Functional Characterization of the Soybean Glycine max Actin Depolymerization Factor GmADF13 for Plant Resistance to Drought Stress
Abstract
:1. Introduction
2. Results
2.1. Cloning and Sequence Analysis of GmADF13
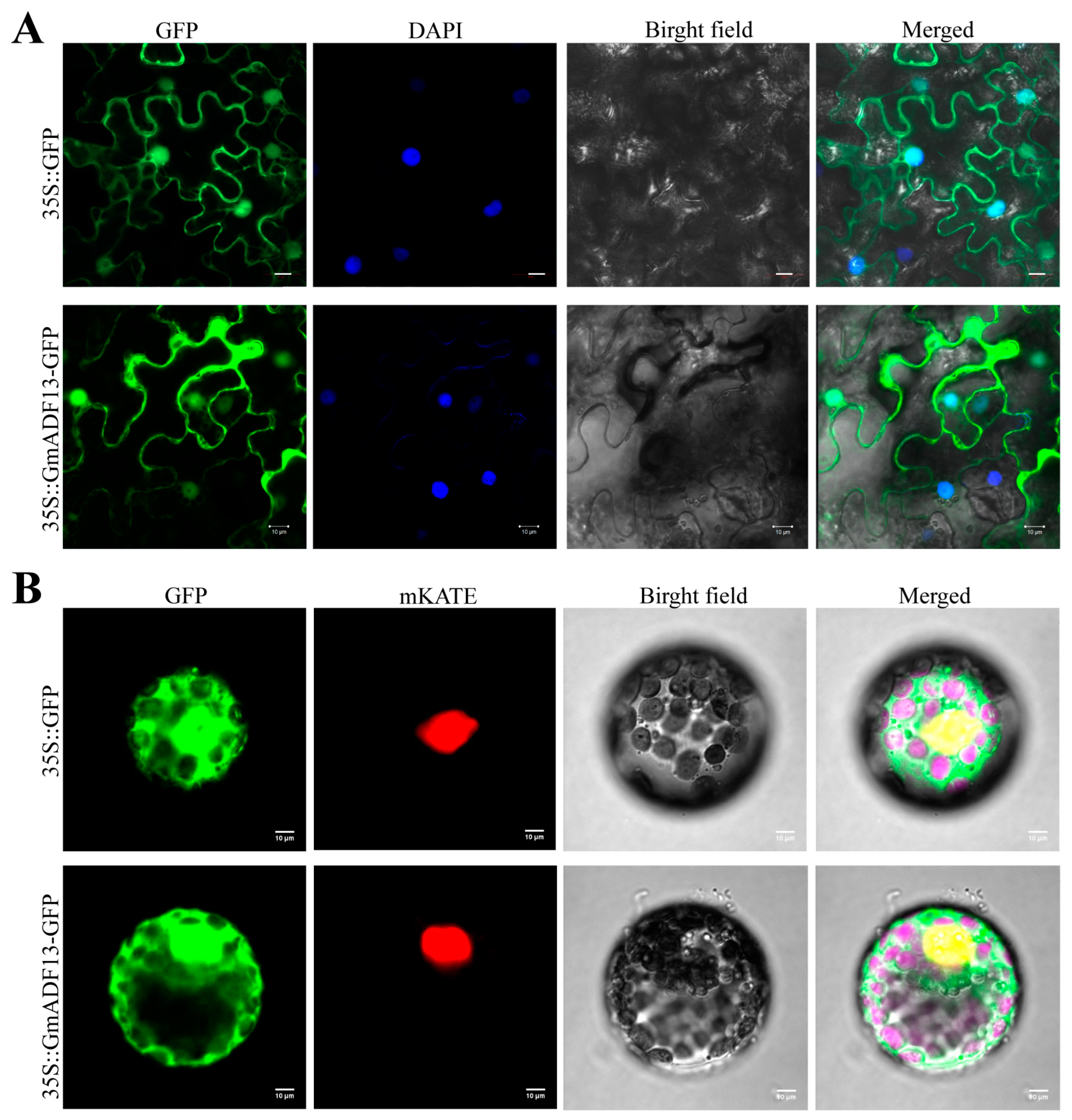
2.2. GmADF13 Encodes Nucleus- and Cytoplasm-Localized ADF Proteins
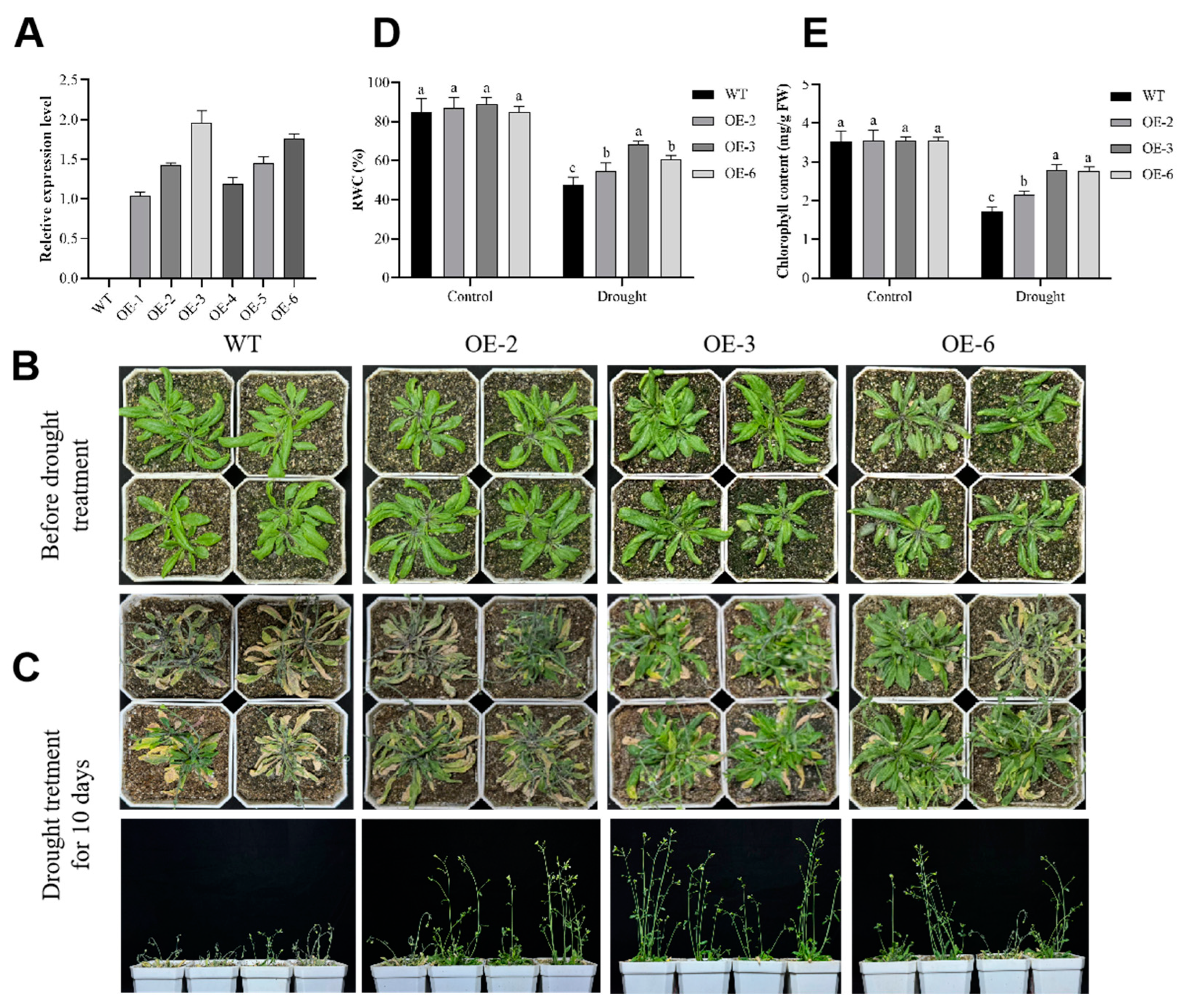
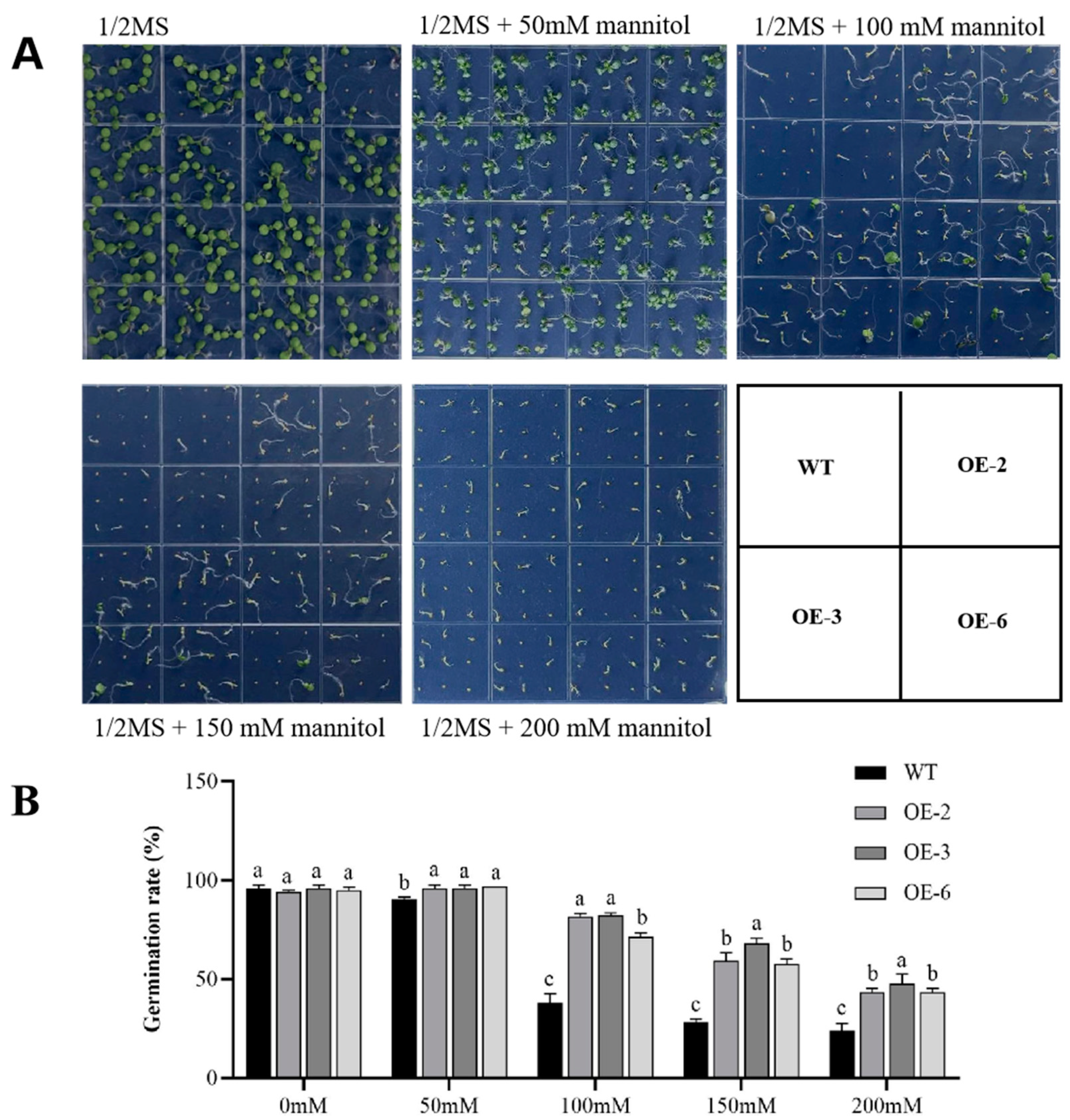
2.3. Arabidopsis Overexpressing GmADF13 Exhibited Enhanced Drought Tolerance
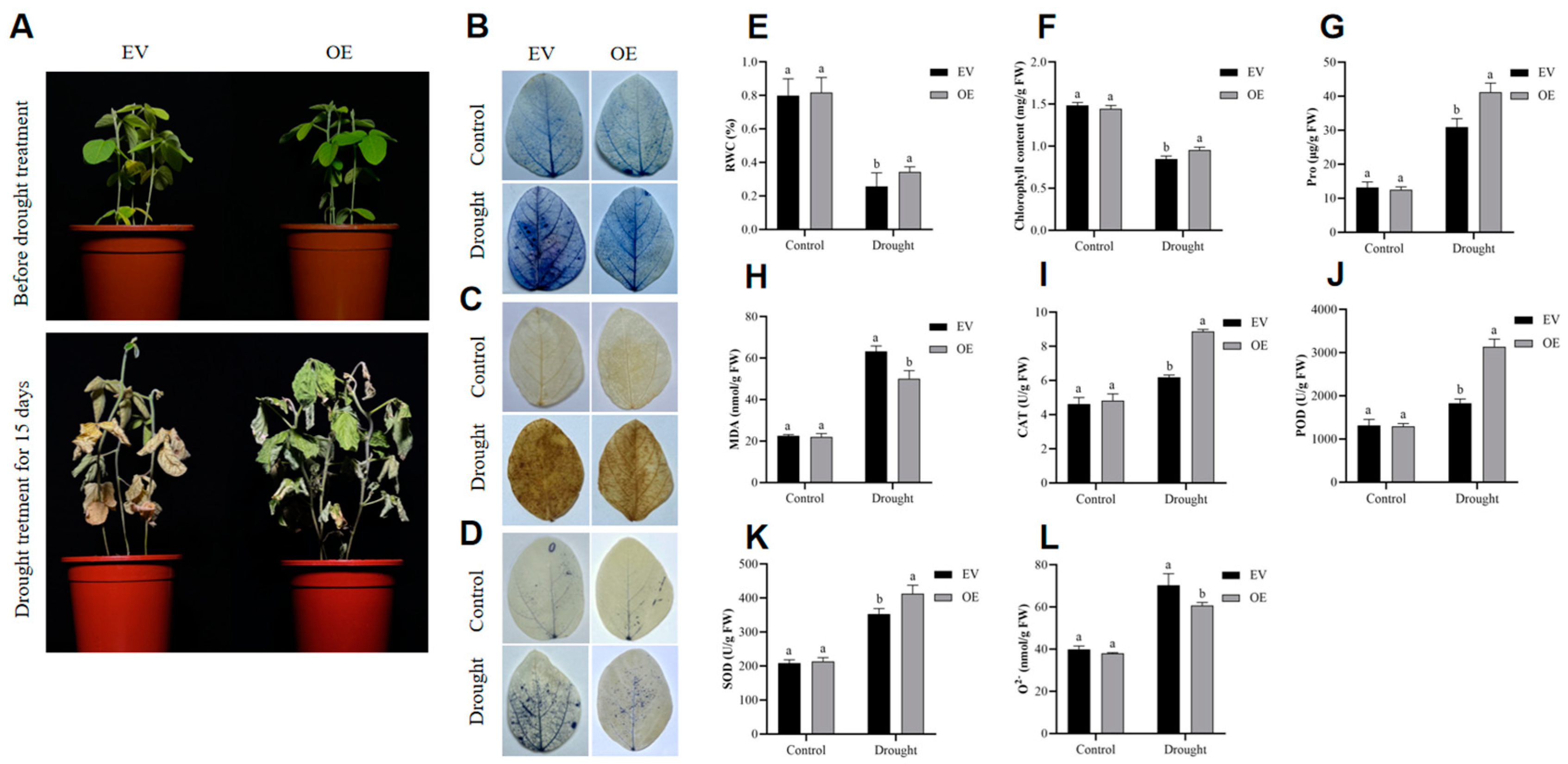
2.4. GmADF13 Improved Drought Tolerance in Transgenic Soybean Hairy Roots
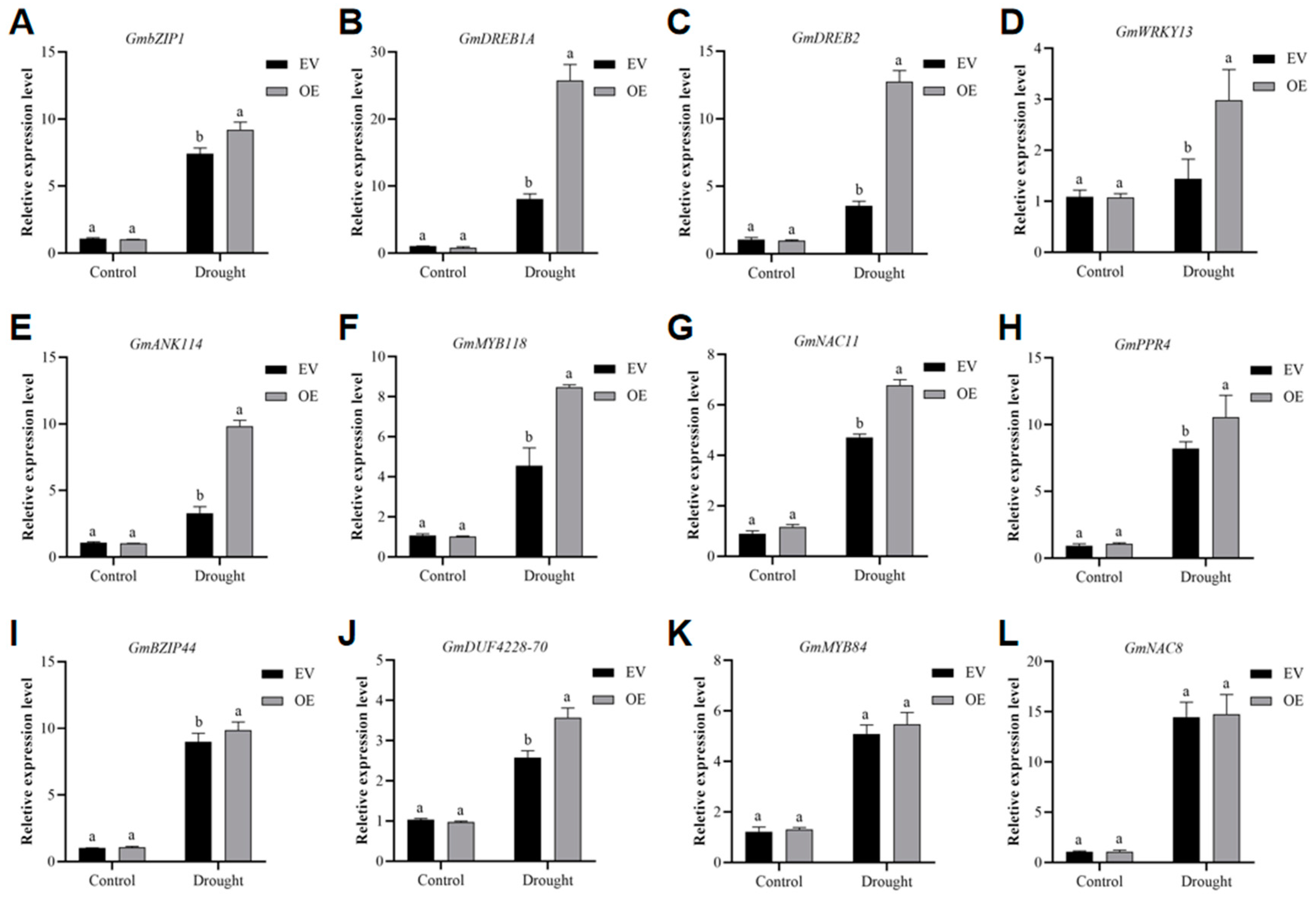
2.5. GmADF13-OE Plants Exhibit Increased Drought-Inducible Gene Transcription
3. Discussion
4. Materials and Methods
4.1. GmADF13-OE Plants Exhibit Increased Drought-Inducible Gene Transcription
4.2. GmADF13 Cloning, RNA Extraction, and qRT-PCR
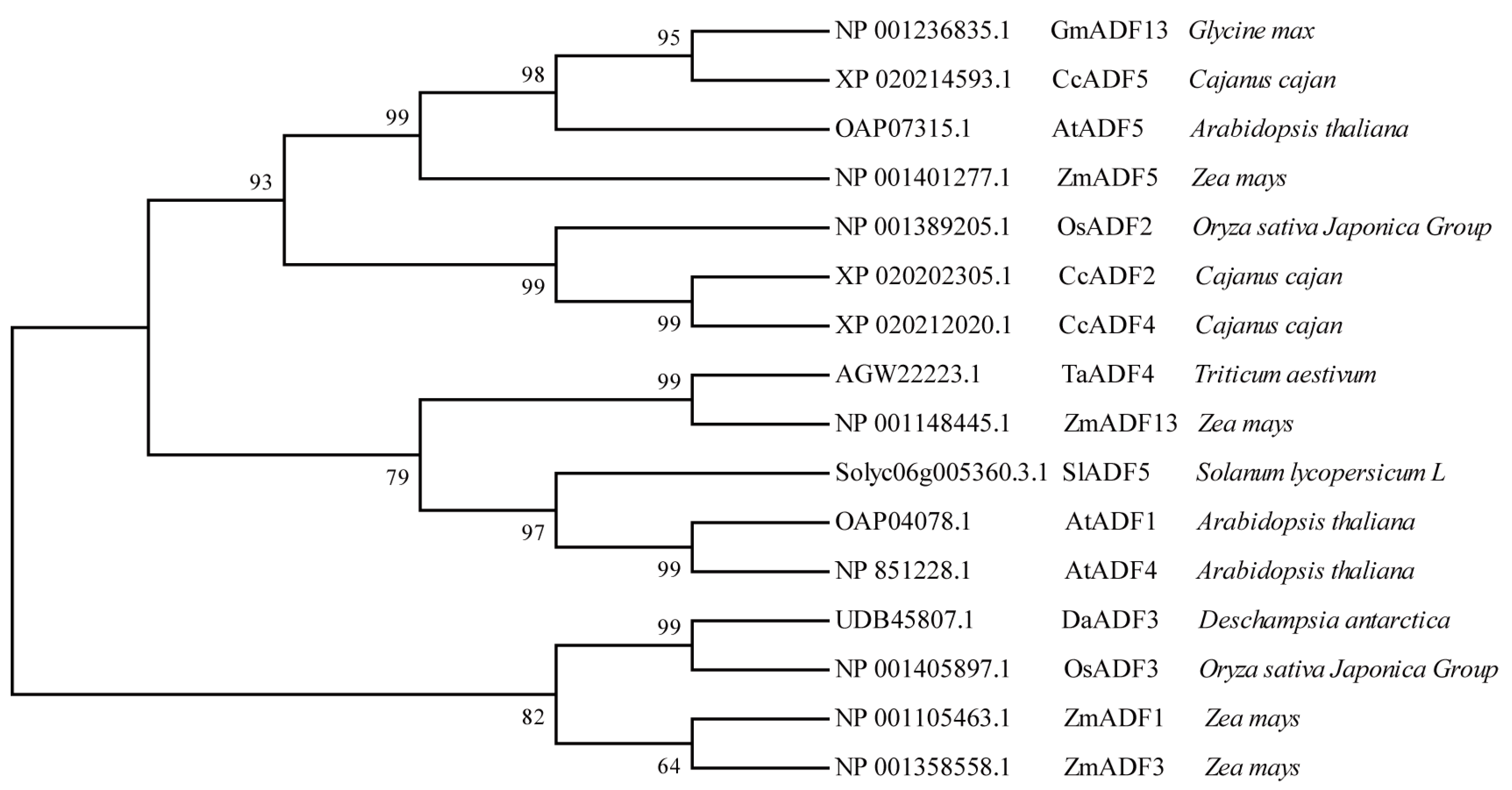
4.3. Phylogenetic Analysis
4.4. Subcellular Localization of GmADF13
4.5. Generation of Transgenic Arabidopsis Plants Overexpressing the GmADF13 Gene and Screening
4.6. Agrobacterium Rhizogenes-Mediated Transformation of Soybean Hairy Roots
4.7. Drought Stress Assays of Transgenic Arabidopsis and Hairy Root Composite Soybeans
4.8. Relative Water and Chlorophyll Content
4.9. Measurement of Proline, Malondialdehyde, Antioxidant Enzymes, and Superoxide Anion Levels
4.10. NBT, DAB, and Trypan blue Staining
4.11. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.; Li, R.F.; Ge, Y.R.; Li, Y.F.; Li, R.L. Plants’ response to abiotic stress: Mechanisms and strategies. Int. J. Mol. Sci. 2023, 24, 915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, S.J.; Ren, T.M.; Niu, M.X.; Liu, X.; Liu, C.; Wang, H.L.; Yin, W.L.; Xia, X.L. Crucial abiotic stress regulatory network of NF-Y transcription factor in plants. Int. J. Mol. Sci. 2023, 24, 4426. [Google Scholar] [CrossRef] [PubMed]
- Miryeganeh, M. Plants’ epigenetic mechanisms and abiotic stress. Genes 2021, 12, 1106. [Google Scholar] [CrossRef] [PubMed]
- González, E.M. Drought stress tolerance in plants. Int. J. Mol. Sci. 2023, 24, 562. [Google Scholar] [CrossRef]
- Shaffique, S.; Hussain, S.; Kang, S.M.; Imran, M.; Injamum-Ul-Hoque, M.; Khan, M.A.; Lee, I.J. Phytohormonal modulation of the drought stress in soybean: Outlook, research progress, and cross-talk. Front. Plant Sci. 2023, 14, 1237295. [Google Scholar] [CrossRef]
- Vurukonda, S.S.; Vardharajula, S.; Shrivastava, M.; Sk, Z.A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Zhu, J.K. Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants. 2018, 4, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Shaffique, S.; Khan, M.A.; Imran, M.; Kang, S.M.; Park, Y.S.; Wani, S.H.; Lee, I.J. Research progress in the field of microbial mitigation of drought stress in plants. Front. Plant Sci. 2022, 13, 870626. [Google Scholar] [CrossRef]
- Pollard, T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef]
- Staiger, C.J. Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 257–288. [Google Scholar] [CrossRef]
- Li, J.; Blanchoin, L.; Staiger, C.J. Signaling to actin stochastic dynamics. Annu. Rev. Plant Biol. 2015, 66, 415–440. [Google Scholar] [CrossRef]
- Porter, K.; Day, B. From filaments to function: The role of the plant actin cytoskeleton in pathogen perception, signaling and immunity. J. Integr. Plant Biol. 2016, 58, 299–311. [Google Scholar] [CrossRef]
- Carlier, M.F.; Laurent, V.; Santolini, J.; Melki, R.; Didry, D.; Xia, G.X.; Hong, Y.; Chua, N.H.; Pantaloni, D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 1997, 136, 1307–1322. [Google Scholar] [CrossRef]
- Bamburg, J.R. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999, 15, 185–230. [Google Scholar] [CrossRef]
- Maciver, S.K.; Hussey, P.J. The ADF/cofilin family: Actin-remodeling proteins. Genome Biol. 2002, 3, reviews3007. [Google Scholar] [CrossRef] [PubMed]
- Staiger, C.J.; Gibbon, B.C.; Kovar, D.R.; Zonia, L.E. Profilin and actin-depolymerizing factor: Modulators of actin organization in plants. Trends Plant Sci. 1997, 2, 275–281. [Google Scholar] [CrossRef]
- Bamburg, J.R.; Harris, H.E.; Weeds, A.G. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 1980, 121, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Inada, N. Plant actin depolymerizing factor: Actin microfilament disassembly and more. J. Plant Res. 2017, 130, 227–238. [Google Scholar] [CrossRef]
- Burgos-Rivera, B.; Ruzicka, D.R.; Deal, R.B.; McKinney, E.C.; King-Reid, L.; Meagher, R.B. Actin depolymerizing factor9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 2008, 68, 619–632. [Google Scholar] [CrossRef]
- Hussey, P.J.; Ketelaar, T.; Deeks, M.J. Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 2006, 57, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Li, X.B.; Xu, D.; Wang, X.L.; Huang, G.Q.; Luo, J.; Li, D.D.; Zhang, Z.T.; Xu, W.L. Three cotton genes preferentially expressed in flower tissues encode actin-depolymerizing factors which are involved in F-actin dynamics in cells. J. Exp. Bot. 2010, 61, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Ortega, Y.; Carrasco-Castilla, J.; Juárez-Verdayes, M.A.; Toscano-Morales, R.; Fonseca-García, C.; Nava, N.; Cárdenas, L.; Quinto, C. Actin depolymerizing factor modulates rhizobial infection and nodule organogenesis in common bean. Int. J. Mol. Sci. 2020, 21, 1970. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.J.; Weeds, A.G.; Hussey, P.J. The maize actin-depolymerizing factor, ZmADF3, redistributes to the growing tip of elongating root hairs and can be induced to translocate into the nucleus with actin. Plant J. 1997, 12, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.Y.; Xie, Y.R.; Jiang, Y.X.; Qu, X.L.; Huang, S.J. Arabidopsis actin-depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 2013, 25, 3405–3423. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.W.; Shi, M.M.; Wang, D.Y.; Gong, Y.J.; Sha, Q.; Lv, P.; Yang, J.; Chu, P.F.; Guo, S.J. Research progress on the roles of actin-depolymerizing factor in plant stress responses. Front. Plant Sci. 2023, 14, 1278311. [Google Scholar] [CrossRef]
- Clément, M.; Ketelaar, T.; Rodiuc, N.; Banora, M.Y.; Smertenko, A.; Engler, G.; Abad, P.; Hussey, P.J.; de Almeida Engler, J. Actin-depolymerizing factor2-mediated actin dynamics are essential for root-knot nematode infection of Arabidopsis. Plant Cell 2009, 21, 2963–2979. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.L.; Deng, L.; Chang, D.; Chen, S.T.; Wang, X.J.; Kang, Z.S. TaADF3, an Actin-depolymerizing factor, negatively modulates wheat resistance against puccinia striiformis. Front. Plant Sci. 2015, 6, 1214. [Google Scholar] [CrossRef]
- Zhang, B.; Hua, Y.; Wang, J.; Huo, Y.; Shimono, M.; Day, B.; Ma, Q. TaADF4, an actin-depolymerizing factor from wheat, is required for resistance to the stripe rust pathogen Puccinia striiformis f. sp. tritici. Plant J. 2017, 89, 1210–1224. [Google Scholar] [CrossRef]
- Fu, Y.P.; Duan, X.Y.; Tang, C.L.; Li, X.R.; Voegele, R.T.; Wang, X.J.; Wei, G.R.; Kang, Z.S. TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. 2014, 78, 16–30. [Google Scholar] [CrossRef]
- Samayoa, L.F.; Malvar, R.A.; Olukolu, B.A.; Holland, J.B.; Butrón, A. Genome-wide association study reveals a set of genes associated with resistance to the Mediterranean corn borer (Sesamia nonagrioides L.) in a maize diversity panel. BMC Plant Biol. 2015, 15, 35. [Google Scholar] [CrossRef] [PubMed]
- Byun, M.Y.; Cui, L.H.; Lee, A.; Oh, H.G.; Yoo, Y.H.; Lee, J.; Kim, W.T.; Lee, H. Abiotic stress-induced actin-depolymerizing factor 3 from deschampsia antarctica enhanced cold tolerance when constitutively expressed in rice. Front. Plant Sci. 2021, 12, 734500. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiu, T.Q.; Yue, J.R.; Guo, N.N.; He, Y.J.; Han, X.P.; Wang, Q.Y.; Jia, P.F.; Wang, H.D.; Li, M.Z.; et al. Arabidopsis ADF1 is regulated by MYB73 and is involved in response to salt stress affecting actin filament organization. Plant Cell Physiol. 2021, 62, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Huang, W.L.; Hong, C.Y.; Lur, H.S.; Chang, M.C. Comprehensive analysis of differentially expressed rice actin depolymerizing factor gene family and heterologous overexpression of OsADF3 confers Arabidopsis thaliana drought tolerance. Rice 2012, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.; Zhang, Z.; He, J.X.; Zhang, P.; Ou, X.B.; Li, T.; Niu, L.P.; Nan, Q.; Niu, Y.; He, W.L.; et al. Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 2019, 70, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.J.; Wang, N.; Yang, R.S.; Wang, X.N.; Luo, P.; Chen, Y.; Wang, F.; Li, M.S.; Weng, J.F.; Zhang, D.G.; et al. ZmADF5, a maize actin-depolymerizing factor conferring enhanced drought tolerance in maize. Plants 2024, 13, 619. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Tran, L.S. Potentials toward genetic engineering of drought-tolerant soybean. Crit. Rev. Biotechnol. 2012, 32, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Zhang, M.; Feng, F.; Tian, Z.X. Toward a “Green Revolution” for Soybean. Mol. Plant. 2020, 13, 688–697. [Google Scholar] [CrossRef]
- Bharti, M.K.; Chalia, S.; Thakur, P.; Sridhara, S.N.; Thakur, A.; Sharma, P.B. Nanoferrites heterogeneous catalysts for biodiesel production from soybean and canola oil: A review. Environ. Chem. Lett. 2021, 19, 3727–3746. [Google Scholar] [CrossRef]
- Nadeem, M.; Li, J.; Yahya, M.; Sher, A.; Ma, C.; Wang, X.; Qiu, L. Research progress and perspective on drought stress in legumes: A review. Int. J. Mol. Sci. 2019, 20, 2541. [Google Scholar] [CrossRef]
- Kunert, K.J.; Vorster, B.J.; Fenta, B.A.; Kibido, T.; Dionisio, G.; Foyer, C.H. Drought stress responses in soybean roots and nodules. Front. Plant Sci. 2016, 7, 1015. [Google Scholar] [CrossRef]
- Guan, R.X.; Qu, Y.; Guo, Y.; Yu, L.L.; Liu, Y.; Jiang, J.H.; Chen, J.G.; Ren, Y.L.; Liu, G.Y.; Tian, L.; et al. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014, 80, 937–950. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, Y.L.; Sun, G.Z.; Liu, Y.X.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Xu, Z.S.; Lan, J.H. Genome-wide analysis of the soybean calmodulin-binding protein 60 family and identification of GmCBP60A-1 responses to drought and salt stresses. Int. J. Mol. Sci. 2021, 22, 3501. [Google Scholar] [CrossRef] [PubMed]
- Shaffique, S.; Farooq, M.; Kang, S.-M.; Lee, I.-J. Recent advances in biochemical reprogramming network under drought stress in soybean. J. Soil Sci. Plant Nutr. 2024. [Google Scholar] [CrossRef]
- Yang, C.F.; Huang, Y.Z.; Lv, W.H.; Zhang, Y.Y.; Bhat, J.A.; Kong, J.J.; Xing, H.; Zhao, J.M.; Zhao, T.J. GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci. 2020, 293, 110442. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Yang, H.L.; Fang, Y.S.; Guo, W.; Chen, H.F.; Zhang, X.J.; Dai, W.J.; Chen, S.L.; Hao, Q.N.; Yuan, S.L.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef]
- Chen, K.; Su, C.; Tang, W.S.; Zhou, Y.B.; Xu, Z.S.; Chen, J.; Li, H.Y.; Chen, M.; Ma, Y.Z. Nuclear transport factor GmNTF2B-1 enhances soybean drought tolerance by interacting with oxidoreductase GmOXR17 to reduce reactive oxygen species content. Plant J. 2021, 107, 740–759. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.W.; Wang, D.Y.; Shi, M.M.; Gong, Y.J.; Yin, S.W.; Jiao, Y.X.; Guo, S.J. Genome-wide identification of actin-depolymerizing factor gene family and their expression patterns under various abiotic stresses in soybean (Glycine max). Front. Plant Sci. 2023, 14, 1236175. [Google Scholar] [CrossRef]
- Ge, Y.; Li, Y.; Zhu, Y.M.; Bai, X.; Lv, D.K.; Guo, D.; Ji, W.; Cai, H. Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol. 2010, 10, 153. [Google Scholar] [CrossRef]
- Zhao, J.Y.; Lu, Z.W.; Sun, Y.; Fang, Z.W.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Xu, Z.S.; Min, D.H. The ankyrin-repeat gene GmANK114 confers drought and salt tolerance in Arabidopsis and soybean. Front. Plant Sci. 2020, 11, 584167. [Google Scholar] [CrossRef]
- Gao, S.Q.; Chen, M.; Xu, Z.S.; Zhao, C.P.; Li, L.; Xu, H.J.; Tang, Y.M.; Zhao, X.; Ma, Y.Z. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 2011, 75, 537–553. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.Y.; Cheng, X.G.; Xu, Z.S.; Li, L.C.; Ye, X.G.; Xia, L.Q.; Ma, Y.Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Su, H.G.; Li, B.; Song, X.Y.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Min, D.H.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the DYW subgroup PPR gene family and identification of GmPPR4 responses to drought stress. Int. J. Mol. Sci. 2019, 20, 5667. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Leng, Z.X.; Liu, Y.; Chen, Z.Y.; Guo, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Xu, Z.S.; Cui, X.Y. Genome-wide analysis of the DUF4228 family in soybean and functional identification of GmDUF4228-70 in response to drought and salt stresses. Front. Plant Sci. 2021, 12, 628299. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, W.X.; Qin, M.Y.; Li, S.; Qiao, M.; Liu, Z.H.; Xiang, F.N. Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017, 58, 1764–1776. [Google Scholar] [CrossRef]
- Anjum, S.A.; Ashraf, U.; Zohaib, A.; Tanveer, M.; Nazir, U. Growth and developmental responses of crop plants under drought stress: A review. Zemdirbyste 2017, 104, 267–276. [Google Scholar] [CrossRef]
- Manavalan, L.P.; Guttikonda, S.K.; Tran, L.S.; Nguyen, H.T. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009, 50, 1260–1276. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, J.X.; Deng, Y.J.; Xu, Z.S.; Xiong, A.S. Overexpression of a carrot BCH gene, DcBCH1, improves tolerance to drought in Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 475. [Google Scholar] [CrossRef] [PubMed]
- Singer, W.M.; Lee, Y.C.; Shea, Z.; Vieira, C.C.; Lee, D.; Li, X.; Cunicelli, M.; Kadam, S.S.; Khan, M.A.W.; Shannon, G.; et al. Soybean genetics, genomics, and breeding for improving nutritional value and reducing antinutritional traits in food and feed. Plant Genome 2023, 16, e20415. [Google Scholar] [CrossRef]
- Yamada, M.; Morishita, H.; Urano, K.; Shiozaki, N.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Yoshiba, Y. Effects of free proline accumulation in petunias under drought stress. J. Exp. Bot. 2005, 56, 1975–1981. [Google Scholar] [CrossRef]
- Abrahám, E.; Hourton-Cabassa, C.; Erdei, L.; Szabados, L. Methods for determination of proline in plants. Methods Mol. Biol. 2010, 639, 317–331. [Google Scholar] [CrossRef]
- Lappalainen, P.; Kessels, M.M.; Cope, M.J.; Drubin, D.G. The ADF homology (ADF-H) domain: A highly exploited actin-binding module. Mol. Biol. Cell 1998, 9, 1951–1959. [Google Scholar] [CrossRef]
- Giehl, R.F.; von Wirén, N. Root nutrient foraging. Plant Physiol. 2014, 166, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Azmat, R.; Moin, S. The remediation of drought stress under VAM inoculation through proline chemical transformation action. J. Photochem. Photobiol. B 2019, 193, 155–161. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Shaffique, S.; Imran, M.; adhikari, A.; Aaqil khan, M.; Rahim, W.; Alomrani, S.O.; Yun, B.-W.; kang, S.-M.; Lee, I.-J. A newly isolated Bacillus pumilus strain SH-9 modulates response to drought stress in soybean via endogenous phytohormones and gene expression (Daegu, South Korea). Plant Stress 2023, 10, 100279. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, Z.; Yao, W.; Gao, Y.; Fan, G.; Guo, Q.; Zhou, B.; Jiang, T. Overexpression of PagERF072 from poplar improves salt tolerance. Int. J. Mol. Sci. 2022, 23, 707. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.M.; Pastori, G.M.; Driscoll, S.; Groten, K.; Bernard, S.; Foyer, C.H. Drought controls on H2O2 accumulation, catalase (CAT) activity and CAT gene expression in wheat. J. Exp. Bot. 2005, 56, 417–423. [Google Scholar] [CrossRef]
- Anjum, N.A.; Sofo, A.; Scopa, A.; Roychoudhury, A.; Gill, S.S.; Iqbal, M.; Lukatkin, A.S.; Pereira, E.; Duarte, A.C.; Ahmad, I. Lipids and proteins—major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. Res. Int. 2015, 22, 4099–4121. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Wang, F.F.; Sun, X.; Shi, X.Y.; Zhai, H.; Tian, C.G.; Kong, F.J.; Liu, B.H.; Yuan, X.H. A global analysis of the polygalacturonase gene family in soybean (Glycine max). PLoS ONE 2016, 11, e0163012. [Google Scholar] [CrossRef]
- Wang, D.Y.; Du, M.X.; Li, J.Y.; Yin, S.W.; Sun, Y.W.; Guo, S.J. Overexpression of GhSWEET42, a SWEET-like gene from cotton, enhances the oil content and seed size. Biotechnol. Biotechnol. Equip. 2023, 37, 2266529. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Teng, C.; Lyu, K.D.; Li, Q.Q.; Li, N.; Lyu, S.H.; Fan, Y.L. An efficient and reproducible method for rroducing composite plants by Agrobacterium rhizogenes-based hairy root transformation. J. Vis. Exp. 2023, 196, e65688. [Google Scholar] [CrossRef]
- Meher; Shivakrishna, P.; Ashok Reddy, K.; Manohar Rao, D. Effect of PEG-6000 imposed drought stress on RNA content, relative water content (RWC), and chlorophyll content in peanut leaves and roots. Saudi J. Biol. Sci. 2018, 25, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Rajewicz, P.A.; Zhang, C.; Atherton, J.; Van, W.S.; Riikonen, A.; Magney, T.; Fernandez-Marin, B.; Plazaola, J.I.G.; Porcar-Castell, A. The photosynthetic response of spectral chlorophyll fluorescence differs across species and light environments in a boreal forest ecosystem. Agric. For. Meteorol. 2023, 334, 109434. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Du, M.; Lyu, P.; Li, J.; Meng, H.; Liu, X.; Shi, M.; Gong, Y.; Sha, Q.; Men, Q.; et al. Functional Characterization of the Soybean Glycine max Actin Depolymerization Factor GmADF13 for Plant Resistance to Drought Stress. Plants 2024, 13, 1651. https://doi.org/10.3390/plants13121651
Wang D, Du M, Lyu P, Li J, Meng H, Liu X, Shi M, Gong Y, Sha Q, Men Q, et al. Functional Characterization of the Soybean Glycine max Actin Depolymerization Factor GmADF13 for Plant Resistance to Drought Stress. Plants. 2024; 13(12):1651. https://doi.org/10.3390/plants13121651
Chicago/Turabian StyleWang, Deying, Mengxue Du, Peng Lyu, Jingyu Li, Huiran Meng, Xinxin Liu, Mengmeng Shi, Yujie Gong, Qi Sha, Qingmei Men, and et al. 2024. "Functional Characterization of the Soybean Glycine max Actin Depolymerization Factor GmADF13 for Plant Resistance to Drought Stress" Plants 13, no. 12: 1651. https://doi.org/10.3390/plants13121651





