Analysis of 26S Proteasome Activity across Arabidopsis Tissues
Abstract
:1. Introduction
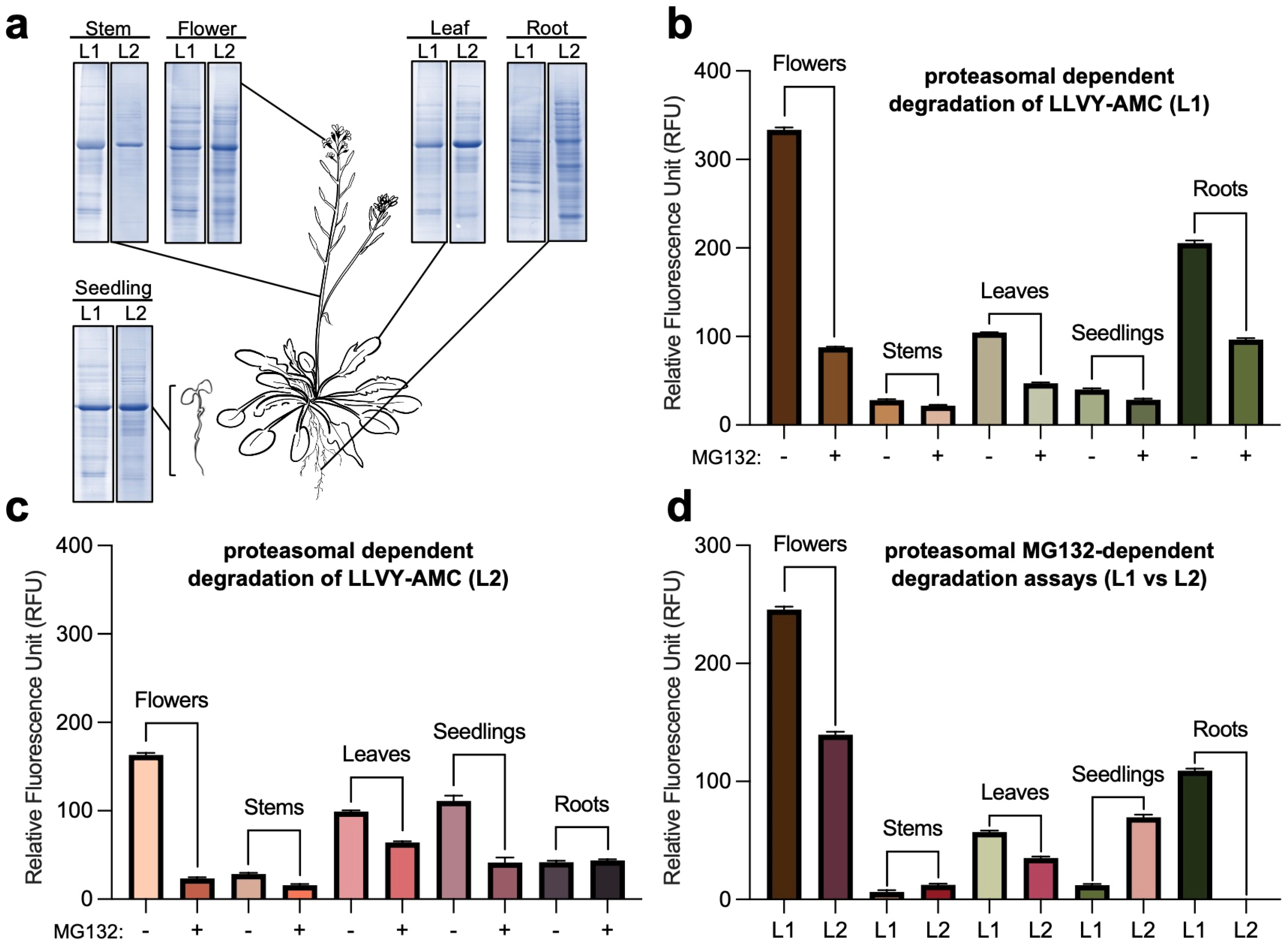
2. Results and Discussion
3. Conclusions
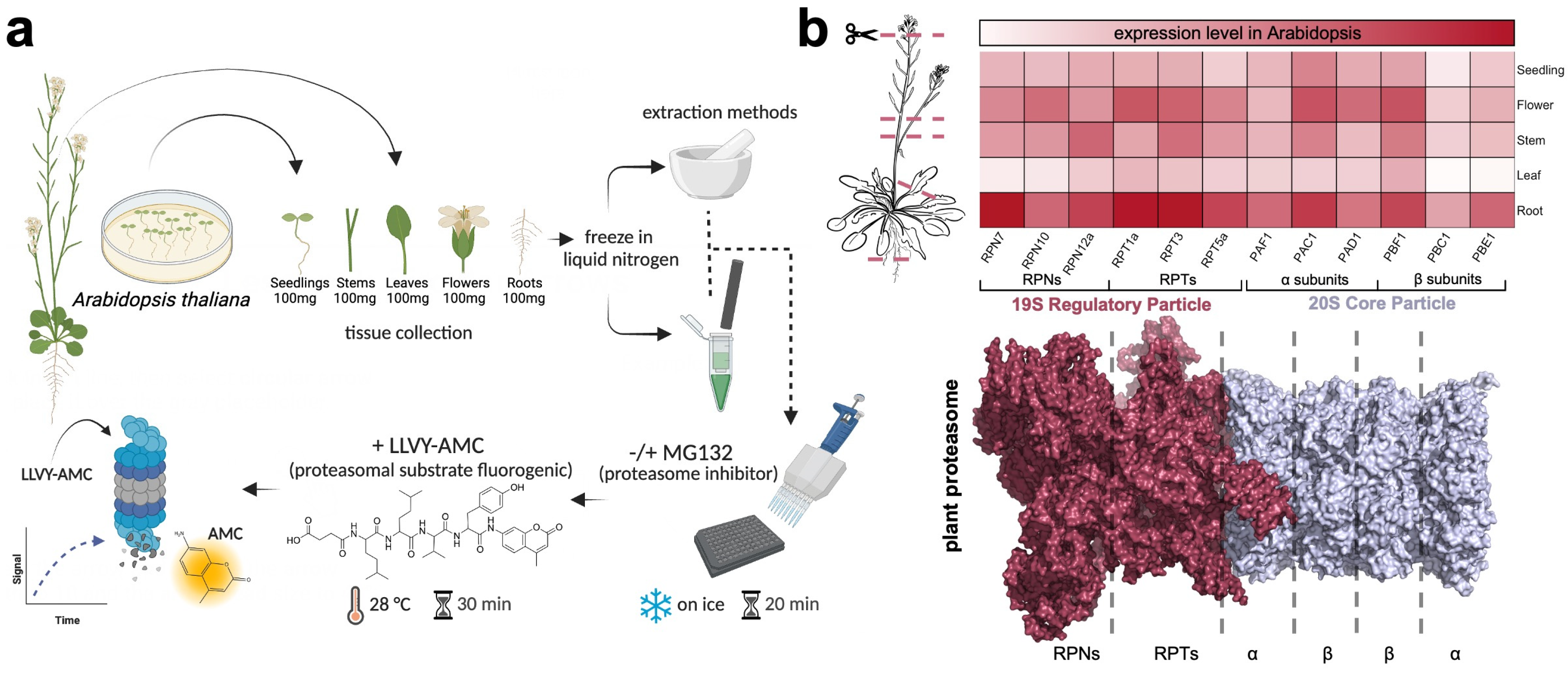
4. Methods
4.1. Materials and Equipment
4.2. Plant Growth Conditions
4.3. Plant Tissue Preparation
4.4. Proteasome Activity Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xu, F.Q.; Xue, H.W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef]
- He, D.; Damaris, R.N.; Li, M.; Khan, I.; Yang, P. Advances on Plant Ubiquitylome-From Mechanism to Application. Int. J. Mol. Sci. 2020, 21, 7909. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef]
- Ling, Q.; Jarvis, P. Plant Signaling: Ubiquitin Pulls the Trigger on Chloroplast Degradation. Curr. Biol. 2016, 26, R38–R40. [Google Scholar] [CrossRef]
- Trujillo, M.; Shirasu, K. Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 2010, 13, 402–408. [Google Scholar] [CrossRef]
- Oh, E.; Akopian, D.; Rape, M. Principles of Ubiquitin-Dependent Signaling. Annu. Rev. Cell Dev. Biol. 2018, 34, 137–162. [Google Scholar] [CrossRef]
- Voges, D.; Zwickl, P.; Baumeister, W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999, 68, 1015–1068. [Google Scholar] [CrossRef]
- Livneh, I.; Cohen-Kaplan, V.; Cohen-Rosenzweig, C.; Avni, N.; Ciechanover, A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016, 26, 869–885. [Google Scholar] [CrossRef]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Shabek, N.; Ciechanover, A. Degradation of ubiquitin: The fate of the cellular reaper. Cell Cycle 2010, 9, 523–530. [Google Scholar] [CrossRef]
- Kurepa, J.; Wang, S.; Li, Y.; Smalle, J. Proteasome regulation, plant growth and stress tolerance. Plant Signal. Behav. 2009, 4, 924–927. [Google Scholar] [CrossRef]
- Kurepa, J.; Smalle, J.A. Structure, function and regulation of plant proteasomes. Biochimie 2008, 90, 324–335. [Google Scholar] [CrossRef]
- Yanagawa, Y.; Kimura, S.; Takase, T.; Sakaguchi, K.; Umeda, M.; Komamine, A.; Tanaka, K.; Hashimoto, J.; Sato, T.; Nakagawa, H. Spatial distribution of the 26S proteasome in meristematic tissues and primordia of rice (Oryza sativa L.). Planta 2002, 214, 703–707. [Google Scholar] [CrossRef]
- Shibahara, T.; Kawasaki, H.; Hirano, H. Mass spectrometric analysis of expression of ATPase subunits encoded by duplicated genes in the 19S regulatory particle of rice 26S proteasome. Arch. Biochem. Biophys. 2004, 421, 34–41. [Google Scholar] [CrossRef]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Guo, X.; Dixon, J.E. The 26S proteasome: A cell cycle regulator regulated by cell cycle. Cell Cycle 2016, 15, 875–876. [Google Scholar] [CrossRef]
- Yu, Z.; Yu, Y.; Wang, F.; Myasnikov, A.G.; Coffino, P.; Cheng, Y. Allosteric coupling between alpha-rings of the 20S proteasome. Nat. Commun. 2020, 11, 4580. [Google Scholar] [CrossRef]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. [Google Scholar] [CrossRef]
- Kim, H.T.; Collins, G.A.; Goldberg, A.L. Measurement of the Multiple Activities of 26S Proteasomes. Methods Mol. Biol. 2018, 1844, 289–308. [Google Scholar] [CrossRef]
- Gan, J.; Leestemaker, Y.; Sapmaz, A.; Ovaa, H. Highlighting the Proteasome: Using Fluorescence to Visualize Proteasome Activity and Distribution. Front. Mol. Biosci. 2019, 6, 14. [Google Scholar] [CrossRef]
- Thibaudeau, T.A.; Smith, D.M. A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 2019, 71, 170–197. [Google Scholar] [CrossRef]
- Yang, P.; Fu, H.; Walker, J.; Papa, C.M.; Smalle, J.; Ju, Y.M.; Vierstra, R.D. Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 2004, 279, 6401–6413. [Google Scholar] [CrossRef]
- Tal, L.; Guercio, A.M.; Varshney, K.; Young, A.; Gutjahr, C.; Shabek, N. C-terminal conformational changes in SCF-D3/MAX2 ubiquitin ligase are required for KAI2-mediated signaling. New Phytol. 2023, 239, 2067–2075. [Google Scholar] [CrossRef]
- Tal, L.; Palayam, M.; Ron, M.; Young, A.; Britt, A.; Shabek, N. A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling. Nat. Plants 2022, 8, 561–573. [Google Scholar] [CrossRef]
- Shabek, N.; Ticchiarelli, F.; Mao, H.; Hinds, T.R.; Leyser, O.; Zheng, N. Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 2018, 563, 652–656. [Google Scholar] [CrossRef]
- Kandolf, S.; Grishkovskaya, I.; Belacic, K.; Bolhuis, D.L.; Amann, S.; Foster, B.; Imre, R.; Mechtler, K.; Schleiffer, A.; Tagare, H.D.; et al. Cryo-EM structure of the plant 26S proteasome. Plant Commun. 2022, 3, 100310. [Google Scholar] [CrossRef]
- Wang, H.; Dongen, J.T.V.; Schippers, J.H. Proteasomal activity is differentially regulated in source and sink tissues of Arabidopsis. bioRxiv 2022. [Google Scholar] [CrossRef]
- Dolde, U.; Muzzopappa, F.; Delesalle, C.; Neveu, J.; Erdel, F.; Vert, G. LEAFY homeostasis is regulated via ubiquitin-dependent degradation and sequestration in cytoplasmic condensates. iScience 2023, 26, 106880. [Google Scholar] [CrossRef]
- Linden, K.J.; Callis, J. The ubiquitin system affects agronomic plant traits. J. Biol. Chem. 2020, 295, 13940–13955. [Google Scholar] [CrossRef]
- Pineiro, M.; Jarillo, J.A. Ubiquitination in the control of photoperiodic flowering. Plant Sci. 2013, 198, 98–109. [Google Scholar] [CrossRef]
- Saleme, M.L.S.; Andrade, I.R.; Eloy, N.B. The Role of Anaphase-Promoting Complex/Cyclosome (APC/C) in Plant Reproduction. Front. Plant Sci. 2021, 12, 642934. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganapathy, J.; Hand, K.A.; Shabek, N. Analysis of 26S Proteasome Activity across Arabidopsis Tissues. Plants 2024, 13, 1696. https://doi.org/10.3390/plants13121696
Ganapathy J, Hand KA, Shabek N. Analysis of 26S Proteasome Activity across Arabidopsis Tissues. Plants. 2024; 13(12):1696. https://doi.org/10.3390/plants13121696
Chicago/Turabian StyleGanapathy, Jagadeesan, Katherine A. Hand, and Nitzan Shabek. 2024. "Analysis of 26S Proteasome Activity across Arabidopsis Tissues" Plants 13, no. 12: 1696. https://doi.org/10.3390/plants13121696




