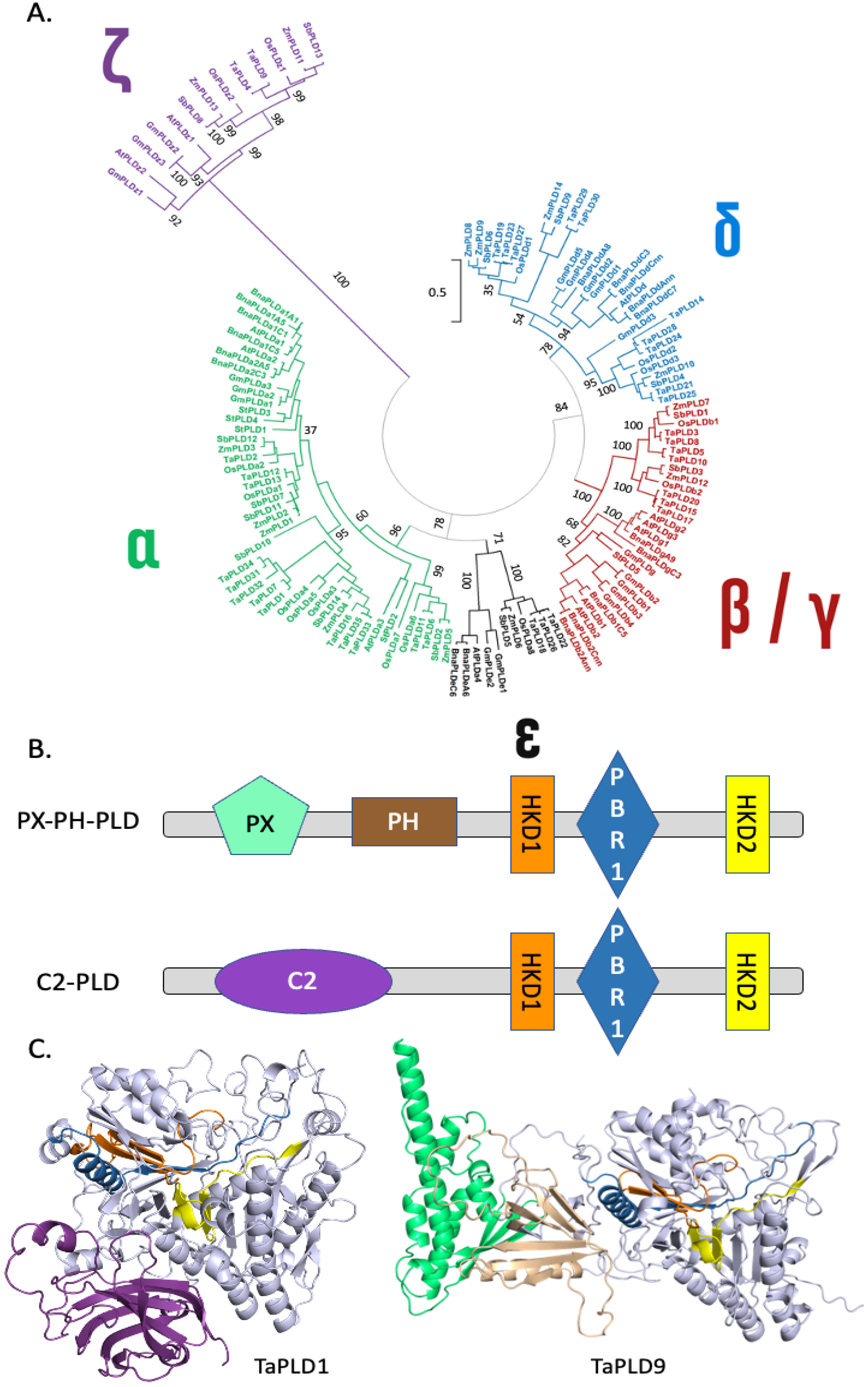
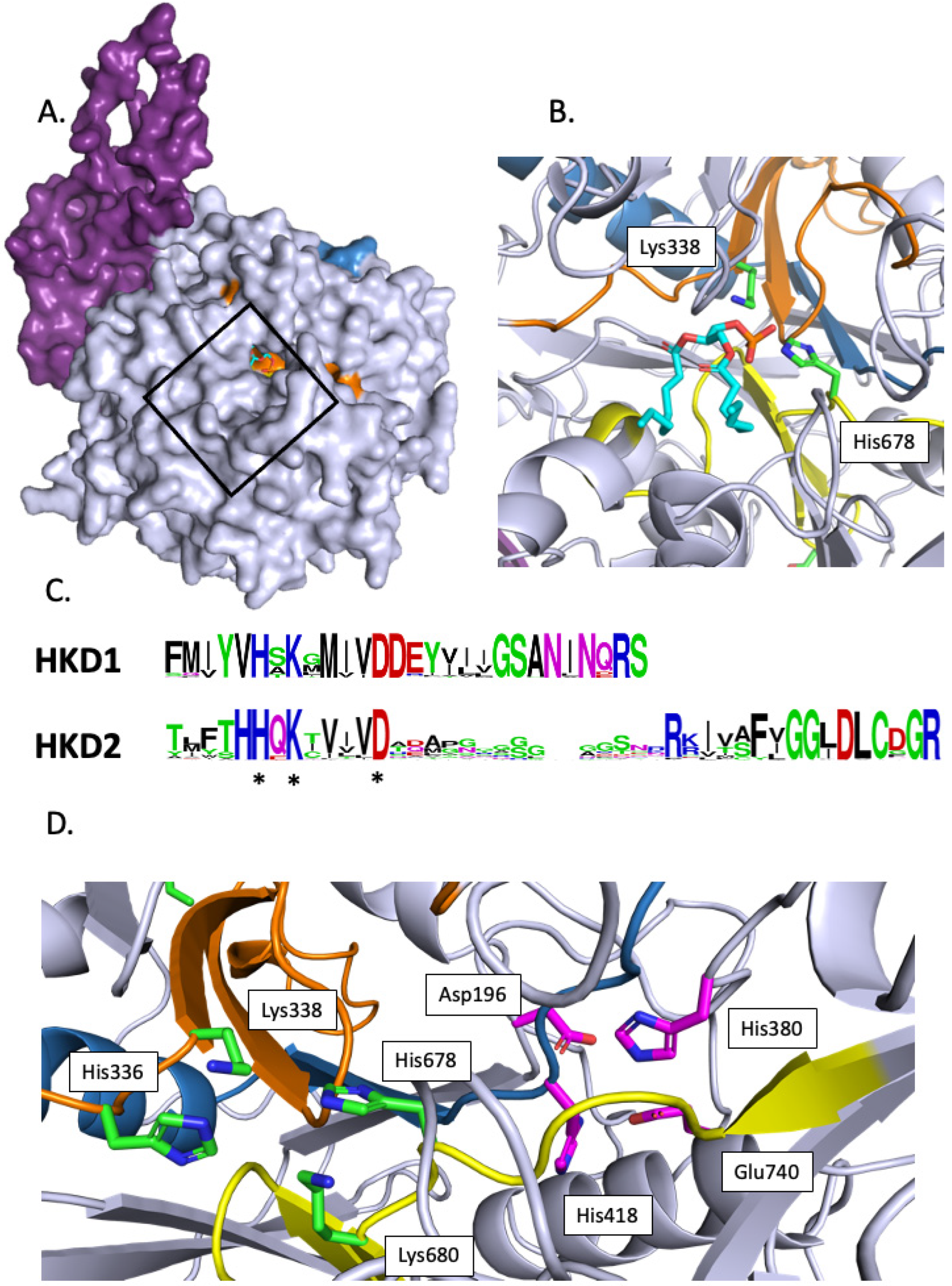
2.1.1. Phospholipases D
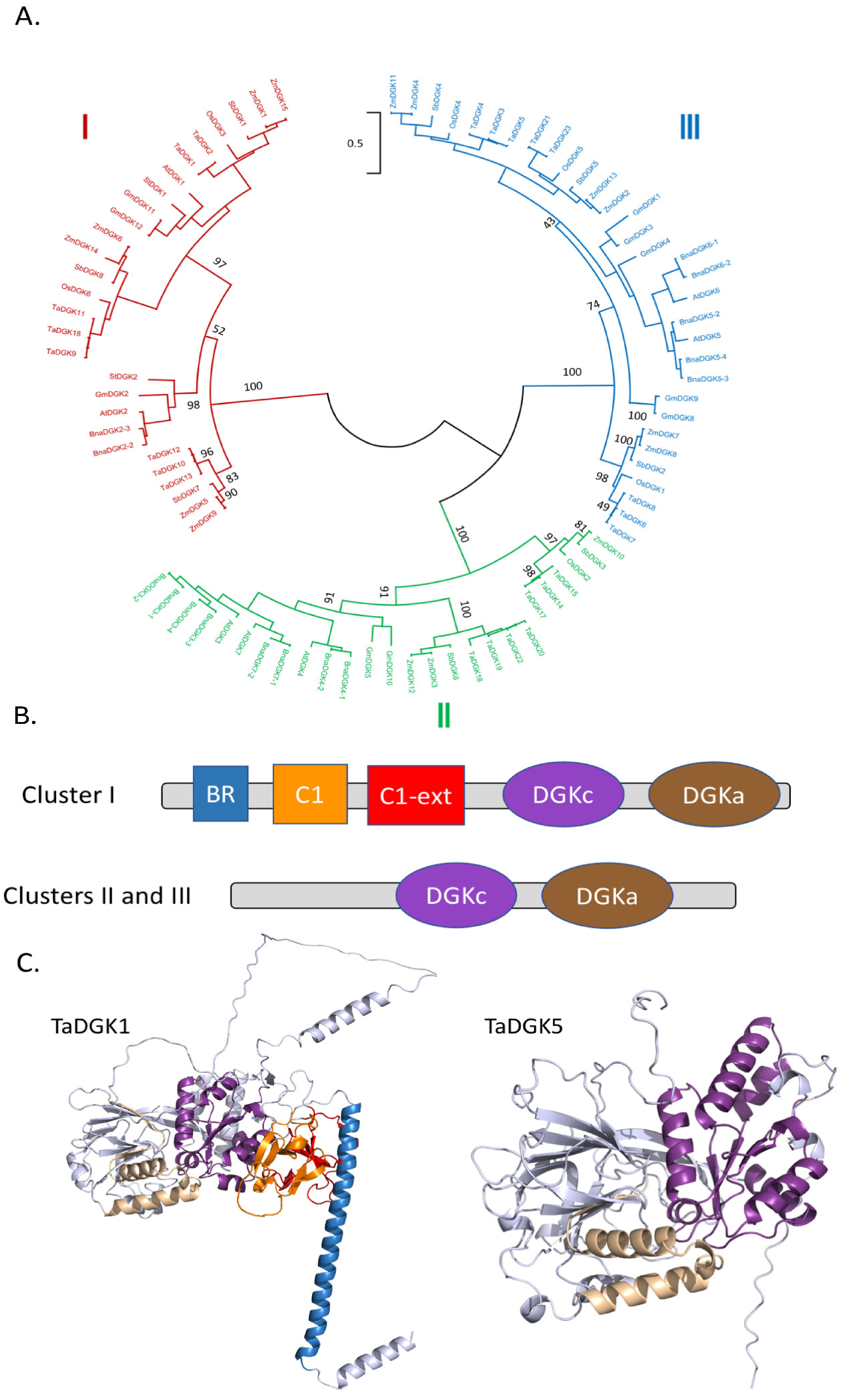
2.1.2. Diacylglycerol Kinases
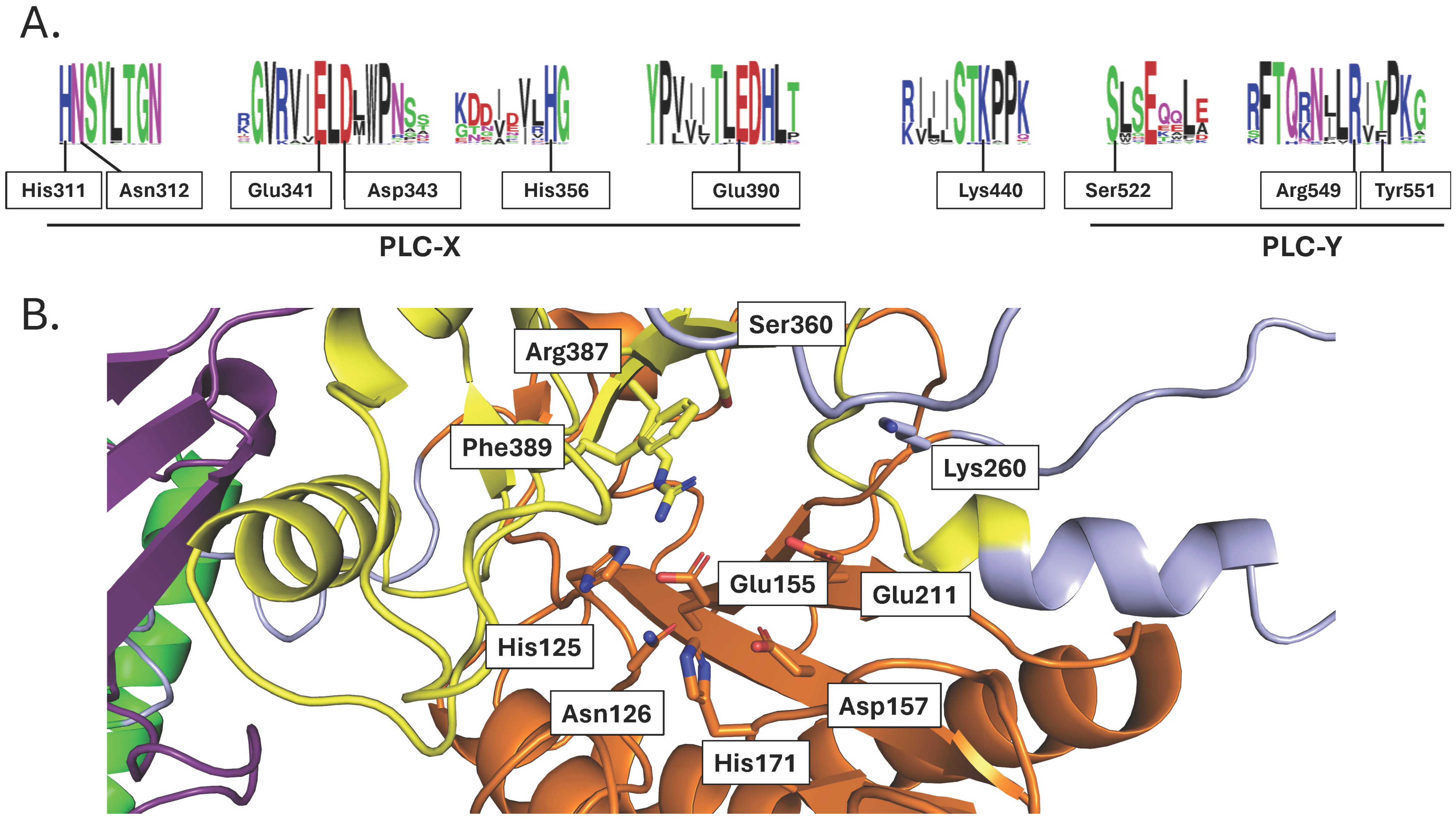
2.1.3. PI-PLCs

2.2. PA Binding Proteins—A Focus on NADPH Oxidase

3. Discussion
3.1. Structural Features of PLDs
3.2. DGKs
3.3. PI-PLCs
3.4. PA Binding Proteins
3.5. Involvement in Crop Responses to Environmental Stresses Responses
4. Conclusions
5. Materials and Methods
5.1. Sequence Alignment, Phylogenetic Analysis, and Consensus
5.2. Prediction of Structures and Visualization of Structure
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furt, F.; Simon-Plas, F.; Mongrand, S. Lipids of the Plant Plasma Membrane. In The Plant Plasma Membrane; Murphy, A.S., Schulz, B., Peer, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3–30. [Google Scholar]
- Uemura, M.; Joseph, R.A.; Steponkus, P.L. Cold Acclimation of Arabidopsis thaliana (Effect on Plasma Membrane Lipid Composition and Freeze-Induced Lesions). Plant Physiol. 1995, 109, 15–30. [Google Scholar] [CrossRef]
- Cacas, J.L.; Buré, C.; Grosjean, K.; Gerbeau-Pissot, P.; Lherminier, J.; Rombouts, Y.; Maes, E.; Bossard, C.; Gronnier, J.; Furt, F.; et al. Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant Physiol. 2016, 170, 367–384. [Google Scholar] [CrossRef]
- Li, H.-m.; Yu, C.-W. Chloroplast Galactolipids: The Link Between Photosynthesis, Chloroplast Shape, Jasmonates, Phosphate Starvation and Freezing Tolerance. Plant Cell Physiol. 2018, 59, 1128–1134. [Google Scholar] [CrossRef]
- Hac-Wydro, K.; Wydro, P. The influence of fatty acids on model cholesterol/phospholipid membranes. Chem. Phys. Lipids 2007, 150, 66–81. [Google Scholar] [CrossRef]
- Ruelland, E.; Vaultier, M.-N.; Zachowski, A.; Hurry, V. Chapter 2 Cold Signalling and Cold Acclimation in Plants. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2009; Volume 49, pp. 35–150. [Google Scholar]
- Urao, T.; Yakubov, B.; Satoh, R.; Yamaguchi-Shinozaki, K.; Seki, M.; Hirayama, T.; Shinozaki, K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 1999, 11, 1743–1754. [Google Scholar] [CrossRef]
- Tran, L.S.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef]
- Takeuchi, J.; Fukui, K.; Seto, Y.; Takaoka, Y.; Okamoto, M. Ligand–receptor interactions in plant hormone signaling. Plant J. 2021, 105, 290–306. [Google Scholar] [CrossRef]
- Gronnier, J.; Franck, C.M.; Stegmann, M.; DeFalco, T.A.; Abarca, A.; von Arx, M.; Dünser, K.; Lin, W.; Yang, Z.; Kleine-Vehn, J.; et al. Regulation of immune receptor kinase plasma membrane nanoscale organization by a plant peptide hormone and its receptors. eLife 2022, 11, e74162. [Google Scholar] [CrossRef]
- Tóth, D.J.; Tóth, J.T.; Damouni, A.; Hunyady, L.; Várnai, P. Effect of hormone-induced plasma membrane phosphatidylinositol 4,5-bisphosphate depletion on receptor endocytosis suggests the importance of local regulation in phosphoinositide signaling. Sci. Rep. 2024, 14, 291. [Google Scholar] [CrossRef]
- Cook, R.; Lupette, J.; Benning, C. The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells 2021, 10, 706. [Google Scholar] [CrossRef]
- Heilmann, M.; Heilmann, I. Regulators regulated: Different layers of control for plasma membrane phosphoinositides in plants. Curr. Opin. Plant Biol. 2022, 67, 102218. [Google Scholar] [CrossRef]
- Pokotylo, I.; Kravets, V.; Martinec, J.; Ruelland, E. The phosphatidic acid paradox: Too many actions for one molecule class? Lessons from plants. Prog. Lipid Res. 2018, 71, 43–53. [Google Scholar] [CrossRef]
- Ruelland, E.; Cantrel, C.; Gawer, M.; Kader, J.C.; Zachowski, A. Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 2002, 130, 999–1007. [Google Scholar] [CrossRef]
- Li, W.; Song, T.; Wallrad, L.; Kudla, J.; Wang, X.; Zhang, W. Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat. Plants 2019, 5, 1012–1021. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef]
- Kong, L.; Ma, X.; Zhang, C.; Kim, S.I.; Li, B.; Xie, Y.; Yeo, I.C.; Thapa, H.; Chen, S.; Devarenne, T.P.; et al. Dual phosphorylation of DGK5-mediated PA burst regulates ROS in plant immunity. Cell 2024, 187, 609–623.e21. [Google Scholar] [CrossRef]
- Bills, B.L.; Knowles, M.K. Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes. Biomolecules 2022, 12, 1707. [Google Scholar] [CrossRef]
- Pokotylo, I.; Kolesnikov, Y.; Kravets, V.; Zachowski, A.; Ruelland, E. Plant phosphoinositide-dependent phospholipases C: Variations around a canonical theme. Biochimie 2014, 96, 144–157. [Google Scholar] [CrossRef]
- Pokotylo, I.; Pejchar, P.; Potocký, M.; Kocourková, D.; Krčková, Z.; Ruelland, E.; Kravets, V.; Martinec, J. The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog. Lipid Res. 2013, 52, 62–79. [Google Scholar] [CrossRef]
- Fan, R.; Zhao, F.; Gong, Z.; Chen, Y.; Yang, B.; Zhou, C.; Zhang, J.; Du, Z.; Wang, X.; Yin, P.; et al. Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C. Nat. Commun. 2023, 14, 194. [Google Scholar] [CrossRef]
- Furt, F.; König, S.; Bessoule, J.J.; Sargueil, F.; Zallot, R.; Stanislas, T.; Noirot, E.; Lherminier, J.; Simon-Plas, F.; Heilmann, I.; et al. Polyphosphoinositides are enriched in plant membrane rafts and form microdomains in the plasma membrane. Plant Physiol. 2010, 152, 2173–2187. [Google Scholar] [CrossRef]
- Ruelland, E.; Kravets, V.; Derevyanchuk, M.; Martinec, J.; Zachowski, A.; Pokotylo, I. Role of phospholipid signalling in plant environmental responses. Environ. Exp. Bot. 2015, 114, 129–143. [Google Scholar] [CrossRef]
- Hallouin, M.; Ghelis, T.; Brault, M.; Bardat, F.; Cornel, D.; Miginiac, E.; Rona, J.P.; Sotta, B.; Jeannette, E. Plasmalemma abscisic acid perception leads to RAB18 expression via phospholipase D activation in Arabidopsis suspension cells. Plant Physiol. 2002, 130, 265–272. [Google Scholar] [CrossRef]
- Krinke, O.; Flemr, M.; Vergnolle, C.; Collin, S.; Renou, J.P.; Taconnat, L.; Yu, A.; Burketová, L.; Valentová, O.; Zachowski, A.; et al. Phospholipase D activation is an early component of the salicylic acid signaling pathway in Arabidopsis cell suspensions. Plant Physiol. 2009, 150, 424–436. [Google Scholar] [CrossRef]
- Cacas, J.L.; Gerbeau-Pissot, P.; Fromentin, J.; Cantrel, C.; Thomas, D.; Jeannette, E.; Kalachova, T.; Mongrand, S.; Simon-Plas, F.; Ruelland, E. Diacylglycerol kinases activate tobacco NADPH oxidase-dependent oxidative burst in response to cryptogein. Plant Cell Environ. 2017, 40, 585–598. [Google Scholar] [CrossRef]
- Kalachova, T.; Škrabálková, E.; Pateyron, S.; Soubigou-Taconnat, L.; Djafi, N.; Collin, S.; Sekereš, J.; Burketová, L.; Potocký, M.; Pejchar, P.; et al. DIACYLGLYCEROL KINASE 5 participates in flagellin-induced signaling in Arabidopsis. Plant Physiol. 2022, 190, 1978–1996. [Google Scholar] [CrossRef]
- Gully, K.; Pelletier, S.; Guillou, M.-C.; Ferrand, M.; Aligon, S.; Pokotylo, I.; Perrin, A.; Vergne, E.; Fagard, M.; Ruelland, E.; et al. The SCOOP12 peptide regulates defense response and root elongation in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 1349–1365. [Google Scholar] [CrossRef]
- Yao, H.-Y.; Xue, H.-W. Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 2018, 60, 851–863. [Google Scholar] [CrossRef]
- Kolesnikov, Y.; Kretynin, S.; Bukhonska, Y.; Pokotylo, I.; Ruelland, E.; Martinec, J.; Kravets, V. Phosphatidic Acid in Plant Hormonal Signaling: From Target Proteins to Membrane Conformations. Int. J. Mol. Sci. 2022, 23, 3227. [Google Scholar] [CrossRef]
- Kim, S.C.; Yao, S.; Zhang, Q.; Wang, X. Phospholipase Dδ and phosphatidic acid mediate heat-induced nuclear localization of glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Plant J. Cell Mol. Biol. 2022, 112, 786–799. [Google Scholar] [CrossRef]
- Lin, J.; Zhao, J.; Du, L.; Wang, P.; Sun, B.; Zhang, C.; Shi, Y.; Li, H.; Sun, H. Activation of MAPK-mediated immunity by phosphatidic acid in response to positive-strand RNA viruses. Plant Commun. 2024, 5, 100659. [Google Scholar] [CrossRef]
- Cao, H.; Liu, Q.; Liu, X.; Ma, Z.; Zhang, J.; Li, X.; Shen, L.; Yuan, J.; Zhang, Q. Phosphatidic acid regulates ammonium uptake by interacting with AMMONIUM TRANSPORTER 1;1 in Arabidopsis. Plant Physiol. 2023, 193, 1954–1969. [Google Scholar] [CrossRef]
- Thaller, D.J.; Tong, D.; Marklew, C.J.; Ader, N.R.; Mannino, P.J.; Borah, S.; King, M.C.; Ciani, B.; Lusk, C.P. Direct binding of ESCRT protein Chm7 to phosphatidic acid-rich membranes at nuclear envelope herniations. J. Cell Biol. 2021, 220, e202004222. [Google Scholar] [CrossRef]
- Li, J.; Shen, L.; Han, X.; He, G.; Fan, W.; Li, Y.; Yang, S.; Zhang, Z.; Yang, Y.; Jin, W.; et al. Phosphatidic acid-regulated SOS2 controls sodium and potassium homeostasis in Arabidopsis under salt stress. EMBO J. 2023, 42, e112401. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Li, G.; Lin, F.; Xue, H.-W. Genome-wide analysis of the phospholipase D family in Oryza sativa and functional characterization of PLDβ1 in seed germination. Cell Res. 2007, 17, 881–894. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Z.; Yue, Y.; Zhao, H.; Fei, X.; E, L.; Liu, C.; Chen, S.; Lai, J.; Song, W. Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants 2021, 7, 1579–1588. [Google Scholar] [CrossRef]
- Lu, S.; Fadlalla, T.; Tang, S.; Li, L.; Ali, U.; Li, Q.; Guo, L. Genome-Wide Analysis of Phospholipase D Gene Family and Profiling of Phospholipids under Abiotic Stresses in Brassica napus. Plant Cell Physiol. 2019, 60, 1556–1566. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, D.; Zhang, Q.; Zhang, W. Genomic analysis of phospholipase D family and characterization of GmPLDαs in soybean (Glycine max). J. Plant Res. 2012, 125, 569–578. [Google Scholar] [CrossRef]
- Eliáš, M.; Potocký, M.; Cvrčková, F.; Žárský, V. Molecular diversity of phospholipase D in angiosperms. BMC Genom. 2002, 3, 2. [Google Scholar] [CrossRef]
- Li, J.; Yu, F.; Guo, H.; Xiong, R.; Zhang, W.; He, F.; Zhang, M.; Zhang, P. Crystal structure of plant PLDα1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D. Cell Res. 2020, 30, 61–69. [Google Scholar] [CrossRef]
- Zheng, L.; Shan, J.; Krishnamoorthi, R.; Wang, X. Activation of Plant Phospholipase Dβ by Phosphatidylinositol 4,5-Bisphosphate: Characterization of Binding Site and Mode of Action. Biochemistry 2002, 41, 4546–4553. [Google Scholar] [CrossRef]
- Sciorra, V.A.; Rudge, S.A.; Prestwich, G.D.; Frohman, M.A.; Engebrecht, J.; Morris, A.J. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 1999, 18, 5911–5921. [Google Scholar] [CrossRef]
- Ge, H.; Chen, C.; Jing, W.; Zhang, Q.; Wang, H.; Wang, R.; Zhang, W. The rice diacylglycerol kinase family: Functional analysis using transient RNA interference. Front. Plant Sci. 2012, 3, 60. [Google Scholar] [CrossRef]
- Gu, Y.; Zhao, C.; He, L.; Yan, B.; Dong, J.; Li, Z.; Yang, K.; Xu, J. Genome-wide identification and abiotic stress responses of DGK gene family in maize. J. Plant Biochem. Biotechnol. 2018, 27, 156–166. [Google Scholar] [CrossRef]
- Tang, F.; Xiao, Z.; Sun, F.; Shen, S.; Chen, S.; Chen, R.; Zhu, M.; Zhang, Q.; Du, H.; Lu, K.; et al. Genome-wide identification and comparative analysis of diacylglycerol kinase (DGK) gene family and their expression profiling in Brassica napus under abiotic stress. BMC Plant Biol. 2020, 20, 473. [Google Scholar] [CrossRef]
- Carther, K.F.I.; Ketehouli, T.; Ye, N.; Yang, Y.H.; Wang, N.; Dong, Y.Y.; Yao, N.; Liu, X.M.; Liu, W.C.; Li, X.W.; et al. Comprehensive Genomic Analysis and Expression Profiling of Diacylglycerol Kinase (DGK) Gene Family in Soybean (Glycine max) under Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 1361. [Google Scholar] [CrossRef]
- Gómez-Merino, F.C.; Brearley, C.A.; Ornatowska, M.; Abdel-Haliem, M.E.F.; Zanor, M.-I.; Mueller-Roeber, B. AtDGK2, a Novel Diacylglycerol Kinase from Arabidopsis thaliana, Phosphorylates 1-Stearoyl-2-arachidonoyl-sn-glycerol and 1,2-Dioleoyl-sn-glycerol and Exhibits Cold-inducible Gene Expression. J. Biol. Chem. 2004, 279, 8230–8241. [Google Scholar] [CrossRef]
- Escobar-Sepúlveda, H.F.; Trejo-Téllez, L.I.; Pérez-Rodríguez, P.; Hidalgo-Contreras, J.V.; Gómez-Merino, F.C. Diacylglycerol Kinases Are Widespread in Higher Plants and Display Inducible Gene Expression in Response to Beneficial Elements, Metal, and Metalloid Ions. Front. Plant Sci. 2017, 8, 129. [Google Scholar] [CrossRef]
- Munnik, T. PI-PLC: Phosphoinositide-Phospholipase C in Plant Signaling. In Phospholipases in Plant Signaling; Wang, X., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 27–54. [Google Scholar]
- Kanemaru, K.; Nakamura, Y. Activation Mechanisms and Diverse Functions of Mammalian Phospholipase C. Biomolecules 2023, 13, 915. [Google Scholar] [CrossRef]
- Delage, E.; Puyaubert, J.; Zachowski, A.; Ruelland, E. Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: Convergences and divergences among eukaryotic kingdoms. Prog. Lipid Res. 2013, 52, 1–14. [Google Scholar] [CrossRef]
- Wang, N.; Shi, Y.; Jiang, Q.; Li, H.; Fan, W.; Feng, Y.; Li, L.; Liu, B.; Lin, F.; Jing, W.; et al. A 14-3-3 protein positively regulates rice salt tolerance by stabilizing phospholipase C1. Plant Cell Environ. 2023, 46, 1232–1248. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, Y.; Li, J.; Li, H. Genome-Wide Investigation of the Phospholipase C Gene Family in Zea mays. Front. Genet. 2021, 11, 611414. [Google Scholar] [CrossRef]
- Iqbal, S.; Ali, U.; Fadlalla, T.; Li, Q.; Liu, H.; Lu, S.; Guo, L. Genome wide characterization of phospholipase A & C families and pattern of lysolipids and diacylglycerol changes under abiotic stresses in Brassica napus L. Plant Physiol. Biochem. 2020, 147, 101–112. [Google Scholar] [CrossRef]
- Wang, F.; Deng, Y.; Zhou, Y.; Dong, J.; Chen, H.; Dong, Y.; Wang, N.; Li, X.; Li, H. Genome-Wide Analysis and Expression Profiling of the Phospholipase C Gene Family in Soybean (Glycine max). PLoS ONE 2015, 10, e0138467. [Google Scholar] [CrossRef]
- Ellis, M.V.; James, S.R.; Perisic, O.; Downes, C.P.; Williams, R.L.; Katan, M. Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of plcdelta1. J. Biol. Chem. 1998, 273, 11650–11659. [Google Scholar] [CrossRef]
- Essen, L.-O.; Perisic, O.; Katan, M.; Wu, Y.; Roberts, M.F.; Williams, R.L. Structural Mapping of the Catalytic Mechanism for a Mammalian Phosphoinositide-Specific Phospholipase C. Biochemistry 1997, 36, 1704–1718. [Google Scholar] [CrossRef]
- Li, G.; Xue, H.-W. Arabidopsis PLDζ2 Regulates Vesicle Trafficking and Is Required for Auxin Response. Plant Cell 2007, 19, 281–295. [Google Scholar] [CrossRef]
- Bowling, F.Z.; Frohman, M.A.; Airola, M.V. Structure and regulation of human phospholipase D. Adv. Biol. Regul. 2021, 79, 100783. [Google Scholar] [CrossRef]
- Qin, W.; Pappan, K.; Wang, X. Molecular Heterogeneity of Phospholipase D (PLD): CLONING OF PLDγ AND REGULATION OF PLANT PLDγ, -β, AND -α BY POLYPHOSPHOINOSITIDES AND CALCIUM. J. Biol. Chem. 1997, 272, 28267–28273. [Google Scholar] [CrossRef]
- Qin, C.; Wang, X. The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol. 2002, 128, 1057–1068. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X. A novel phospholipase D of arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 2001, 127, 1102–1112. [Google Scholar] [CrossRef]
- Hirano, Y.; Gao, Y.-G.; Stephenson, D.J.; Vu, N.T.; Malinina, L.; Simanshu, D.K.; Chalfant, C.E.; Patel, D.J.; Brown, R.E. Structural basis of phosphatidylcholine recognition by the C2–domain of cytosolic phospholipase A2α. eLife 2019, 8, e44760. [Google Scholar] [CrossRef]
- Zheng, L.; Krishnamoorthi, R.; Zolkiewski, M.; Wang, X. Distinct Ca2+ Binding Properties of Novel C2 Domains of Plant Phospholipase Dα and β. J. Biol. Chem. 2000, 275, 19700–19706. [Google Scholar] [CrossRef]
- Chandra, M.; Chin, Y.K.Y.; Mas, C.; Feathers, J.R.; Paul, B.; Datta, S.; Chen, K.-E.; Jia, X.; Yang, Z.; Norwood, S.J.; et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 2019, 10, 1528. [Google Scholar] [CrossRef]
- Sung, T.-C.; Zhang, Y.; Morris, A.J.; Frohman, M.A. Structural Analysis of Human Phospholipase D1. J. Biol. Chem. 1999, 274, 3659–3666. [Google Scholar] [CrossRef]
- Singh, N.; Reyes-Ordoñez, A.; Compagnone, M.A.; Moreno, J.F.; Leslie, B.J.; Ha, T.; Chen, J. Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 2021, 12, 4339. [Google Scholar] [CrossRef]
- Hodgkin, M.N.; Masson, M.R.; Powner, D.; Saqib, K.M.; Ponting, C.P.; Wakelam, M.J. Phospholipase D regulation and localisation is dependent upon a phosphatidylinositol 4,5-biphosphate-specific PH domain. Curr. Biol. CB 2000, 10, 43–46. [Google Scholar] [CrossRef]
- Simon, M.L.; Platre, M.P.; Assil, S.; van Wijk, R.; Chen, W.Y.; Chory, J.; Dreux, M.; Munnik, T.; Jaillais, Y. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. Cell Mol. Biol. 2014, 77, 322–337. [Google Scholar] [CrossRef]
- Martin, T.F.J. PHOSPHOINOSITIDE LIPIDS AS SIGNALING MOLECULES: Common Themes for Signal Transduction, Cytoskeletal Regulation, and Membrane Trafficking. Annu. Rev. Cell Dev. Biol. 1998, 14, 231–264. [Google Scholar] [CrossRef]
- Vaultier, M.-N.; Cantrel, C.; Guerbette, F.; Boutté, Y.; Vergnolle, C.; Çiçek, D.; Bolte, S.; Zachowski, A.; Ruelland, E. The hydrophobic segment of Arabidopsis thaliana cluster I diacylglycerol kinases is sufficient to target the proteins to cell membranes. FEBS Lett. 2008, 582, 1743–1748. [Google Scholar] [CrossRef]
- Colón-González, F.; Kazanietz, M.G. C1 domains exposed: From diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta 2006, 1761, 827–837. [Google Scholar] [CrossRef]
- Gao, K.; Liu, Y.-L.; Li, B.; Zhou, R.-G.; Sun, D.-Y.; Zheng, S.-Z. Arabidopsis thaliana Phosphoinositide-Specific Phospholipase C Isoform 3 (AtPLC3) and AtPLC9 have an Additive Effect on Thermotolerance. Plant Cell Physiol. 2014, 55, 1873–1883. [Google Scholar] [CrossRef]
- van Hooren, M.; van Wijk, R.; Vaseva, I.I.; Van Der Straeten, D.; Haring, M.; Munnik, T. Ectopic Expression of Distinct PLC Genes Identifies ‘Compactness’ as a Possible Architectural Shoot Strategy to Cope with Drought Stress. Plant Cell Physiol. 2023. [Google Scholar] [CrossRef]
- Otterhag, L.; Sommarin, M.; Pical, C. N-terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C activity in Arabidopsis thaliana. FEBS Lett. 2001, 497, 165–170. [Google Scholar] [CrossRef]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, D.M.; Yu, W.W.; Shi, L.L.; Zhang, Y.; Lai, Y.X.; Huang, L.P.; Qi, H.; Chen, Q.F.; Yao, N.; et al. Phosphatidic acid modulates MPK3- and MPK6-mediated hypoxia signaling in Arabidopsis. Plant Cell 2022, 34, 889–909. [Google Scholar] [CrossRef]
- Wang, P.; Shen, L.; Guo, J.; Jing, W.; Qu, Y.; Li, W.; Bi, R.; Xuan, W.; Zhang, Q.; Zhang, W. Phosphatidic Acid Directly Regulates PINOID-Dependent Phosphorylation and Activation of the PIN-FORMED2 Auxin Efflux Transporter in Response to Salt Stress. Plant Cell 2019, 31, 250–271. [Google Scholar] [CrossRef]
- Guo, L.; Mishra, G.; Taylor, K.; Wang, X. Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases. J. Biol. Chem. 2011, 286, 13336–13345. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, S.; Liu, S.; Takano, T. The Arabidopsis sucrose non-fermenting-1-related protein kinase AtSnRK2.4 interacts with a transcription factor, AtMYB21, that is involved in salt tolerance. Plant Sci. 2021, 303, 110685. [Google Scholar] [CrossRef]
- Yao, H.; Wang, G.; Guo, L.; Wang, X. Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis. Plant Cell 2013, 25, 5030–5042. [Google Scholar] [CrossRef]
- Shen, L.; Tian, Q.; Yang, L.; Zhang, H.; Shi, Y.; Shen, Y.; Zhou, Z.; Wu, Q.; Zhang, Q.; Zhang, W. Phosphatidic acid directly binds with rice potassium channel OsAKT2 to inhibit its activity. Plant J. Cell Mol. Biol. 2020, 102, 649–665. [Google Scholar] [CrossRef]
- Cao, H.; Gong, R.; Yuan, S.; Su, Y.; Lv, W.; Zhou, Y.; Zhang, Q.; Deng, X.; Tong, P.; Liang, S.; et al. Phospholipase Dα6 and phosphatidic acid regulate gibberellin signaling in rice. EMBO Rep. 2021, 22, e51871. [Google Scholar] [CrossRef]
- Pandit, S.; Goel, R.; Mishra, G. Phosphatidic acid binds to and stimulates the activity of ARGAH2 from Arabidopsis. Plant Physiol. Biochem. 2022, 185, 344–355. [Google Scholar] [CrossRef]
- Yoon, M.Y.; Kim, M.Y.; Shim, S.; Kim, K.D.; Ha, J.; Shin, J.H.; Kang, S.; Lee, S.-H. Transcriptomic Profiling of Soybean in Response to High-Intensity UV-B Irradiation Reveals Stress Defense Signaling. Front. Plant Sci. 2016, 7, 1917. [Google Scholar] [CrossRef]
- Yeken, M.Z.; Özer, G.; Çiftçi, V. Genome-Wide Identification and Expression Analysis of DGK (Diacylglycerol Kinase) Genes in Common Bean. J. Plant Growth Regul. 2023, 42, 2558–2569. [Google Scholar] [CrossRef]
- Wang, C.-R.; Yang, A.-F.; Yue, G.-D.; Gao, Q.; Yin, H.-Y.; Zhang, J.-R. Enhanced expression of phospholipase C 1 (ZmPLC1) improves drought tolerance in transgenic maize. Planta 2008, 227, 1127–1140. [Google Scholar] [CrossRef]
- Lu, S.; Yao, S.; Wang, G.; Guo, L.; Zhou, Y.; Hong, Y.; Wang, X. Phospholipase Dε enhances Braasca napus growth and seed production in response to nitrogen availability. Plant Biotechnol. J. 2016, 14, 926–937. [Google Scholar] [CrossRef]
- Yao, S.; Wang, G.; Wang, X. Effects of Phospholipase Dε Overexpression on Soybean Response to Nitrogen and Nodulation. Front. Plant Sci. 2022, 13, 852923. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, J.; Chen, X.; Zhao, D.; Zhou, X.; Zhang, Y.; Wang, X.; Zhao, J. Phospholipase D- and phosphatidic acid-mediated phospholipid metabolism and signaling modulate symbiotic interaction and nodulation in soybean (Glycine max). Plant J. Cell Mol. Biol. 2021, 106, 142–158. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kuroda, M.; Yamakawa, H.; Ashizawa, T.; Hirayae, K.; Kurimoto, L.; Shinya, T.; Shibuya, N. Suppression of a phospholipase D gene, OsPLDbeta1, activates defense responses and increases disease resistance in rice. Plant Physiol. 2009, 150, 308–319. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, X.; Ge, S.; Zhang, H.; Che, X.; Liu, S.; Liu, D.; Li, H.; Gu, X.; He, L.; et al. Involvement of Phospholipase C in Photosynthesis and Growth of Maize Seedlings. Genes 2022, 13, 1011. [Google Scholar] [CrossRef]
- Pappan, K.; Zheng, S.; Wang, X. Identification and Characterization of a Novel Plant Phospholipase D That Requires Polyphosphoinositides and Submicromolar Calcium for Activity in Arabidopsis. J. Biol. Chem. 1997, 272, 7048–7054. [Google Scholar] [CrossRef]
- Wang, N.-N.; Ni, P.; Wei, Y.-L.; Hu, R.; Li, Y.; Li, X.-B.; Zheng, Y. Phosphatidic acid interacts with an HD-ZIP transcription factor GhHOX4 to influence its function in fiber elongation of cotton (Gossypium hirsutum). Plant J. 2024, 118, 423–436. [Google Scholar] [CrossRef]
- McLaughlin, S.; Wang, J.; Gambhir, A.; Murray, D. PIP2 and Proteins: Interactions, Organization, and Information Flow. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 151–175. [Google Scholar] [CrossRef]
- Kalachova, T.; Puga-Freitas, R.; Kravets, V.; Soubigou-Taconnat, L.; Repellin, A.; Balzergue, S.; Zachowski, A.; Ruelland, E. The inhibition of basal phosphoinositide-dependent phospholipase C activity in Arabidopsis suspension cells by abscisic or salicylic acid acts as a signalling hub accounting for an important overlap in transcriptome remodelling induced by these hormones. Environ. Exp. Bot. 2016, 123, 37–49. [Google Scholar] [CrossRef]
- Šašek, V.; Janda, M.; Delage, E.; Puyaubert, J.; Guivarc’h, A.; López Maseda, E.; Dobrev, P.I.; Caius, J.; Bóka, K.; Valentová, O.; et al. Constitutive salicylic acid accumulation in pi4kIIIβ1β2 Arabidopsis plants stunts rosette but not root growth. New Phytol. 2014, 203, 805–816. [Google Scholar] [CrossRef]
- Kalachova, T.; Janda, M.; Šašek, V.; Ortmannová, J.; Nováková, P.; Dobrev, I.P.; Kravets, V.; Guivarc’h, A.; Moura, D.; Burketová, L.; et al. Identification of salicylic acid-independent responses in an Arabidopsis phosphatidylinositol 4-kinase beta double mutant. Ann. Bot. 2020, 125, 775–784. [Google Scholar] [CrossRef]
- Starodubtseva, A.; Kalachova, T.; Retzer, K.; Jelínková, A.; Dobrev, P.; Lacek, J.; Pospíchalová, R.; Angelini, J.; Guivarc’h, A.; Pateyron, S.; et al. An Arabidopsis mutant deficient in phosphatidylinositol-4-phosphate kinases ß1 and ß2 displays altered auxin-related responses in roots. Sci. Rep. 2022, 12, 6947. [Google Scholar] [CrossRef]
- Jang, J.H.; Nguyen, N.Q.; Légeret, B.; Beisson, F.; Kim, Y.-J.; Sim, H.-J.; Lee, O.R. Phospholipase pPLAIIIα Increases Germination Rate and Resistance to Turnip Crinkle Virus when Overexpressed. Plant Physiol. 2020, 184, 1482–1498. [Google Scholar] [CrossRef]
- Morin, H.; Chételat, A.; Stolz, S.; Marcourt, L.; Glauser, G.; Wolfender, J.-L.; Farmer, E.E. Wound-response jasmonate dynamics in the primary vasculature. New Phytol. 2023, 240, 1484–1496. [Google Scholar] [CrossRef]
- Liu, N.-J.; Hou, L.-P.; Bao, J.-J.; Wang, L.-J.; Chen, X.-Y. Sphingolipid metabolism, transport, and functions in plants: Recent progress and future perspectives. Plant Commun. 2021, 2, 100214. [Google Scholar] [CrossRef]
- Yang, D.; Liu, X.; Yin, X.; Dong, T.; Yu, M.; Wu, Y. Rice Non-Specific Phospholipase C6 Is Involved in Mesocotyl Elongation. Plant Cell Physiol. 2021, 62, 985–1000. [Google Scholar] [CrossRef]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef]
- Pixley, K.V.; Falck-Zepeda, J.B.; Paarlberg, R.L.; Phillips, P.W.B.; Slamet-Loedin, I.H.; Dhugga, K.S.; Campos, H.; Gutterson, N. Genome-edited crops for improved food security of smallholder farmers. Nat. Genet. 2022, 54, 364–367. [Google Scholar] [CrossRef]
- Wolabu, T.W.; Mahmood, K.; Jerez, I.T.; Cong, L.; Yun, J.; Udvardi, M.; Tadege, M.; Wang, Z.; Wen, J. Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality. Plant Biotechnol. J. 2023, 21, 1383–1392. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 2004, 21, 951–960. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. PyMOL. Available online: http://www.pymol.org/pymol (accessed on 29 May 2024).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amokrane, L.; Pokotylo, I.; Acket, S.; Ducloy, A.; Troncoso-Ponce, A.; Cacas, J.-L.; Ruelland, E. Phospholipid Signaling in Crop Plants: A Field to Explore. Plants 2024, 13, 1532. https://doi.org/10.3390/plants13111532
Amokrane L, Pokotylo I, Acket S, Ducloy A, Troncoso-Ponce A, Cacas J-L, Ruelland E. Phospholipid Signaling in Crop Plants: A Field to Explore. Plants. 2024; 13(11):1532. https://doi.org/10.3390/plants13111532
Chicago/Turabian StyleAmokrane, Lucas, Igor Pokotylo, Sébastien Acket, Amélie Ducloy, Adrian Troncoso-Ponce, Jean-Luc Cacas, and Eric Ruelland. 2024. "Phospholipid Signaling in Crop Plants: A Field to Explore" Plants 13, no. 11: 1532. https://doi.org/10.3390/plants13111532
APA StyleAmokrane, L., Pokotylo, I., Acket, S., Ducloy, A., Troncoso-Ponce, A., Cacas, J.-L., & Ruelland, E. (2024). Phospholipid Signaling in Crop Plants: A Field to Explore. Plants, 13(11), 1532. https://doi.org/10.3390/plants13111532









