Exploring Aluminum Tolerance Mechanisms in Plants with Reference to Rice and Arabidopsis: A Comprehensive Review of Genetic, Metabolic, and Physiological Adaptations in Acidic Soils
Abstract
:1. Introduction
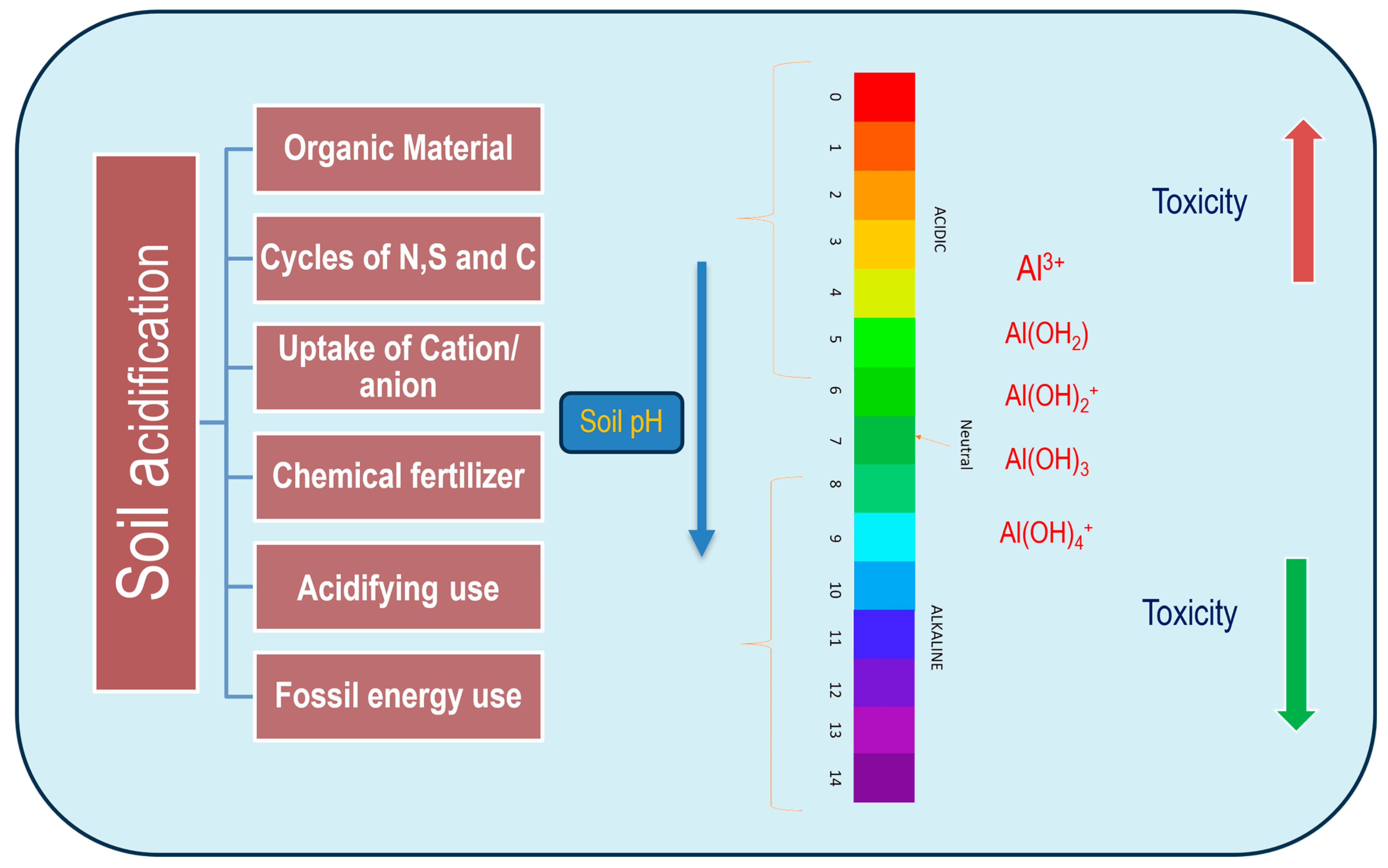
2. Aluminum Bioavailability in Different Forms to Sensitize the Plants
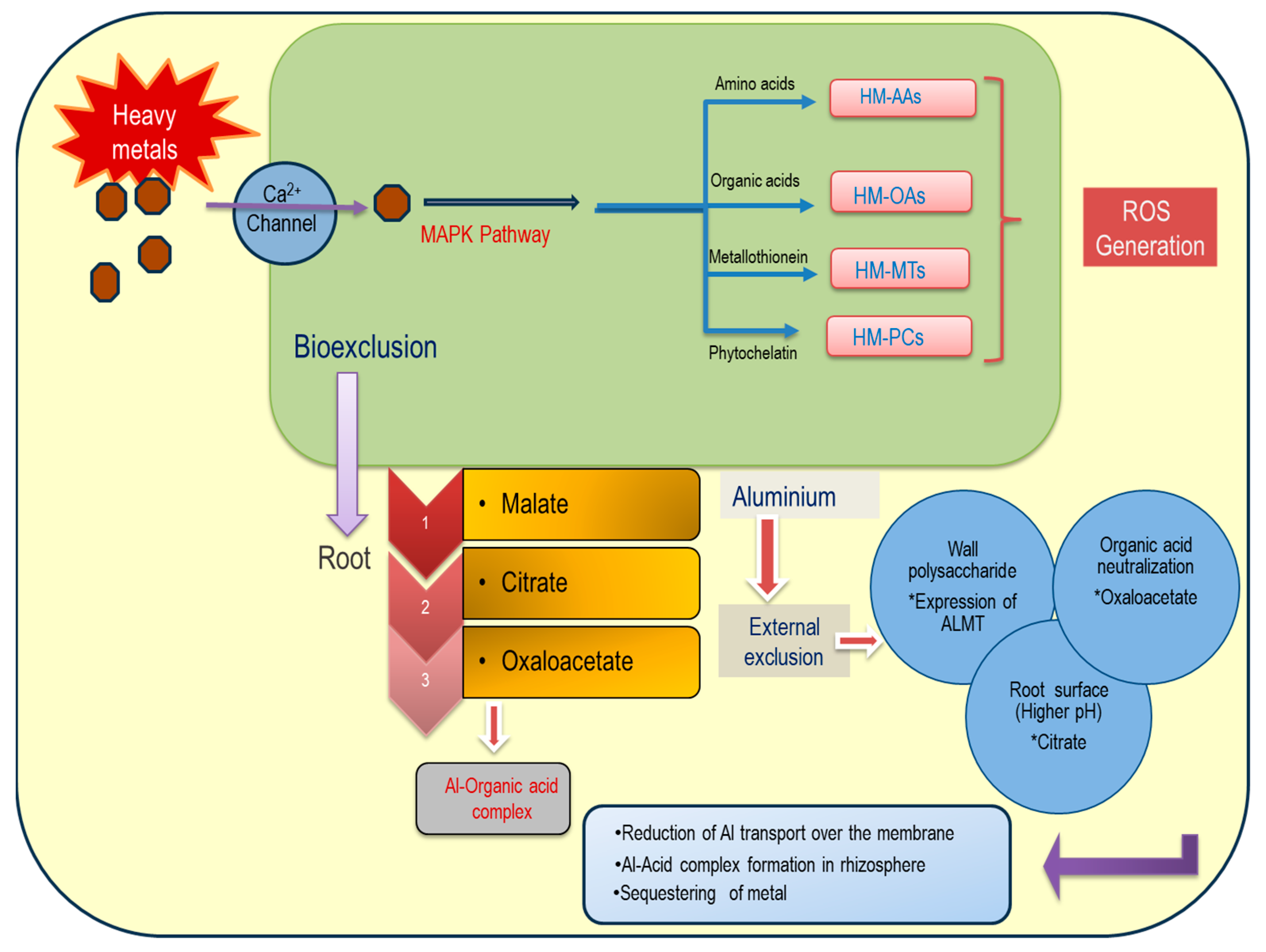
3. Aluminum-Induced Oxidative Stress and Metabolic Alterations
4. Metabolome Induction to Aluminum Responses in Plants
5. Root Phenotypes and Quantitative Trait Loci for Aluminum Toxicity
6. Metabolite Shifting from Central Carbon Metabolism
7. Aluminum-Induced Signaling for Reactive Oxygen Species Development
8. Comprehensive Genomics for Aluminum Toxicity
9. Special Metabolomic Pathways to Register Aluminum Toxicity
10. Special Metabolites and Their Contribution to Al Tolerance in Plants
11. Metabolomes under Regulation of Signal Transduction and Protein Turnover
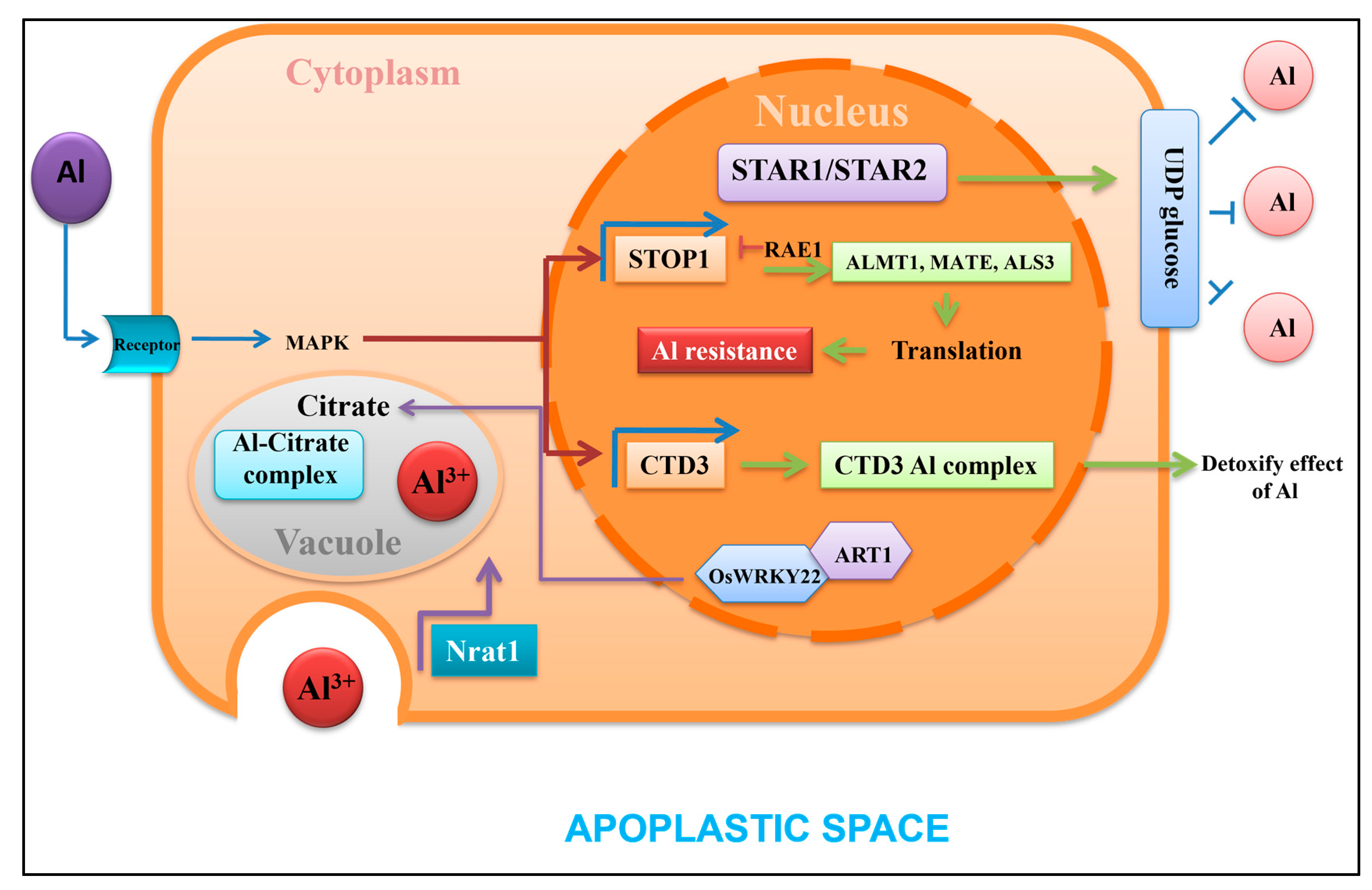
12. Transcriptional Control of Aluminum Tolerance in Roots under Acidic Cytosol
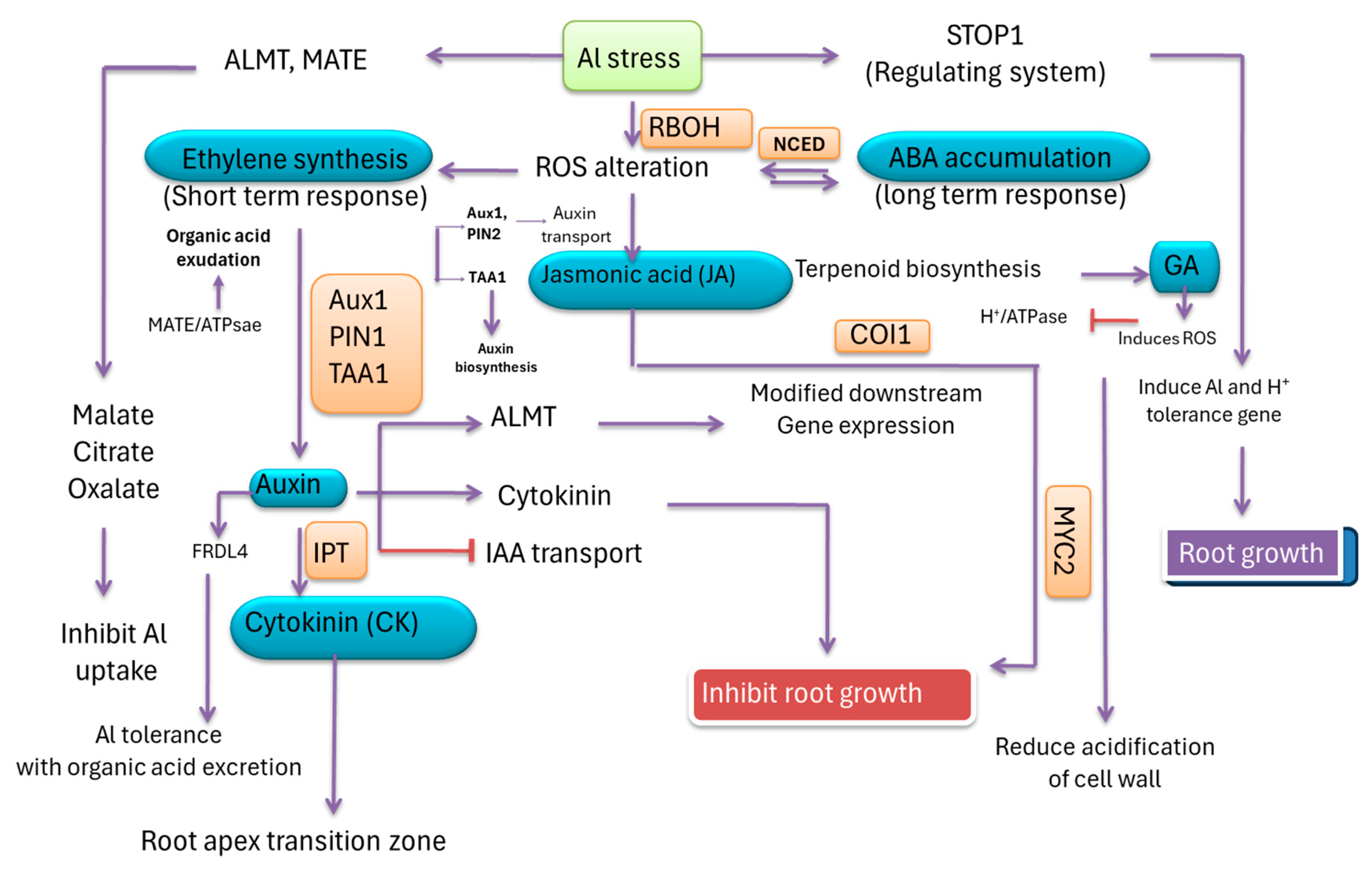
13. Phytohormone Signaling and Functioning for Aluminum Tolerance
14. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tyagi, W.; Yumnam, J.S.; Sen, D.; Rai, M. Root transcriptome reveals efficient cell signaling and energy conservation key to aluminum toxicity tolerance in acidic soil adapted rice genotype. Sci. Rep. 2020, 10, 4580. [Google Scholar] [CrossRef] [PubMed]
- Banet, T.; Massey, M.S.; Zohar, I.; Litaor, M.I.; Ippolito, J.A. Assessing modified aluminum-based water treatment residuals as a plant-available phosphorus source. Chemosphere 2020, 247, 125949. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, K.; AlKahtani, M.; Attia, K.; Hafez, Y.; Király, L.; Künstler, A. The role of plant growth-promoting bacteria in alleviating the adverse effects of drought on plants. Biology 2021, 10, 520. [Google Scholar] [CrossRef] [PubMed]
- Kar, D.; Pradhan, A.A.; Datta, S. The role of solute transporters in aluminum toxicity and tolerance. Physiol. Plant. 2021, 171, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Haghighi, T.M.; Saharkhiz, M.J.; Kavoosi, G.; Jowkar, A. Monitoring amino acid profile and protein quality of licorice (Glycyrrhiza glabra L.) under drought stress, silicon nutrition and mycorrhiza inoculation. Sci. Hortic. 2022, 295, 110808. [Google Scholar] [CrossRef]
- Rahman, R.; Upadhyaya, H. Aluminium toxicity and its tolerance in plant: A review. J. Plant. Biol. 2021, 64, 101–121. [Google Scholar] [CrossRef]
- Dhiman, S.; Khanna, K.; Kumar, P.; Bhardwaj, T.; Devi, K.; Sharma, N.; Sharma, P.; Arora, P.; Kapoor, N.; Sharma, A.; et al. Divulging molecular perspectives of plant defense machinery under heavy metal toxicity. J. Plant Growth Regul. 2023, 9, 1–37. [Google Scholar] [CrossRef]
- Ofoe, R.; Thomas, R.H.; Asiedu, S.K.; Wang-Pruski, G.; Fofana, B.; Abbey, L. Aluminum in plant: Benefits, toxicity and tolerance mechanisms. Front. Plant Sci. 2023, 13, 1085998. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.A.; Silva, M.F.; Wakin, T.; Nunes-Nesi, A.; Araújo, W.L. Metabolic and DNA checkpoints for the enhancement of Al tolerance. J. Hazard. Mater. 2022, 430, 128366. [Google Scholar] [CrossRef]
- Zarattini, M.; Corso, M.; Kadowaki, M.A.; Monclaro, A.; Magri, S.; Milanese, I.; Jolivet, S.; de Godoy, M.O.; Hermans, C.; Fagard, M.; et al. LPMO-oxidized cellulose oligosaccharides evoke immunity in Arabidopsis conferring resistance towards necrotrophic fungus B. cinerea. CommunBiol 2021, 4, 727. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Ashraf, M. Antioxidants as modulators of arsenic-induced oxidative stress tolerance in plants: An overview. J. Hazard. Mater. 2022, 427, 127891. [Google Scholar] [CrossRef] [PubMed]
- Che-Othman, M.H.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat mitochondrial respiration shifts from the tricarboxylic acid cycle to the GABA shunt under salt stress. New Phytol. 2020, 225, 1166–1180. [Google Scholar] [CrossRef] [PubMed]
- Szurman-Zubrzycka, M.; Chwiałkowska, K.; Niemira, M.; Kwaśniewski, M.; Nawrot, M.; Gajecka, M.; Larsen, P.B.; Szarejko, I. Aluminum or low pH–which is the bigger enemy of barley? transcriptome analysis of barley root meristem under Al and low pH stress. Front. Genet. 2021, 12, 675260. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.; Yang, Z.; Zhan, M.; Zheng, M.; You, J.; Meng, X.; Li, H.; Gao, J. Two Sweet Sorghum (Sorghum bicolor L.) WRKY Transcription factors promote aluminum tolerance via the reduction in callose deposition. Int. J. Mol. Sci. 2023, 24, 10288. [Google Scholar] [CrossRef] [PubMed]
- Shetty, R.; Vidya, C.S.N.; Prakash, N.B.; Lux, A.; Vaculik, M. Aluminum toxicity in plants and its possible mitigation in acid soils by biochar: A review. Sci. Total Environ. 2021, 765, 142744. [Google Scholar] [CrossRef] [PubMed]
- Schaller, J.; Puppe, D.; Kaczorek, D.; Ellerbrock, R.; Sommer, M. Silicon cycling in soils revisited. Plants 2021, 10, 295. [Google Scholar] [CrossRef] [PubMed]
- Johnston, N.R.; Strobel, S.A. Principles of fluoride toxicity and the cellular response: A review. Arch. Toxicol. 2020, 94, 1051–1069. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Jogawat, A.; Lal, S.K.; Lakra, N.; Mehta, S.; Shabek, N.; Narayan, O.P. Plant mineral transport systems and the potential for crop improvement. Planta 2021, 253, 45. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.K.; Yadav, V.; Vaculík, M.; Gassmann, W.; Pike, S.; Arif, N.; Tripathi, D.K. Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 2021, 41, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Caparros, P.; De Filippis, L.; Gul, A.; Hasanuzzaman, M.; Ozturk, M.; Altay, V.; Lao, M.T. Oxidative stress and antioxidant metabolism under adverse environmental conditions: A review. Bot. Rev. 2021, 87, 421–466. [Google Scholar] [CrossRef]
- Dahiya, V. Heavy metal toxicity of drinking water: A silent killer. GSC Biol. Pharm. Sci. 2022, 19, 020–025. [Google Scholar] [CrossRef]
- Mansoor, S.; Wani, O.A.; Lone, J.K.; Manhas, S.; Kour, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive oxygen species in plants: From source to sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Worthington, M.; Perez, J.G.; Mussurova, S.; Silva-Cordoba, A.; Castiblanco, V.; Cardoso Arango, J.A.; Jones, C.; Fernandez-Fuentes, N.; Skot, L.; Dyer, S.; et al. A new genome allows the identification of genes associated with natural variation in aluminium tolerance in Brachiaria grasses. J. Exp. Bot. 2021, 72, 302–319. [Google Scholar] [CrossRef] [PubMed]
- Jardim-Messeder, D.; Margis-Pinheiro, M.; Sachetto-Martins, G. Salicylic acid and adenine nucleotides regulate the electron transport system and ROS production in plant mitochondria. Biochim. Biophys. Acta. Bioenerg. 2022, 1863, 148559. [Google Scholar] [CrossRef] [PubMed]
- Vera-Villalobos, H.; Lunario-Delgado, L.; Pérez-Retamal, D.; Román, D.; Leiva, J.C.; Zamorano, P.; Wulff-Zottele, C. Sulfate nutrition improves short-term Al3+-stress tolerance in roots of Lolium perenne L. Plant. Physiol. Biochem. 2020, 148, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, B.; Bandyopadhyay, P.; Das, A.; Pal, S.; Hasanuzzaman, M.; Adak, M.K. Abscisic acid priming confers salt tolerance in maize seedlings by modulating osmotic adjustment, bond energies, ROS homeostasis, and organic acid metabolism. Plant Physiol. Biochem. 2023, 202, 107980. [Google Scholar] [CrossRef] [PubMed]
- Ahammed, G.J.; Li, Z.; Chen, J.; Dong, Y.; Qu, K.; Guo, T.; Li, X. Reactive oxygen species signaling in melatonin-mediated plant stress response. Plant Physiol. Biochem. 2024, 207, 108398. [Google Scholar] [CrossRef] [PubMed]
- Dourmap, C.; Roque, S.; Morin, A.; Caubrière, D.; Kerdiles, M.; Béguin, K.; Couée, I. Stress signalling dynamics of the mitochondrial electron transport chain and oxidative phosphorylation system in higher plants. Ann. Bot. 2020, 125, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Shen, R.F. Towards sustainable use of acidic soils: Deciphering aluminum-resistant mechanisms in plants. Fundam. Res. 2023, 14, 41. [Google Scholar] [CrossRef]
- Peng, X.; Wang, N.; Sun, S.; Geng, L.; Guo, N.; Liu, A.; Ahammed, G.J. Reactive oxygen species signaling is involved in melatonin-induced reduction of chlorothalonil residue in tomato leaves. J. Hazard. Mater. 2023, 443, 130212. [Google Scholar] [CrossRef]
- Han, X.; Zhao, Y.; Chen, Y.; Xu, J.; Jiang, C.; Wang, X.; Zhuo, R.; Lu, M.-Z.; Zhang, J. Lignin biosynthesis and accumulation in response to abiotic stresses in woody plants. J. For. Res. 2022, 2, 9. [Google Scholar] [CrossRef]
- Adamiec, M.; Dobrogojski, J.; Wojtyla, Ł.; Luciński, R. Stress-related expression of the chloroplast EGY3 pseudoprotease and its possible impact on chloroplasts’ proteome composition. Front Plant Sci. 2022, 13, 965143. [Google Scholar] [CrossRef] [PubMed]
- Angulo-Bejarano, P.I.; Puente-Rivera, J.; Cruz-Ortega, R. Metal and metalloid toxicity in plants: An overview on molecular aspects. Plants 2021, 10, 635. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, B.; Rizvi, A.; Syed, A.; Rajput, V.D.; Elgorban, A.M.; Al-Rejaie, S.S.; Minkina, T.; Khan, M.S.; Lee, J. Understanding the phytotoxic impact of Al3+, nano-size, and bulk Al2O3 on growth and physiology of maize (Zea mays L.) in aqueous and soil media. Chemosphere 2022, 300, 134555. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Wang, Y.; Bao, Z.; Cui, P.; Wang, S.; Lin, S. Calcium binding to herring egg phosphopeptides: Binding characteristics, conformational structure and intermolecular forces. Food Chem. 2020, 310, 125867. [Google Scholar] [CrossRef]
- Nivetha, N.; Srivarshine, B.; Sowmya, B.; Rajendiran, M.; Saravanan, P.; Rajeshkannan, R.; Dragoi, E.-N. A comprehensive review on bio-stimulation and bio-enhancement towards remediation of heavy metals degeneration. Chemosphere 2023, 312, 137099. [Google Scholar] [CrossRef]
- Sah, R.P.; Chakraborty, M.; Prasad, K.; Pandit, M.; Tudu, V.K.; Chakravarty, M.K.; Moharana, D. Impact of water deficit stress in maize: Phenology and yield components. Sci. Rep. 2020, 10, 2944. [Google Scholar] [CrossRef]
- Miftahudin, M.; Roslim, D.I.; Fendiyanto, M.H.; Satrio, R.D.; Zulkifli, A.; Umaiyah, E.I.; Chikmawati, T.; Sulistyaningsih, Y.-C.; Suharsono, S.; Hartana, A.; et al. OsGERLP: A novel aluminum tolerance rice gene isolated from a local cultivar in Indonesia. Plant Physiol. Biochem. 2021, 162, 86–99. [Google Scholar] [CrossRef]
- Pereira-Lima, Í.A.; Batista-Silva, W.; Siqueira, J.A.; Silva, M.F.; Medeiros, D.B.; Cavalcanti, J.H.; de Carvalho Gonçalves, J.F.; Ribeiro, D.M.; Fernie, A.R.; Nunes-Nesi, A.; et al. Differential Aluminum tolerance in Arabidopsis thaliana ecotypes is seemingly related to metabolite changes. Environ. Exp. Bot. 2023, 214, 105472. [Google Scholar] [CrossRef]
- Chandra, J.; Keshavkant, S. Mechanisms underlying the phytotoxicity and genotoxicity of aluminum and their alleviation strategies: A review. Chemosphere 2021, 278, 130384. [Google Scholar] [CrossRef]
- Saha, I.; Ghosh, A.; Dolui, D.; Fujita, M.; Hasanuzzaman, M.; Adak, M.K. Differential impact of nitric oxide and abscisic acid on the cellular and physiological functioning of sub1A QTL bearing rice genotype under salt stress. Plants 2022, 11, 1084. [Google Scholar] [CrossRef] [PubMed]
- Kaya, O.; Kose, C.; Esıtken, A.; Turan, M.; Utku, O. Can organic acid and sugar compositions be used to predict cell death point limits? Receptacle and pistil organs of apricot (Prunus armeniaca L.). Rend. Lincei. Sci. Fis. Nat. 2021, 32, 493–509. [Google Scholar] [CrossRef]
- Moreno, A.; Morsali, M.; Sipponen, M.H. Catalyst-free synthesis of lignin vitrimers with tunable mechanical properties: Circular polymers and recoverable adhesives. ACS Appl. Mater. Interfaces 2021, 13, 57952–57961. [Google Scholar] [CrossRef] [PubMed]
- Karumanchi, A.R.; Sivan, P.; Kummari, D.; Rajasheker, G.; Kumar, S.A.; Reddy, P.S.; Kishor, P.K. Root and leaf anatomy, ion accumulation, and transcriptome pattern under salt stress conditions in contrasting genotypes of sorghum bicolor. Plants 2023, 12, 2400. [Google Scholar] [CrossRef] [PubMed]
- Mansinhos, I.; Gonçalves, S.; Rodríguez-Solana, R.; Ordóñez-Díaz, J.L.; Moreno-Rojas, J.M.; Romano, A. Impact of temperature on phenolic and osmolyte contents in in vitro cultures and micropropagated plants of two mediterranean plant species, Lavandula viridis and Thymus lotocephalus. Plants 2022, 11, 3516. [Google Scholar] [CrossRef]
- Pappi, P.; Nikoloudakis, N.; Fanourakis, D.; Zambounis, A.; Delis, C.; Tsaniklidis, G. Differential triggering of the phenylpropanoid biosynthetic pathway key genes transcription upon cold stress and viral infection in tomato leaves. Horticulturae 2021, 7, 448. [Google Scholar] [CrossRef]
- Zhou, Y.; Neuhäuser, B.; Neumann, G.; Ludewig, U. LaALMT1 mediates malate release from phosphorus-deficient white lupin root tips and metal root to shoot translocation. Plant Cell Environ. 2020, 43, 1691–1706. [Google Scholar] [CrossRef] [PubMed]
- Wiley, C.D.; Campisi, J. The metabolic roots of senescence: Mechanisms and opportunities for intervention. Nat. Metab. 2021, 3, 1290–1301. [Google Scholar] [CrossRef] [PubMed]
- Vasilev, F.; Ezhova, Y.; Chun, J.T. Signaling enzymes and ion channels being modulated by the actin cytoskeleton at the plasma membrane. Int. J. Mol. Sci. 2021, 22, 10366. [Google Scholar] [CrossRef]
- Bhattacharjee, B.; Ali, A.; Tuteja, N.; Gill, S.; Pattanayak, A. Identification and expression pattern of aluminium-responsive genes in roots of rice genotype with reference to Al-sensitivity. Sci. Rep. 2023, 13, 12184. [Google Scholar] [CrossRef]
- Ap Rees, T.; Hill, S.A. Metabolic control analysis of plant metabolism. Plant Cell Environ. 1994, 17, 587–599. [Google Scholar] [CrossRef]
- Guo, M.; Li, C.; Huang, R.; Qu, L.; Liu, J.; Zhang, C.; Ge, Y. Ferulic acid enhanced resistance against blue mold of Malus domestica by regulating reactive oxygen species and phenylpropanoid metabolism. Postharvest Biol. Technol. 2023, 202, 112378. [Google Scholar] [CrossRef]
- Tang, J.; Ding, Y.; Nan, J.; Yang, X.; Sun, L.; Zhao, X.; Jiang, L. Transcriptome sequencing and ITRAQ reveal the detoxification mechanism of Bacillus GJ1, a potential biocontrol agent for Huanglongbing. PLoS ONE 2018, 13, 0200427. [Google Scholar] [CrossRef] [PubMed]
- Dahuja, A.; Kumar, R.R.; Sakhare, A.; Watts, A.; Singh, B.; Goswami, S.; Praveen, S. Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant 2021, 171, 785–801. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Das, S.; Bansal, S.; Singh, G.; Sardar, S.; Dhar, H.; Ram, H. Heavy metal stress in rice: Uptake, transport, signaling, and tolerance mechanisms. Physiol. Plant. 2021, 173, 430–448. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Baquy, M.A.A.; Guan, P.; Yan, J.; Wang, R.; Xu, R.; Xie, L. Effect of soil acidification on the growth and nitrogen use efficiency of maize in Ultisols. J. Soils. Sediments 2020, 20, 1435–1445. [Google Scholar] [CrossRef]
- Ranjan, A.; Sinha, R.; Lal, S.K.; Bishi, S.K.; Singh, A.K. Phytohormone signalling and cross-talk to alleviate aluminium toxicity in plants. Plant Cell Rep. 2021, 40, 1331–1343. [Google Scholar] [CrossRef]
- Jamla, M.; Khare, T.; Joshi, S.; Patil, S.; Penna, S.; Kumar, V. Omics approaches for understanding heavy metal responses and tolerance in plants. Curr. Plant. Biol. 2021, 27, 100213. [Google Scholar] [CrossRef]
- Buanafina, M.M.D.O.; Morris, P. The impact of cell wall feruloylation on plant growth, responses to environmental stress, plant pathogens and cell wall degradability. Agronomy 2022, 12, 1847. [Google Scholar] [CrossRef]
- Horst, W.J.; Wang, Y.; Eticha, D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: A review. Ann. Bot. 2010, 106, 185–197. [Google Scholar] [CrossRef]
- Šípošová, K.; Labancová, E.; Hačkuličová, D.; Kollárová, K.; Vivodová, Z. The changes in the maize root cell walls after exogenous application of auxin in the presence of cadmium. Environ. Sci. Pollut. Res. 2023, 30, 87102–87117. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Ahmed, Z.; Eldin, S.M.; Ali, I.; Usman, M.; Iqbal, R.; Tariq, A. Biochar as a green sorbent for remediation of polluted soils and associated toxicity risks. A critical review. Separations 2023, 10, 197. [Google Scholar] [CrossRef]
- Adak, M.K.; Das, A.; Kundu, A.; Chatterjee, M.; Hasanuzzaman, M. Molecular mechanisms in understanding anoxia tolerance in rice seeds under submergence and their implication in rice biotechnology. Seeds 2023, 2, 246–258. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Piater, L.A.; Steenkamp, P.A.; Dubery, I.A.; Tugizimana, F.; Mhlongo, M.I. Comparative metabolite profiling of wheat cultivars (Triticum aestivum) reveals signatory markers for resistance and susceptibility to stripe rust and aluminium (Al3+) toxicity. Metabolites 2022, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, M.Y.A.; de Oliveira, A.L.M.; Valentinuzzi, F.; Jayme, N.S.; Monterisi, S.; Fattorini, R.; Pii, Y. An insight into the role of the organic acids produced by Enterobacter sp. strain 15S in solubilizing tricalcium phosphate: In situ study on cucumber. BMC Microbiol. 2023, 23, 184. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.; Singh, A.; Banerjee, A.; Roychoudhury, A. Exogenous supplementation of melatonin alters representative organic acids and enzymes of respiratory cycle as well as sugar metabolism during arsenic stress in two contrasting indica rice cultivars. J. Biotechnol. 2020, 324, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Sade, H.; Meriga, B.; Surapu, V.; Gadi, J.; Sunita, M.S.L.; Suravajhala, P.; Kishor, P.B.K. Toxicity and tolerance of aluminum in plants: Tailoring plants to suit to acid soils. Biometals 2016, 29, 187–210. [Google Scholar] [CrossRef] [PubMed]
- Halim, M.A.; Rahman, M.M.; Megharaj, M.; Naidu, R. Cadmium immobilization in the rhizosphere and plant cellular detoxification: Role of plant-growth-promoting rhizobacteria as a sustainable solution. J. Agric. Food. Chem. 2020, 68, 13497–13529. [Google Scholar] [CrossRef]
- Lyu, J.; Jin, L.; Meng, X.; Jin, N.; Wang, S.; Hu, L.; Yu, J. Exogenous Si mitigates the effects of cinnamic-acid-induced stress by regulating carbon metabolism and photosynthetic pigments in cucumber seedlings. Agronomy 2022, 12, 1569. [Google Scholar] [CrossRef]
- Yadav, B.; Jogawat, A.; Rahman, M.S.; Narayan, O.P. Secondary metabolites in the drought stress tolerance of crop plants: A review. Gene Rep. 2021, 23, 101040. [Google Scholar] [CrossRef]
- Pinto, V.B.; Almeida, V.C.; Pereira-Lima, Í.A.; Vale, E.M.; Araújo, W.L.; Silveira, V.; Viana, J.M.S. Deciphering the major metabolic pathways associated with aluminum tolerance in popcorn roots using label-free quantitative proteomics. Planta 2021, 254, 132. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Akhter, Y. A review on enzyme complexes of electron transport chain from Mycobacterium tuberculosis as promising drug targets. Int. J. Biol. Macromol. 2022, 212, 474–494. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, C.V.; Costa, A.C.; Müller, C.; Castoldi, G.; Costa, A.M.; de Paula Barbosa, K.; Da Silva, A.A. Potential of calcium nitrate to mitigate the aluminum toxicity in Phaseolus vulgaris: Effects on morphoanatomical traits, mineral nutrition and photosynthesis. Ecotoxicology 2020, 29, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Bano, K.; Kumar, B.; Tenguria, R.K.; Alsahli, A.A.; Chen, Y. Salicylic acid and sulfur synergism ameliorates arsenic toxicity in Brassica napus through regulating carbohydrate accumulation and ethylene production. S. Afr. J. Bot. 2023, 160, 246–259. [Google Scholar] [CrossRef]
- Ambrosino, L.; Colantuono, C.; Diretto, G.; Fiore, A.; Chiusano, M.L. Bioinformatics resources for plant abiotic stress responses: State of the art and opportunities in the fast evolving-omics era. Plants 2020, 9, 591. [Google Scholar] [CrossRef]
- Botté, A.; Zaidi, M.; Guery, J.; Fichet, D.; Leignel, V. Aluminium in aquatic environments: Abundance and ecotoxicological impacts. Aquat. Ecol. 2022, 56, 751–773. [Google Scholar] [CrossRef]
- Jalili, S.; Ehsanpour, A.A.; Javadirad, S.M. The role of melatonin on caspase-3-like activity and expression of the genes involved in programmed cell death (PCD) induced by in vitro salt stress in alfalfa (Medicago sativa L.) roots. Bot. Stud. 2022, 63, 19. [Google Scholar] [CrossRef] [PubMed]
- Podar, D.; Maathuis, F.J. The role of roots and rhizosphere in providing tolerance to toxic metals and metalloids. Plant Cell Environ. 2022, 45, 719–736. [Google Scholar] [CrossRef]
- Munir, R.; Yasin, M.U.; Afzal, M.; Jan, M.; Muhammad, S.; Jan, N.; Gan, Y. Melatonin alleviated cadmium accumulation and toxicity by modulating phytohormonal balance and antioxidant metabolism in rice. Chemosphere 2024, 346, 140590. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant. Sci. 2015, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Sychta, K.; Słomka, A.; Kuta, E. Insights into plant programmed cell death induced by heavy metals—Discovering a terra incognita. Cells 2021, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Mandzhieva, S. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Kumar, A.; Kaur, N. ROS and oxidative burst: Roots in plant development. Plant Divers. 2020, 42, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Jeyasri, R.; Muthuramalingam, P.; Satish, L.; Pandian, S.K.; Chen, J.T.; Ahmar, S.; Ramesh, M. An overview of abiotic stress in cereal crops: Negative impacts, regulation, biotechnology and integrated omics. Plants 2021, 10, 1472. [Google Scholar] [CrossRef] [PubMed]
- Devanna, B.N.; Mandlik, R.; Raturi, G.; Sudhakaran, S.S.; Sharma, Y.; Sharma, S.; Deshmukh, R. Versatile role of silicon in cereals: Health benefits, uptake mechanism, and evolution. Plant Physiol. Biochem. 2021, 165, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Tai, B.; Hussain, A.; Jahan, I.; Yang, B.; Xing, F. Genome-wide identification and expression analysis of the basic leucine zipper (bZIP) transcription factor gene family in Fusarium graminearum. Genes 2022, 13, 607. [Google Scholar] [CrossRef] [PubMed]
- Jan, N.; Rather, A.M.U.D.; John, R.; Chaturvedi, P.; Ghatak, A.; Weckwerth, W.; Mir, R.R. Proteomics for abiotic stresses in legumes: Present status and future directions. Crit. Rev. Biotechnol. 2023, 43, 171–190. [Google Scholar] [CrossRef]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 2020, 11, 552969. [Google Scholar] [CrossRef]
- Xie, L.; Li, H.; Zhong, Z.; Guo, J.; Hu, G.; Gao, Y.; Zhang, P. Metabolome analysis under aluminum toxicity between aluminum-tolerant and-sensitive rice (Oryza sativa L.). Plants 2022, 11, 1717. [Google Scholar] [CrossRef]
- Park, J.C.; Yoo, Y.; Lim, H.; Yun, S.; Win, K.T.Y.S.; Kim, K.M.; Lee, S.W. Intracellular Ca2+ accumulation triggered by caffeine provokes resistance against a broad range of biotic stress in rice. Plant Cell Environ. 2022, 45, 1049–1064. [Google Scholar] [CrossRef]
- Kour, J.; Khanna, K.; Singh, A.D.; Dhiman, S.; Bhardwaj, T. Calcium’s multifaceted functions: From nutrient to secondary messenger during stress. S. Afr. J. Bot. 2023, 152, 247–263. [Google Scholar] [CrossRef]
- Valenti, M.; Molina, M.; Cid, V.J. Heterologous expression and auto-activation of human pro-inflammatory caspase-1 in saccharomyces cerevisiae and comparison to caspase-8. Front. Immunol. 2021, 12, 668602. [Google Scholar] [CrossRef]
- Kuroyanagi, M.; Yamada, K.; Hatsugai, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 2005, 280, 32914–32920. [Google Scholar] [CrossRef]
- Van Aken, O. Mitochondrial redox systems as central hubs in plant metabolism and signaling. Plant Physiol. 2021, 186, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Samperna, S.; Masi, M.; Vurro, M.; Evidente, A.; Marra, M. Cyclopaldic acid, the main phytotoxic metabolite of Diplodia cupressi, induces programmed cell death and autophagy in Arabidopsis thaliana. Toxins 2022, 14, 474. [Google Scholar] [CrossRef] [PubMed]
- Labudda, M.; Różańska, E.; Prabucka, B.; Muszyńska, E.; Marecka, D.; Kozak, M.; Sobczak, M. Activity profiling of barley vacuolar processing enzymes provides new insights into the plant and cyst nematode interaction. Mol. Plant Pathol. 2020, 21, 38–52. [Google Scholar] [CrossRef]
- Amintas, S.; Dupin, C.; Boutin, J.; Beaumont, P.; Moreau-Gaudry, F.; Bedel, A.; Dabernat, S. Bioactive food components for colorectal cancer prevention and treatment: A good match. Crit. Rev. Food Sci. Nutr. 2023, 63, 6615–6629. [Google Scholar] [CrossRef] [PubMed]
- Challabathula, D.; Analin, B.; Mohanan, A.; Bakka, K. Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and-tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport. J. Plant Physiol. 2022, 268, 153583. [Google Scholar] [CrossRef]
- Thome, T.; Kim, K.; Dong, G.; Ryan, T.E. The role of mitochondrial and redox alterations in the skeletal myopathy associated with chronic kidney disease. Antioxid Redox Signal 2023, 38, 318–337. [Google Scholar] [CrossRef]
- Akbari, B.; Baghaei-Yazdi, N.; Bahmaie, M.; Mahdavi, A.F. The role of plant-derived natural antioxidants in reduction of oxidative stress. BioFactors 2022, 48, 611–633. [Google Scholar] [CrossRef]
- Hernansanz-Agustín, P.; Enríquez, J.A. Generation of reactive oxygen species by mitochondria. Antioxidants 2021, 10, 415. [Google Scholar] [CrossRef]
- Hassan, N.; Krieg, T.; Zinser, M.; Schröder, K.; Kröger, N. An overview of scaffolds and biomaterials for skin expansion and soft tissue regeneration: Insights on zinc and magnesium as new potential key elements. Polymers 2023, 15, 3854. [Google Scholar] [CrossRef] [PubMed]
- Mittal, I.; Jhanji, S.; Dhatt, K.K. Efficacy of sodium nitroprusside, a nitric oxide donor, on vase life and postharvest attributes of gladiolus spikes. Acta. Physiol. Plant 2021, 43, 108. [Google Scholar] [CrossRef]
- Şirin, S.; Aslım, B. Determination of antioxidant capacity, phenolic acid composition and antiproliferative effect associated with phenylalanine ammonia lyase (PAL) activity in some plants naturally growing under salt stress. Med. Chem. Res. 2019, 28, 229–238. [Google Scholar] [CrossRef]
- Wani, A.K.; Akhtar, N.; Mir, T.U.G.; Singh, R.; Jha, P.K.; Mallik, S.K.; Prakash, A. Targeting apoptotic pathway of cancer cells with phytochemicals and plant-based nanomaterials. Biomolecules 2023, 13, 194. [Google Scholar] [CrossRef]
- Ahmad, B.; Dar, T.A.; Khan, M.; Ahmad, A.; Rinklebe, J.; Chen, Y.; Ahmad, P. Oligochitosan fortifies antioxidative and photosynthetic metabolism and enhances secondary metabolite accumulation in arsenic-stressed peppermint. Front Plant Sci. 2022, 13, 987746. [Google Scholar] [CrossRef]
- Goncharuk, E.A.; Zagoskina, N.V. Heavy metals, their phytotoxicity, and the role of phenolic antioxidants in plant stress responses with focus on cadmium. Molecules 2023, 28, 3921. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Riaz, M.; Liu, J.; Yu, M.; Cuncang, J. The aluminum tolerance and detoxification mechanisms in plants; recent advances and prospects. Crit. Rev. Environ. Sci. Technol. 2022, 52, 1491–1527. [Google Scholar] [CrossRef]
- Niedziela, A.; Domżalska, L.; Dynkowska, W.M.; Pernisová, M.; Rybka, K. Aluminum stress induces irreversible proteomic changes in the roots of the sensitive but not the tolerant genotype of triticale seedlings. Plants 2022, 11, 165. [Google Scholar] [CrossRef]
- Conde, A.; Chaves, M.M.; Gerós, H. Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol. 2011, 52, 1583–1602. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, M.; Vishwakarma, K.; Kumar, V.; Arif, N.; Das, S.; Johnson, R.; Hasanuzzaman, M. Metal/metalloid-based nanomaterials for plant abiotic stress tolerance: An overview of the mechanisms. Plants 2022, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Kosakivska, I.V.; Babenko, L.M.; Romanenko, K.O.; Korotka, I.Y.; Potters, G. Molecular mechanisms of plant adaptive responses to heavy metals stress. Cell. Biol. Int. 2021, 45, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Yang, J.; Sun, T.; Wang, X.; Li, Y. Evidence that synergism between potassium and nitrate enhances the alleviation of ammonium toxicity in rice seedling roots. PLoS ONE 2021, 16, 0248796. [Google Scholar] [CrossRef]
- Ojeda-Rivera, J.O.; Oropeza-Aburto, A.; Herrera-Estrella, L. Dissection of root transcriptional responses to low pH, aluminum toxicity and iron excess under Pi-limiting conditions in Arabidopsis wild-type and stop1 seedlings. Front. Plant Sci. 2020, 11, 01200. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Su, C.; Cui, Z.; Huang, L.; Gu, S.; Jiang, S.; Zhao, M. Transcriptomics and metabolomics reveal tolerance new mechanism of rice roots to Al stress. Front. Genet. 2023, 13, 1063984. [Google Scholar] [CrossRef]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef]
- Rasheed, A.; Fahad, S.; Hassan, M.U.; Tahir, M.M.; Aamer, M.; Wu, Z.M. A review on aluminum toxicity and quantitative trait loci mapping in rice (Oryza sativa L.). Appl. Ecol. Environ. Res. 2020, 18, 3951. [Google Scholar] [CrossRef]
- Moussa, H.; Quezada, E.; Viña, D.; Riadi, H.; Gil-Longo, J. Redox-Active Phenolic Compounds Mediate the Cytotoxic and Antioxidant Effects of Carpodesmia tamariscifolia (Cystoseira tamariscifolia). Chem. Biodivers. 2020, 17, 2000121. [Google Scholar] [CrossRef]
- Sterckeman, T.; Thomine, S. Mechanisms of cadmium accumulation in plants. Crit. Rev. Plant Sci. 2020, 39, 322–359. [Google Scholar] [CrossRef]
- Jin, J.F.; He, Q.Y.; Li, P.F.; Lou, H.Q.; Chen, W.W.; Yang, J.L. Genome-wide identification and gene expression analysis of acyl-activating enzymes superfamily in tomato (Solanum lycopersicum) under aluminum stress. Front. Plant Sci. 2021, 12, 754147. [Google Scholar] [CrossRef]
- Liu, F.; Ma, D.; Yu, J.; Meng, R.; Wang, Z.; Zhang, B.; Xia, J. Overexpression of an ART1-Interacting Gene OsNAC016 Improves Al Tolerance in Rice. Int. J. Mol. Sci. 2023, 24, 17036. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Komatsu, S. Subcellular proteomics to elucidate soybean response to abiotic stress. Plants 2023, 12, 2865. [Google Scholar] [CrossRef]
- Tan, R.J.; Yang, Y.; Yan, Y.W.; Mao, D.D.; Yuan, H.M.; Wang, C.; Zhao, F.G.; Luan, S. Two transporters mobilize magnesium from vacuolar stores to enable plant acclimation to magnesium deficiency. Plant Physiol. 2022, 190, 1307–1320. [Google Scholar]
- Sun, L.M.; Che, J.; Ma, J.F.; Shen, R.F. Expression level of transcription factor ART1 is responsible for differential aluminum tolerance in Indica rice. Plants 2021, 10, 634. [Google Scholar] [CrossRef]
- Maheen, N.; Shafiq, M.; Sadiq, S.; Farooq, M.; Ali, Q.; Habib, U.; Ali, F. Genome Identification and Characterization of WRKY Transcription Factor Gene Family in Mandarin (Citrus reticulata). Agriculture 2023, 13, 1182. [Google Scholar] [CrossRef]
- Canellas, L.P.; Canellas, N.O.A.; Irineu, L.E.S.D.S.; Olivares, F.L.; Piccolo, A. Plant chemical priming by humic acids. Chem. Biol. Technol. Agric. 2020, 7, 12. [Google Scholar] [CrossRef]
- Jingguang, C.; Qi, L.; Baiquan, Z.; Longbiao, G.; Guoyou, Y. Progress on molecular mechanism of aluminum resistance in rice. Rice Sci. 2020, 27, 454–467. [Google Scholar] [CrossRef]
- Feng, Y.; Bayaer, E.; Qi, Y. Advances in the biological functions of auxin transporters in rice. Agriculture 2022, 12, 989. [Google Scholar] [CrossRef]
- Arif, Y.; Sami, F.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salicylic acid in relation to other phytohormones in plant: A study towards physiology and signal transduction under challenging environment. Environ. Exp. Bot. 2020, 175, 104040. [Google Scholar] [CrossRef]
- Wei, H.; Jing, Y.; Zhang, L.; Kong, D. Phytohormones and their crosstalk in regulating stomatal development and patterning. J. Exp. Bot. 2021, 72, 2356–2370. [Google Scholar] [CrossRef]
- Le Poder, L.; Mercier, C.; Février, L.; Duong, N.; David, P.; Pluchon, S.; Desnos, T. Uncoupling aluminum toxicity from aluminum signals in the STOP1 pathway. Front. Plant Sci. 2022, 13, 785791. [Google Scholar] [CrossRef]
- Kopittke, M.P. Role of phytohormones in aluminium rhizotoxicity. Plant Cell Environ. 2016, 39, 2319–2328. [Google Scholar] [CrossRef]
- Khan, S.; Sehar, Z.; Albaqami, M.; Khan, N.A. Ethylene crosstalk with isoprenoid-derived signaling molecules in the context of salinity tolerance. Environ. Exp. Bot. 2023, 212, 105379. [Google Scholar] [CrossRef]
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef]
- Baek, D.; Kim, W.Y.; Cha, J.Y.; Park, H.J.; Shin, G.; Park, J.; Yun, D.J. The GIGANTEA-ENHANCED EM LEVEL complex enhances drought tolerance via regulation of abscisic acid synthesis. Plant Physiol. 2020, 184, 443–458. [Google Scholar] [CrossRef]
- Das, S.; Ganesan, M. Aluminum induced malate transporter (ALMT1) is regulating the Aluminum stress tolerance responses of mungbean seedlings. Plant Gene 2022, 32, 100388. [Google Scholar] [CrossRef]
- Gavassi, M.A.; Silva, G.S.; da Silva, C.D.M.S.; Thompson, A.J.; Macleod, K.; Oliveira, P.M.R.; Habermann, G. NCED expression is related to increased ABA biosynthesis and stomatal closure under aluminum stress. Environ. Exp. Bot. 2021, 185, 104404. [Google Scholar] [CrossRef]
- Cáceres, C.; Quintana, J.; Nunes-Nesi, A.; Cohen, J.D.; Delgado, M.; Ribera-Fonseca, A.; Reyes-Díaz, M. Interplay of phytohormone signaling with aluminum and drought-stress resistance mechanisms: An integrated perspective amidst climate change. Environ. Expl. Bot. 2023, 23, 105575. [Google Scholar] [CrossRef]
- Kabir, A.H.; Tahura, S.; Elseehy, M.M.; El-Shehawi, A.M. Molecular characterization of Fe-acquisition genes causing decreased Fe uptake and photosynthetic inefficiency in Fe-deficient sunflower. Sci. Rep. 2021, 11, 5537. [Google Scholar] [CrossRef]
- Fan, Y.; Ouyang, Y.; Pan, Y.; Hong, T.; Wu, C.; Lin, H. Effect of aluminum stress on the absorption and transportation of aluminum and macronutrients in roots and leaves of Aleurites montana. For. Ecol. Manag. 2020, 458, 117813. [Google Scholar] [CrossRef]
- Mukherjee, A.; Gaurav, A.K.; Singh, S.; Yadav, S.; Bhowmick, S.; Abeysinghe, S.; Verma, J.P. The bioactive potential of phytohormones: A review. Biotechnol. Rep. 2022, 35, 00748. [Google Scholar] [CrossRef]
- Jin, J.; Essemine, J.; Xu, Z.; Duan, J.; Shan, C.; Mei, Z.; Cai, W. Arabidopsis Ethylene Insensitive 3 directly regulates the expression of PG1β-like family genes in response to aluminum stress. J. Exp. Bot. 2022, 73, 4923–4940. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Gantait, S.; Mitra, M.; Yang, Y.; Li, X. Role of ethylene crosstalk in seed germination and early seedling development: A review. Plant Physiol. Biochem. 2020, 151, 124–131. [Google Scholar] [CrossRef]
- Ulhassan, Z.; Bhat, J.A.; Zhou, W.; Senan, A.M.; Alam, P.; Ahmad, P. Attenuation mechanisms of arsenic induced toxicity and its accumulation in plants by engineered nanoparticles: A review. Environ. Pollut. 2022, 302, 119038. [Google Scholar] [CrossRef] [PubMed]
- Shaffique, S.; Kang, S.-M.; Hoque, M.I.U.; Imran, M.; Aaqil Khan, M.; Lee, I.-J. Research Progress in Soybean by Phytohormone Modulation and Metal Chelation over the Past Decade. Agriculture 2023, 13, 1325. [Google Scholar] [CrossRef]
- Tu, T.; Zheng, S.; Ren, P.; Meng, X.; Zhao, J.; Chen, Q.; Li, C. Coordinated cytokinin signaling and auxin biosynthesis mediates arsenate-induced root growth inhibition. Plant Physiol. 2021, 185, 1166–1181. [Google Scholar] [CrossRef]
- Yuan, J.; Cheng, L.; Li, H.; An, C.; Wang, Y.; Zhang, F. Physiological and protein profiling analysis provides insight into the underlying molecular mechanism of potato tuber development regulated by jasmonic acid in vitro. BMC Plant Biol. 2022, 22, 481. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, N.; Das, A.; Pal, S.; Roy, S.; Sil, S.K.; Adak, M.K.; Hasanuzzaman, M. Exploring Aluminum Tolerance Mechanisms in Plants with Reference to Rice and Arabidopsis: A Comprehensive Review of Genetic, Metabolic, and Physiological Adaptations in Acidic Soils. Plants 2024, 13, 1760. https://doi.org/10.3390/plants13131760
Chakraborty N, Das A, Pal S, Roy S, Sil SK, Adak MK, Hasanuzzaman M. Exploring Aluminum Tolerance Mechanisms in Plants with Reference to Rice and Arabidopsis: A Comprehensive Review of Genetic, Metabolic, and Physiological Adaptations in Acidic Soils. Plants. 2024; 13(13):1760. https://doi.org/10.3390/plants13131760
Chicago/Turabian StyleChakraborty, Nilakshi, Abir Das, Sayan Pal, Soumita Roy, Sudipta Kumar Sil, Malay Kumar Adak, and Mirza Hasanuzzaman. 2024. "Exploring Aluminum Tolerance Mechanisms in Plants with Reference to Rice and Arabidopsis: A Comprehensive Review of Genetic, Metabolic, and Physiological Adaptations in Acidic Soils" Plants 13, no. 13: 1760. https://doi.org/10.3390/plants13131760
APA StyleChakraborty, N., Das, A., Pal, S., Roy, S., Sil, S. K., Adak, M. K., & Hasanuzzaman, M. (2024). Exploring Aluminum Tolerance Mechanisms in Plants with Reference to Rice and Arabidopsis: A Comprehensive Review of Genetic, Metabolic, and Physiological Adaptations in Acidic Soils. Plants, 13(13), 1760. https://doi.org/10.3390/plants13131760






