Overexpression of Wild Soybean Expansin Gene GsEXLB14 Enhanced the Tolerance of Transgenic Soybean Hairy Roots to Salt and Drought Stresses
Abstract
:1. Introduction
2. Results
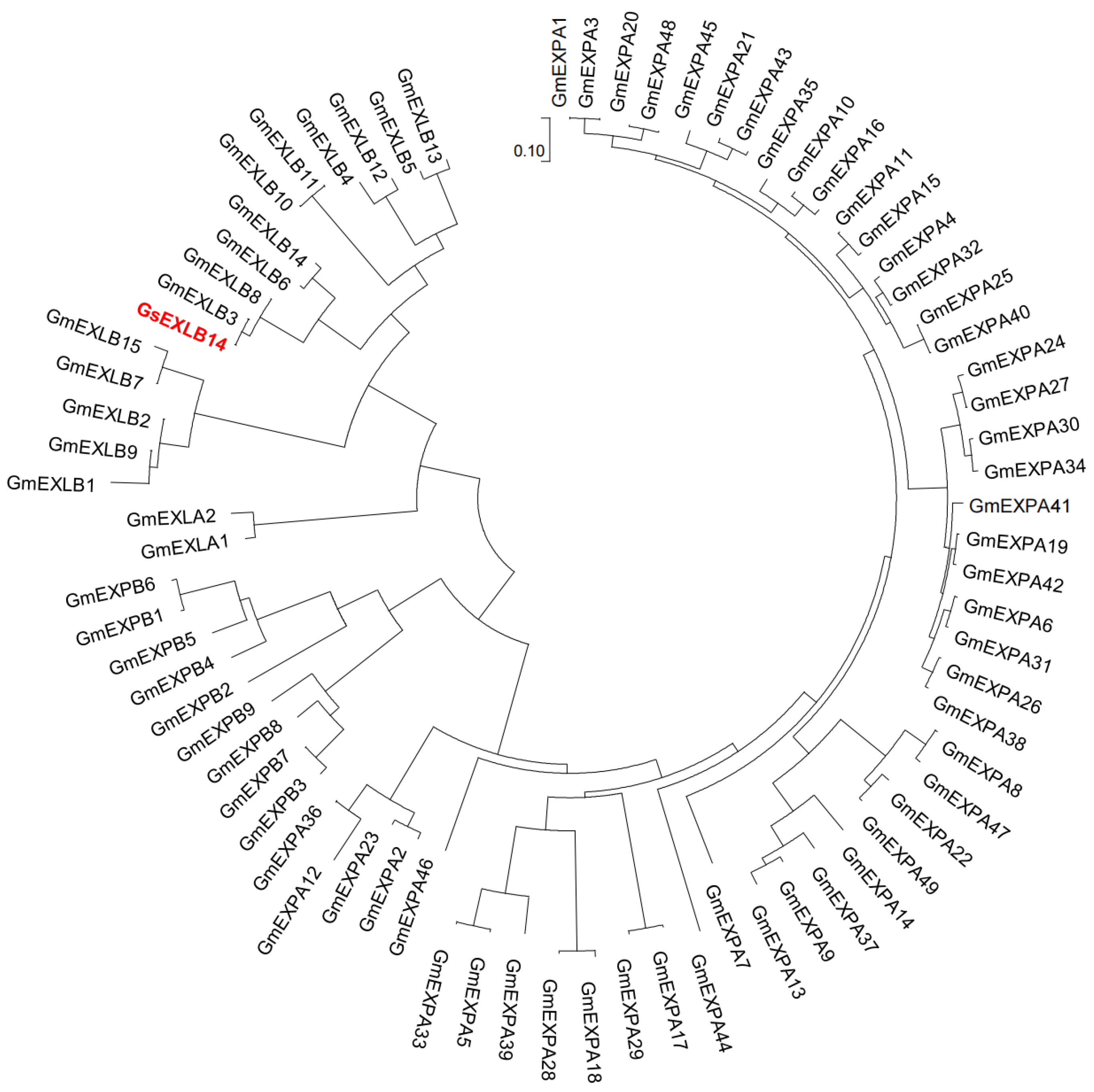
2.1. Basic Information of GsEXLB14 Amino Acid Sequence and Construction of Evolutionary Tree
2.2. Transcriptional Pattern of GsEXLB14 Gene under Salt and Drought Stress
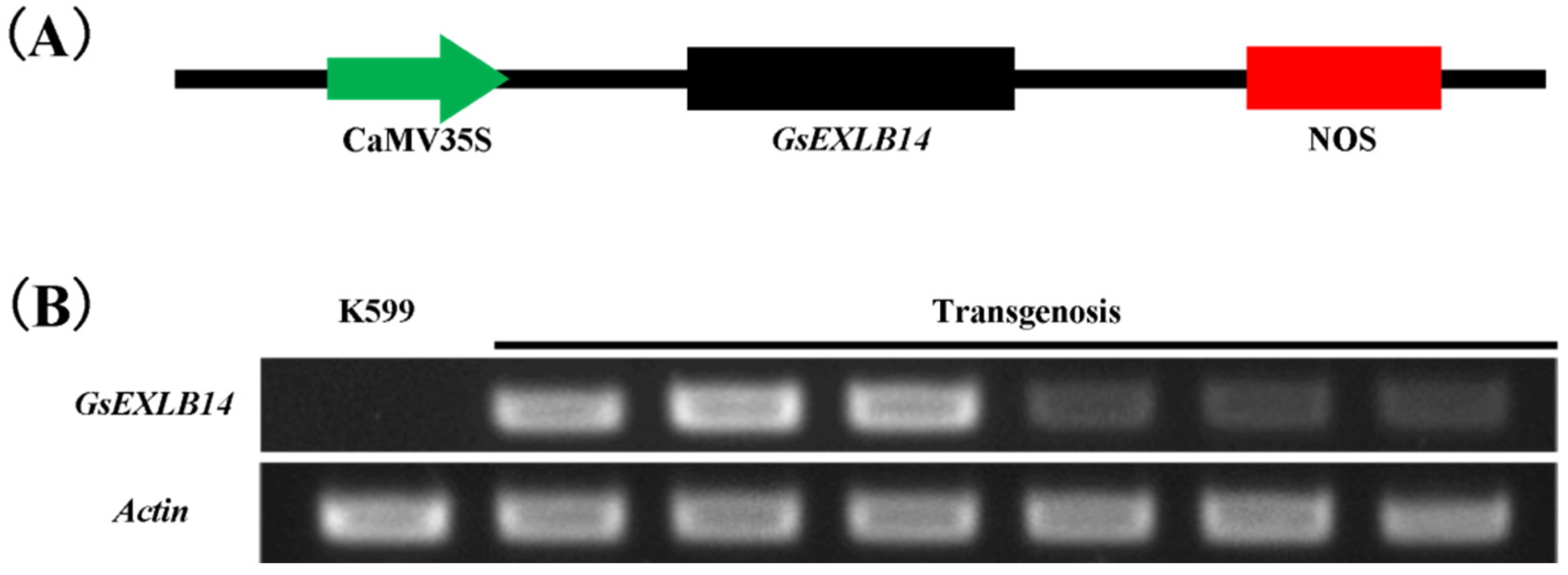
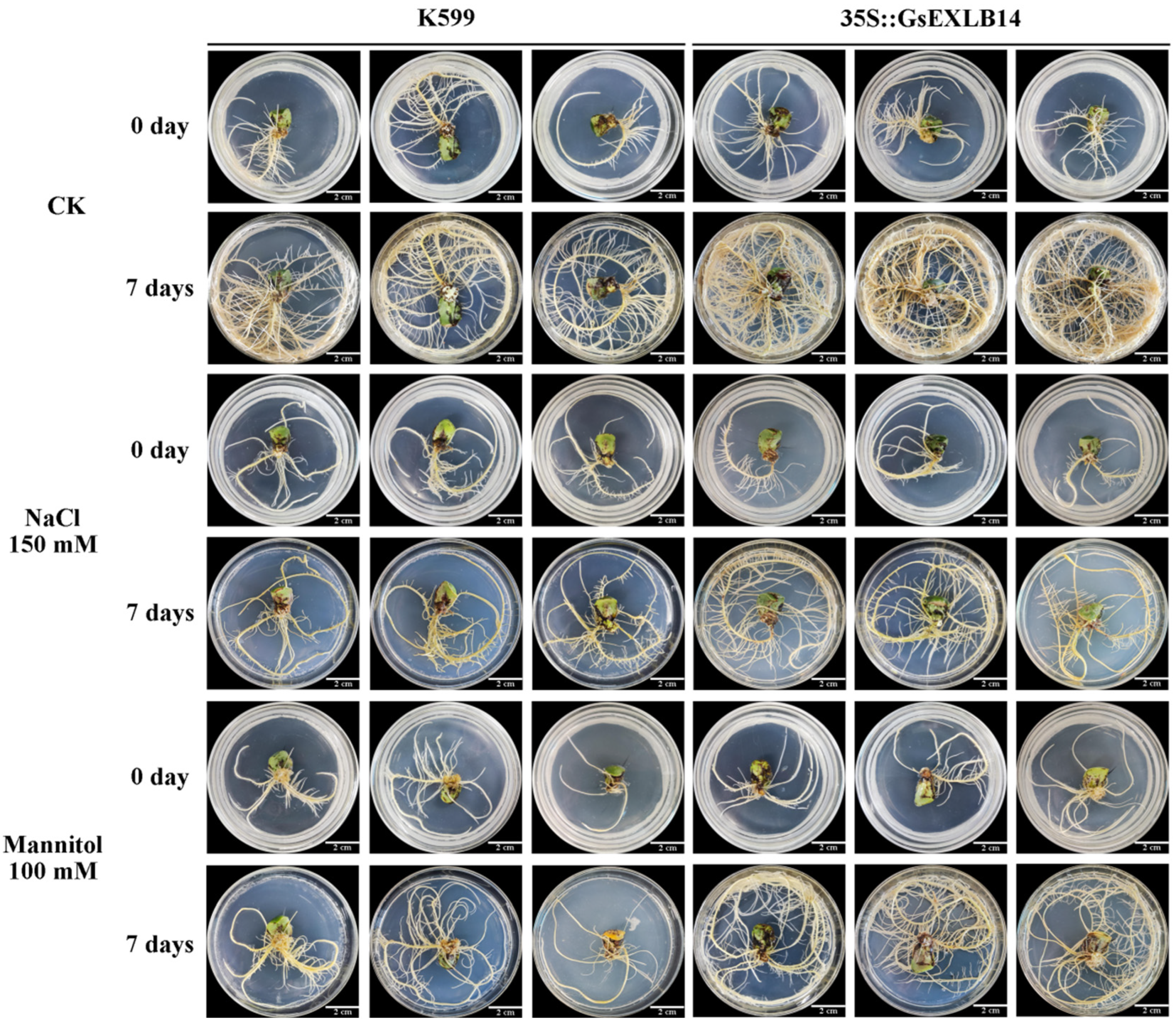
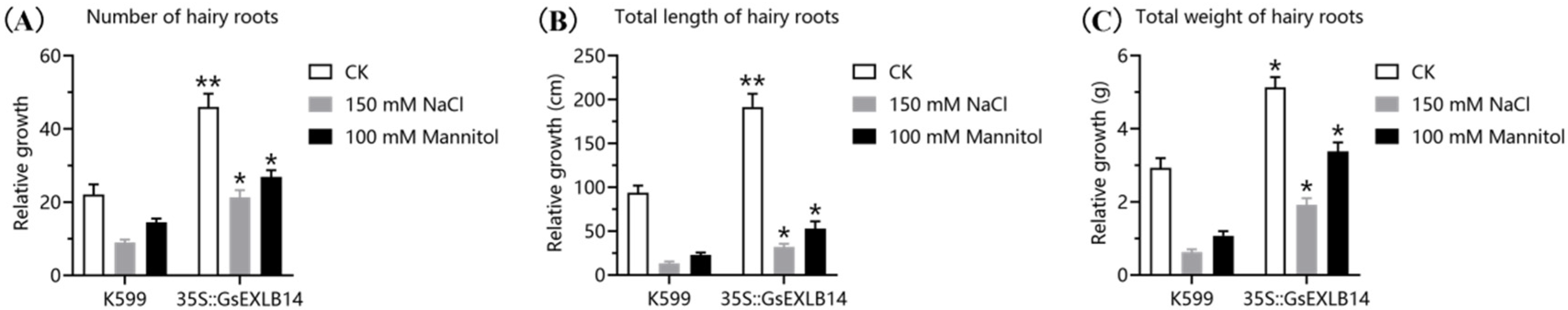
2.3. Observation on the Phenotype of Soybean Hairy Roots Overexpressing GsEXLB14 Gene
2.4. Transcriptome Analysis of Soybean Hairy Roots Overexpressing GsEXLB14 Gene
3. Discussion
3.1. Transcription Patterns of GsEXLB14
3.2. Potential Functions of GsEXLB14
4. Materials and Methods
4.1. Cloning of GsEXLB14 Gene
4.2. Transcriptional Pattern Analysis of GsEXLB14 Gene under Salt and Drought Stress
4.3. Construction of Overexpression Vector of GsEXLB14 Gene
4.4. GsEXLB14 Overexpression through Soybean Hairy Roots
4.5. Phenotypic Observation on Hairy Roots of Soybean Overexpressing GsEXLB14 Gene
4.6. Transcriptome Assay of Hairy Roots
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Guo, B.; Sun, L.; Jiang, S.; Ren, H.; Sun, R.; Wei, Z.; Hong, H.; Luan, X.; Wang, J.; Wang, X.; et al. Soybean genetic resources contributing to sustainable protein production. Theor. Appl. Genet. 2022, 135, 4095–4121. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef]
- Bisht, A.; Saini, D.K.; Kaur, B.; Batra, R.; Kaur, S.; Kaur, I.; Jindal, S.; Malik, P.; Sandhu, P.K.; Kaur, A.; et al. Multi-omics assisted breeding for biotic stress resistance in soybean. Mol. Biol. Rep. 2023, 50, 3787–3814. [Google Scholar] [CrossRef]
- Kofsky, J.; Zhang, H.; Song, B.H. The Untapped Genetic Reservoir: The Past, Current, and Future Applications of the Wild Soybean (Glycine soja). Front. Plant Sci. 2018, 9, 949. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Jia, B.; Sun, M.; Sun, X. Insights into the regulation of wild soybean tolerance to salt-alkaline stress. Front. Plant Sci. 2022, 13, 1002302. [Google Scholar] [CrossRef]
- You, H.; Liu, Y.; Minh, T.N.; Lu, H.; Zhang, P.; Li, W.; Xiao, J.; Ding, X.; Li, Q. Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja). J. Appl. Genet. 2020, 61, 489–501. [Google Scholar] [CrossRef] [PubMed]
- McQueen-Mason, S.; Durachko, D.M.; Cosgrove, D.J. Two endogenous proteins that induce cell wall extension in plants. Plant Cell 1992, 4, 1425–1433. [Google Scholar] [PubMed]
- Kök, B.Ö.; Celik Altunoglu, Y.; Öncül, A.B.; Karaci, A.; Cengiz Baloglu, M. Expansin gene family database: A comprehensive bioinformatics resource for plant expansin multigene family. J. Bioinform. Comput. Biol. 2023, 21, 2350015. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Yennawar, N.H.; Li, L.C.; Dudzinski, D.M.; Tabuchi, A.; Cosgrove, D.J. Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc. Natl. Acad. Sci. USA 2006, 103, 14664–14671. [Google Scholar] [CrossRef] [PubMed]
- Samalova, M.; Gahurova, E.; Hejatko, J. Expansin-mediated developmental and adaptive responses: A matter of cell wall biomechanics? Quant. Plant Biol. 2022, 3, e11. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Bradford, K.J. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 2000, 124, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.T.; Cosgrove, D.J. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 2002, 14, 3237–3253. [Google Scholar] [CrossRef] [PubMed]
- Boron, A.K.; Van Loock, B.; Suslov, D.; Markakis, M.N.; Verbelen, J.P.; Vissenberg, K. Over-expression of AtEXLA2 alters etiolated arabidopsis hypocotyl growth. Ann. Bot. 2015, 115, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zheng, J.; Zhou, H.; Chen, S.; Gao, Z.; Yang, Y.; Li, X.; Liao, H. The soybean β-expansin gene GmINS1 contributes to nodule development in response to phosphate starvation. Physiol. Plant. 2021, 172, 2034–2047. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, J.; Walk, T.C.; Liao, H. Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment. Appl. Microbiol. Biotechnol. 2014, 98, 2805–2817. [Google Scholar] [CrossRef]
- Green, P.B. Expression of pattern in plants: Combining molecular and calculus-based biophysical paradigms. Am. J. Bot. 1999, 86, 1059–1076. [Google Scholar] [CrossRef]
- Wei, P.C.; Zhang, X.Q.; Zhao, P.; Wang, X.C. Regulation of stomatal opening by the guard cell expansin AtEXPA1. Plant Signal. Behav. 2011, 6, 740–742. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Lee, Y.; Cho, H.T.; Kende, H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 2003, 15, 1386–1398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yan, H.; Chen, W.; Liu, J.; Jiang, C.; Jiang, H.; Zhu, S.; Cheng, B. Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol. Genet. Genom. 2014, 289, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.K.; Lee, H.H.; Bennett, A.B. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc. Natl. Acad. Sci. USA 1997, 94, 5955–5960. [Google Scholar] [CrossRef] [PubMed]
- Minoia, S.; Boualem, A.; Marcel, F.; Troadec, C.; Quemener, B.; Cellini, F.; Petrozza, A.; Vigouroux, J.; Lahaye, M.; Carriero, F.; et al. Induced mutations in tomato SlExp1 alter cell wall metabolism and delay fruit softening. Plant Sci. 2016, 242, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Calderini, D.F.; Castillo, F.M.; Arenas-M, A.; Molero, G.; Reynolds, M.P.; Craze, M.; Bowden, S.; Milner, M.J.; Wallington, E.J.; Dowle, A.; et al. Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential. New Phytol. 2021, 230, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, H.; Li, J.; Cao, Z.; Deng, B.; Liu, X.; Qin, G. Identification of Candidate Expansin Genes Associated with Seed Weight in Pomegranate (Punica granatum L.). Genes 2024, 15, 212. [Google Scholar] [CrossRef] [PubMed]
- Lü, P.; Kang, M.; Jiang, X.; Dai, F.; Gao, J.; Zhang, C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 2013, 237, 1547–1559. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Wu, D.; Zhao, H.; Gong, L.; Xu, J. Regulation of SmEXPA13 expression by SmMYB1R1-L enhances salt tolerance in Salix matsudana Koidz. Int. J. Biol. Macromol. 2024, 270 Pt 1, 132292. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, B.; Li, C.; Lei, C.; Kong, C.; Yang, Y.; Gong, M. A comprehensive expression analysis of the expansin gene family in potato (Solanum tuberosum) discloses stress-responsive expansin-like B genes for drought and heat tolerances. PLoS ONE 2019, 14, e0219837. [Google Scholar] [CrossRef]
- Feng, X.; Xu, Y.; Peng, L.; Yu, X.; Zhao, Q.; Feng, S.; Zhao, Z.; Li, F.; Hu, B. TaEXPB7-B, a β-expansin gene involved in low-temperature stress and abscisic acid responses, promotes growth and cold resistance in Arabidopsis thaliana. J. Plant Physiol. 2019, 240, 153004. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Chen, Y.; An, J.; Zhao, Z.; Zhang, G.; Wang, Y.; Wang, W. Wheat expansin gene TaEXPA2 is involved in conferring plant tolerance to Cd toxicity. Plant Sci. 2018, 270, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xie, J.; Liao, H.; Wang, X. Overexpression of β-expansin gene GmEXPB2 improves phosphorus efficiency in soybean. Physiol. Plant. 2014, 150, 194–204. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, G.; An, J.; Li, Q.; Chen, Y.; Zhao, X.; Wu, J.; Wang, Y.; Hao, Q.; Wang, W.; et al. Expansin gene TaEXPA2 positively regulates drought tolerance in transgenic wheat (Triticum aestivum L.). Plant science 2020, 298, 110596. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Li, C.; He, F.; Xu, Y.; Li, L.; Wang, X.; Chen, Q.; Li, F. Genome-Wide Identification of Expansin Genes in Wild Soybean (Glycine soja) and Functional Characterization of Expansin B1 (GsEXPB1) in Soybean Hair Root. Int. J. Mol. Sci. 2022, 23, 5407. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Lin, Y.; Wang, C.; Lu, J.; Liu, Z.; He, Z.; Shu, X.; Chen, W.; Wu, R.; Li, B.; et al. Expansin SlExp1 and endoglucanase SlCel2 synergistically promote fruit softening and cell wall disassembly in tomato. Plant Cell 2024, 36, 709–726. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, W.; Miao, Y.; Peters, M.; Schultze-Kraft, R.; Liu, G.; Chen, Z. Development of transgenic composite Stylosanthes plants to study root growth regulated by a β-expansin gene, SgEXPB1, under phosphorus deficiency. Plant Cell Rep. 2023, 42, 575–585. [Google Scholar] [CrossRef]
- Chen, S.; Luo, Y.; Wang, G.; Feng, C.; Li, H. Genome-wide identification of expansin genes in Brachypodium distachyon and functional characterization of BdEXPA27. Plant Sci. 2020, 296, 110490. [Google Scholar] [CrossRef]
- Won, S.K.; Choi, S.B.; Kumari, S.; Cho, M.; Lee, S.H.; Cho, H.T. Root hair-specific EXPANSIN B genes have been selected for Graminaceae root hairs. Mol. Cells 2010, 30, 369–376. [Google Scholar] [CrossRef]
- He, X.; Zeng, J.; Cao, F.; Ahmed, I.M.; Zhang, G.; Vincze, E.; Wu, F. HvEXPB7, a novel β-expansin gene revealed by the root hair transcriptome of Tibetan wild barley, improves root hair growth under drought stress. J. Exp. Bot. 2015, 66, 7405–7419. [Google Scholar] [CrossRef]
- Zou, X.; Liu, L.; Hu, Z.; Wang, X.; Zhu, Y.; Zhang, J.; Li, X.; Kang, Z.; Lin, Y.; Yin, C. Salt-induced inhibition of rice seminal root growth is mediated by ethylene-jasmonate interaction. J. Exp. Bot. 2021, 72, 5656–5672. [Google Scholar] [CrossRef]
- Kong, Y.; Wang, B.; Du, H.; Li, W.; Li, X.; Zhang, C. GmEXLB1, a Soybean Expansin-Like B Gene, Alters Root Architecture to Improve Phosphorus Acquisition in Arabidopsis. Front. Plant Sci. 2019, 10, 808. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, J.; Tan, Z.; Zeng, R.; Liao, H. GmEXPB2, a Cell Wall β-Expansin, Affects Soybean Nodulation through Modifying Root Architecture and Promoting Nodule Formation and Development. Plant Physiol. 2015, 169, 2640–2653. [Google Scholar] [CrossRef]
- Basso, M.F.; Lourenço-Tessutti, I.T.; Moreira-Pinto, C.E.; Mendes, R.A.G.; Pereira, D.G.; Grandis, A.; Macedo, L.L.P.; Macedo, A.F.; Gomes, A.C.M.M.; Arraes, F.B.M.; et al. Overexpression of the GmEXPA1 gene reduces plant susceptibility to Meloidogyne incognita. Plant Cell Rep. 2023, 42, 137–152. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Structure and growth of plant cell walls. Nat. Rev. Mol. Cell Biol. 2024, 25, 340–358. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Ahn, J.H.; Song, S.K.; Choi, Y.D.; Lee, J.S. Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol. 2003, 131, 985–997. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Kende, H.; Bradford, K.; Brummell, D.; Cho, H.T.; Cosgrove, D.; Fleming, A.; Gehring, C.; Lee, Y.; McQueen-Mason, S.; Rose, J.; et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 2004, 55, 311–314. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Expanding wheat yields with expansin. New Phytol. 2021, 230, 403–405. [Google Scholar] [CrossRef]
- Chen, L.J.; Zou, W.S.; Fei, C.Y.; Wu, G.; Li, X.Y.; Lin, H.H.; Xi, D.H. α-Expansin EXPA4 Positively Regulates Abiotic Stress Tolerance but Negatively Regulates Pathogen Resistance in Nicotiana tabacum. Plant Cell Physiol. 2018, 59, 2317–2330. [Google Scholar] [CrossRef]
- Chen, Y.; Han, Y.; Kong, X.; Kang, H.; Ren, Y.; Wang, W. Ectopic expression of wheat expansin gene TaEXPA2 improved the salt tolerance of transgenic tobacco by regulating Na+ /K+ and antioxidant competence. Physiol. Plant. 2017, 159, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Jadamba, C.; Kang, K.; Paek, N.C.; Lee, S.I.; Yoo, S.C. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chang, L.; Sun, W.; Ullah, A.; Yang, X. Overexpression of an expansin-like gene, GhEXLB2 enhanced drought tolerance in cotton. Plant Physiol. Biochem. 2021, 162, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, A.; Malekpour, M.; Sobhanverdi, S. The Arabidopsis expansin gene (AtEXPA18) is capable to ameliorate drought stress tolerance in transgenic tobacco plants. Mol. Biol. Rep. 2021, 48, 5913–5922. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, M.; Kim, J.Y.; Yoon, E.K.; Kim, J.A.; Lee, S.I. BrEXLB1, a Brassica rapa Expansin-Like B1 Gene is Associated with Root Development, Drought Stress Response, and Seed Germination. Genes 2020, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Maia, R.T.; Silva, I.S.D.S.; Fernandes de Souza, A.; Frazão, N.F.; de Lima, R.M.; Campos, M.A. Miraculin-based sweeteners in the protein-engineering era: An alternative for developing more efficient and safer products. J. Biomol. Struct. Dyn. 2023, 1–9, advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef]
- Li, C.; Zhang, H.; Wang, X.; Liao, H. A comparison study of Agrobacterium-mediated transformation methods for root-specific promoter analysis in soybean. Plant Cell Rep. 2014, 33, 1921–1932. [Google Scholar] [CrossRef]

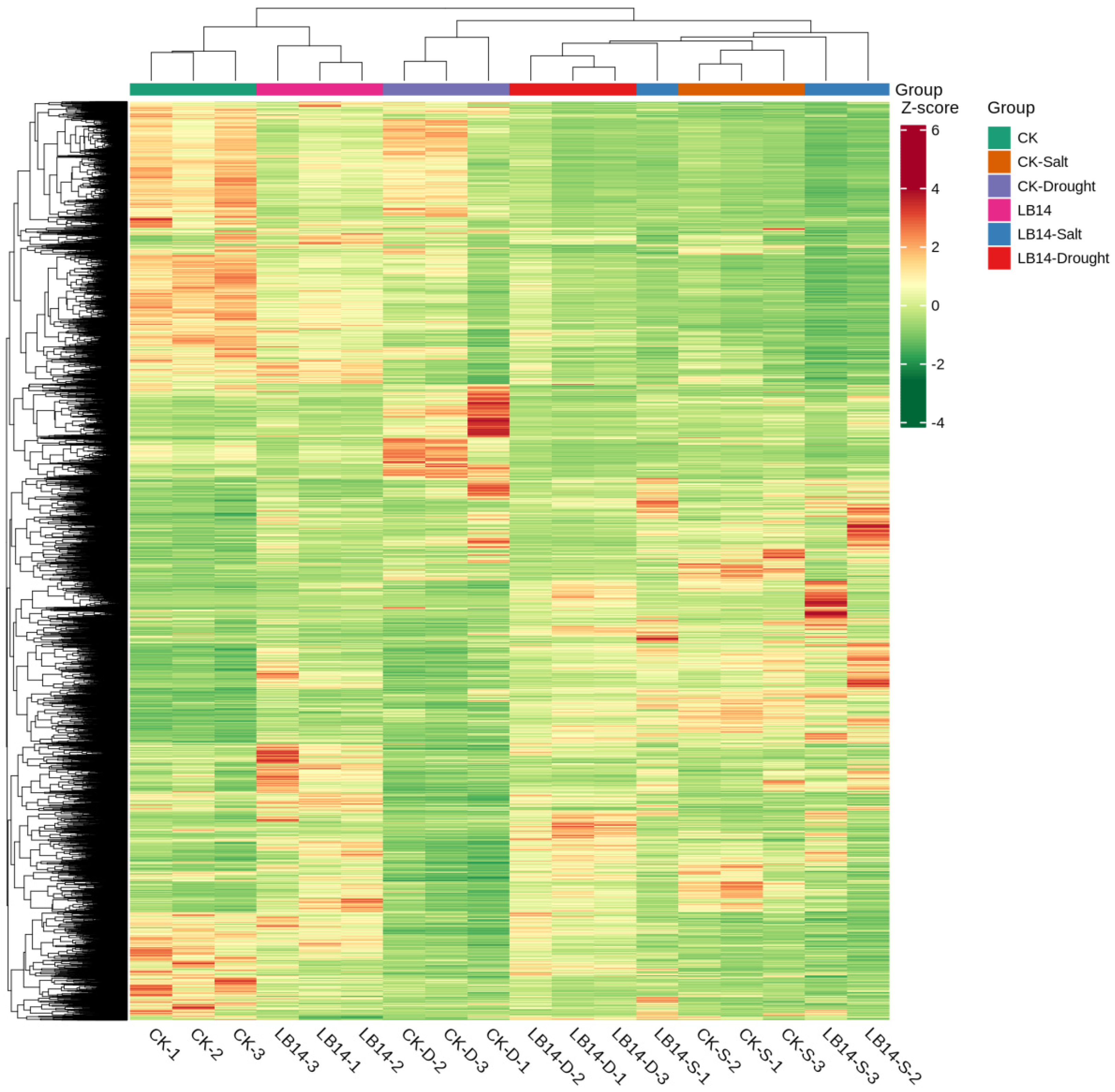
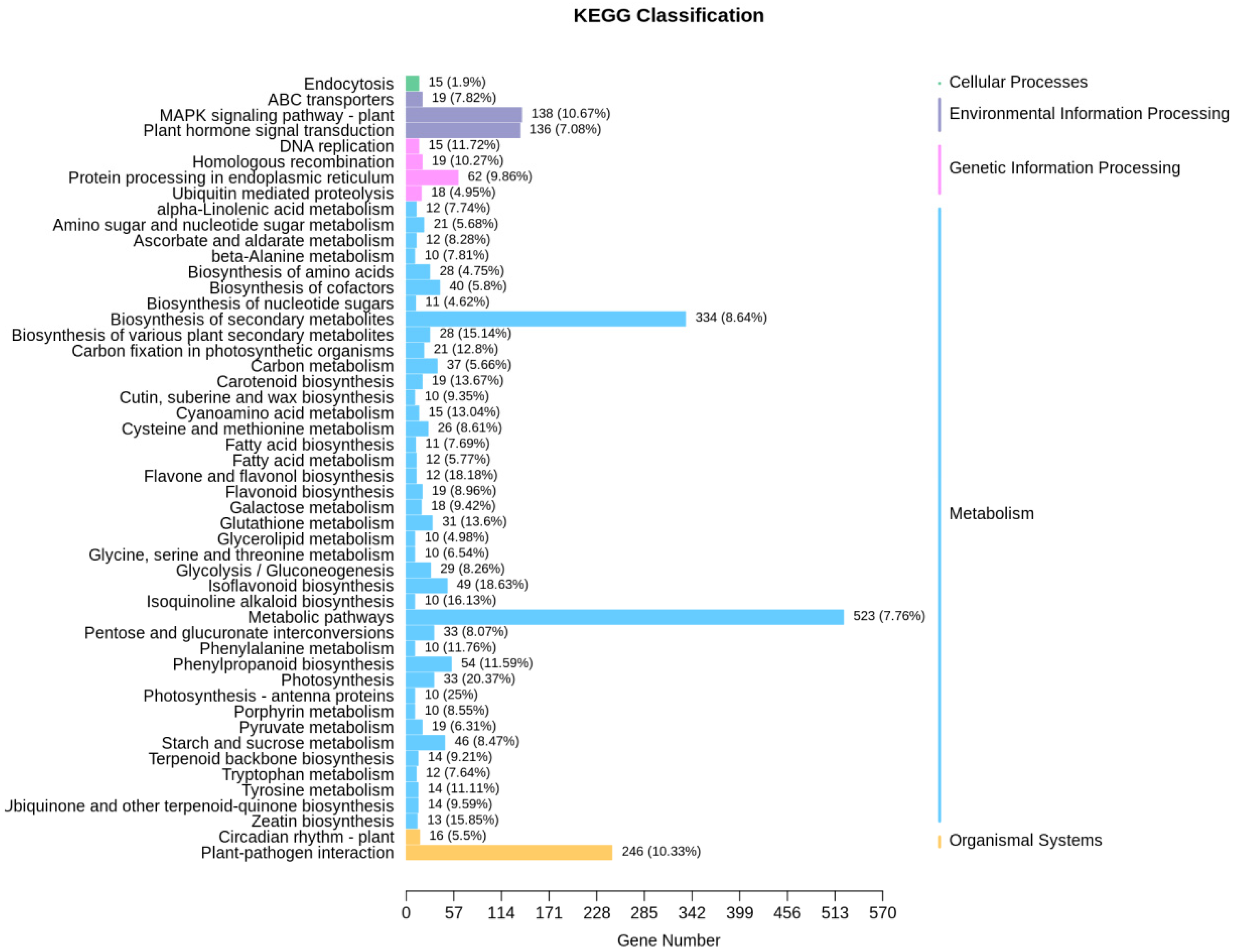
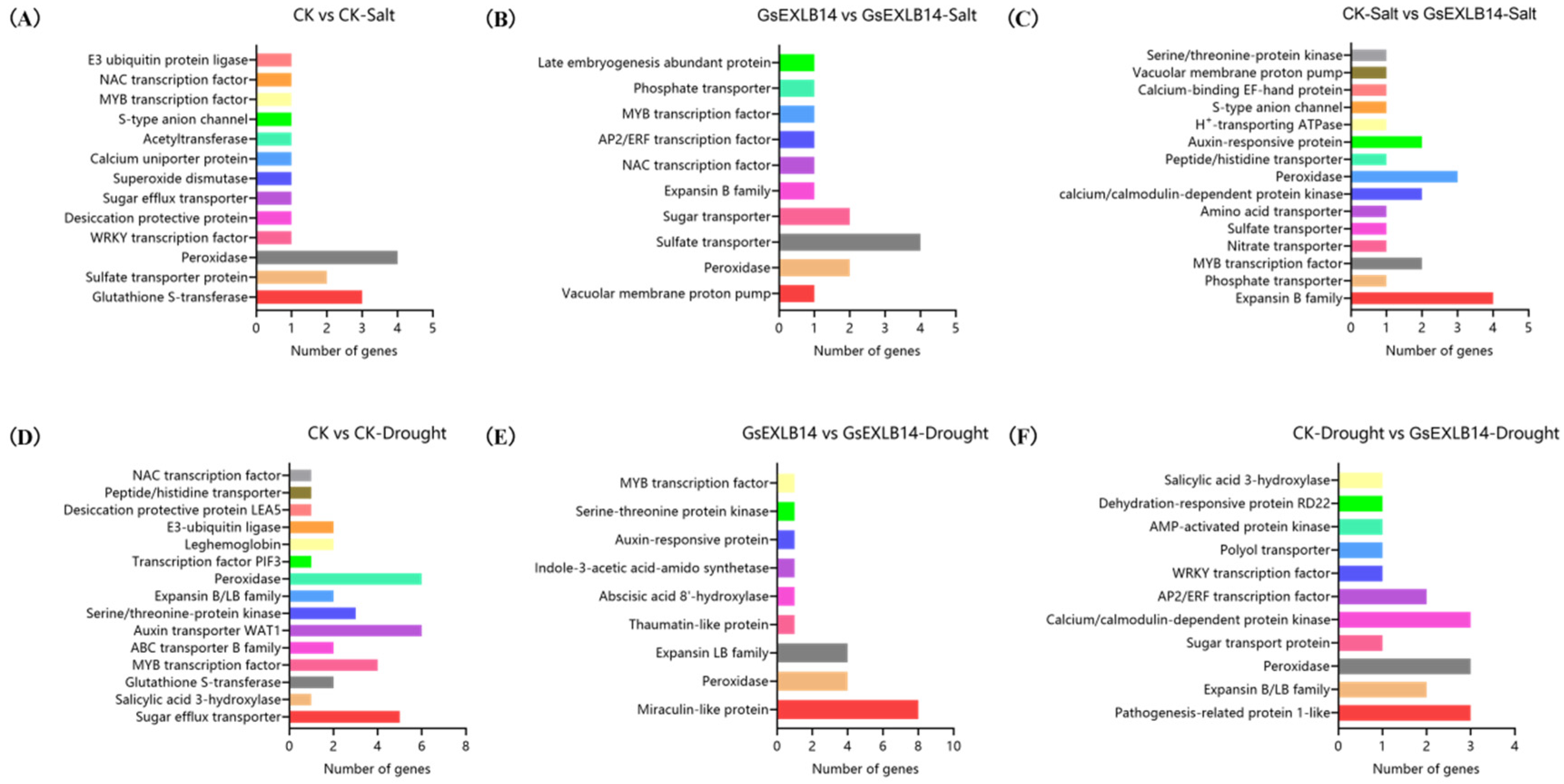
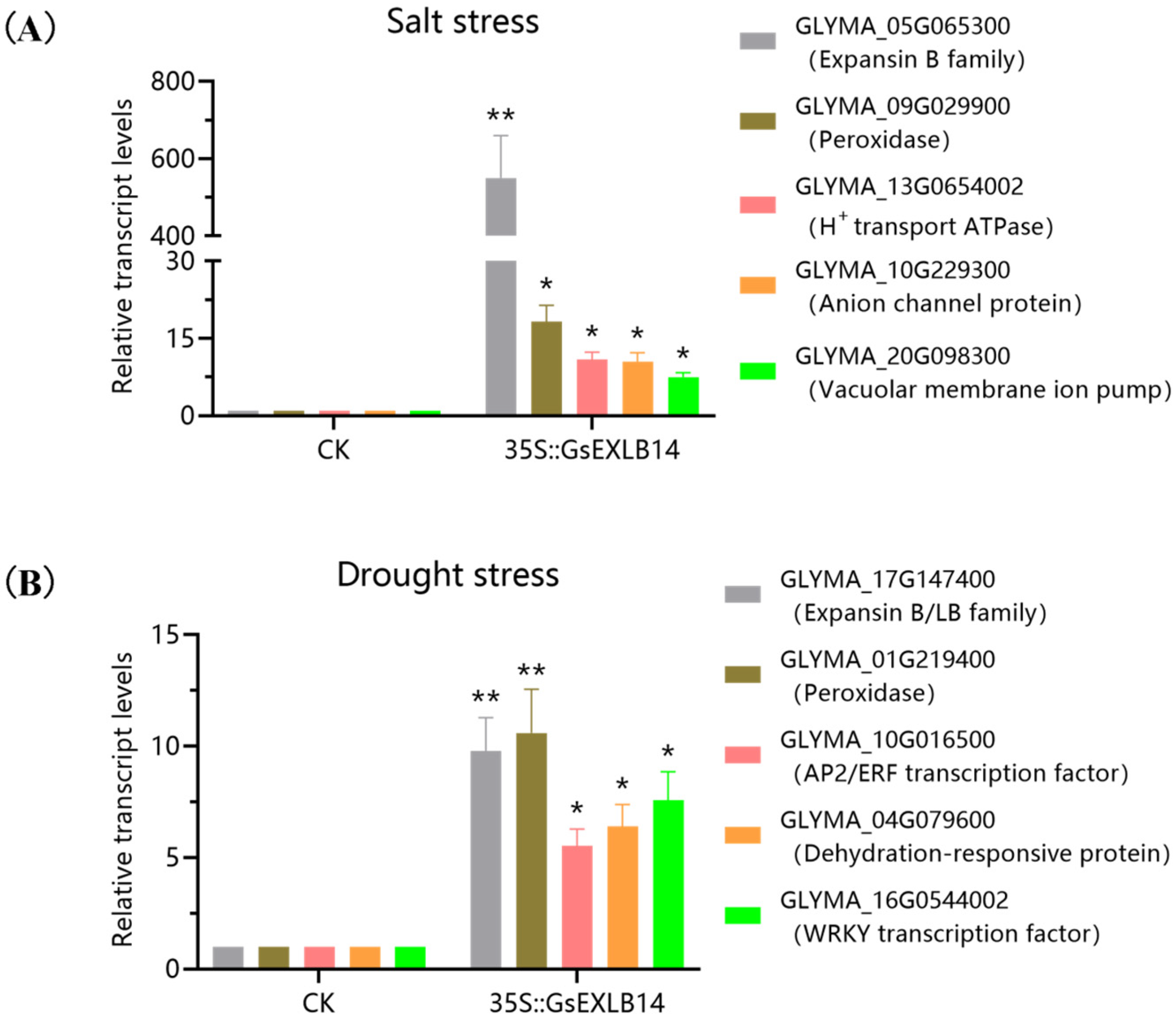
| Group | Total | Down | Up |
|---|---|---|---|
| CK_vs_CK-Drought | 4496 | 2836 | 1660 |
| CK_vs_CK-Salt | 7314 | 4054 | 3260 |
| CK_vs_LB14 | 3314 | 827 | 2487 |
| LB14_vs_LB14-Drought | 2412 | 1551 | 861 |
| LB14_vs_LB14-Salt | 4434 | 3307 | 1127 |
| CK-Drought_vs_LB14-Drought | 6551 | 2790 | 3761 |
| CK-Salt_vs_LB14-Salt | 3580 | 2231 | 1349 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhang, T.; Li, C.; Zhou, C.; Liu, B.; Wu, Y.; He, F.; Xu, Y.; Li, F.; Feng, X. Overexpression of Wild Soybean Expansin Gene GsEXLB14 Enhanced the Tolerance of Transgenic Soybean Hairy Roots to Salt and Drought Stresses. Plants 2024, 13, 1656. https://doi.org/10.3390/plants13121656
Wang L, Zhang T, Li C, Zhou C, Liu B, Wu Y, He F, Xu Y, Li F, Feng X. Overexpression of Wild Soybean Expansin Gene GsEXLB14 Enhanced the Tolerance of Transgenic Soybean Hairy Roots to Salt and Drought Stresses. Plants. 2024; 13(12):1656. https://doi.org/10.3390/plants13121656
Chicago/Turabian StyleWang, Linlin, Tong Zhang, Cuiting Li, Changjun Zhou, Bing Liu, Yaokun Wu, Fumeng He, Yongqing Xu, Fenglan Li, and Xu Feng. 2024. "Overexpression of Wild Soybean Expansin Gene GsEXLB14 Enhanced the Tolerance of Transgenic Soybean Hairy Roots to Salt and Drought Stresses" Plants 13, no. 12: 1656. https://doi.org/10.3390/plants13121656





