Forest Orchids under Future Climate Scenarios: Habitat Suitability Modelling to Inform Conservation Strategies
Abstract
1. Introduction
2. Results
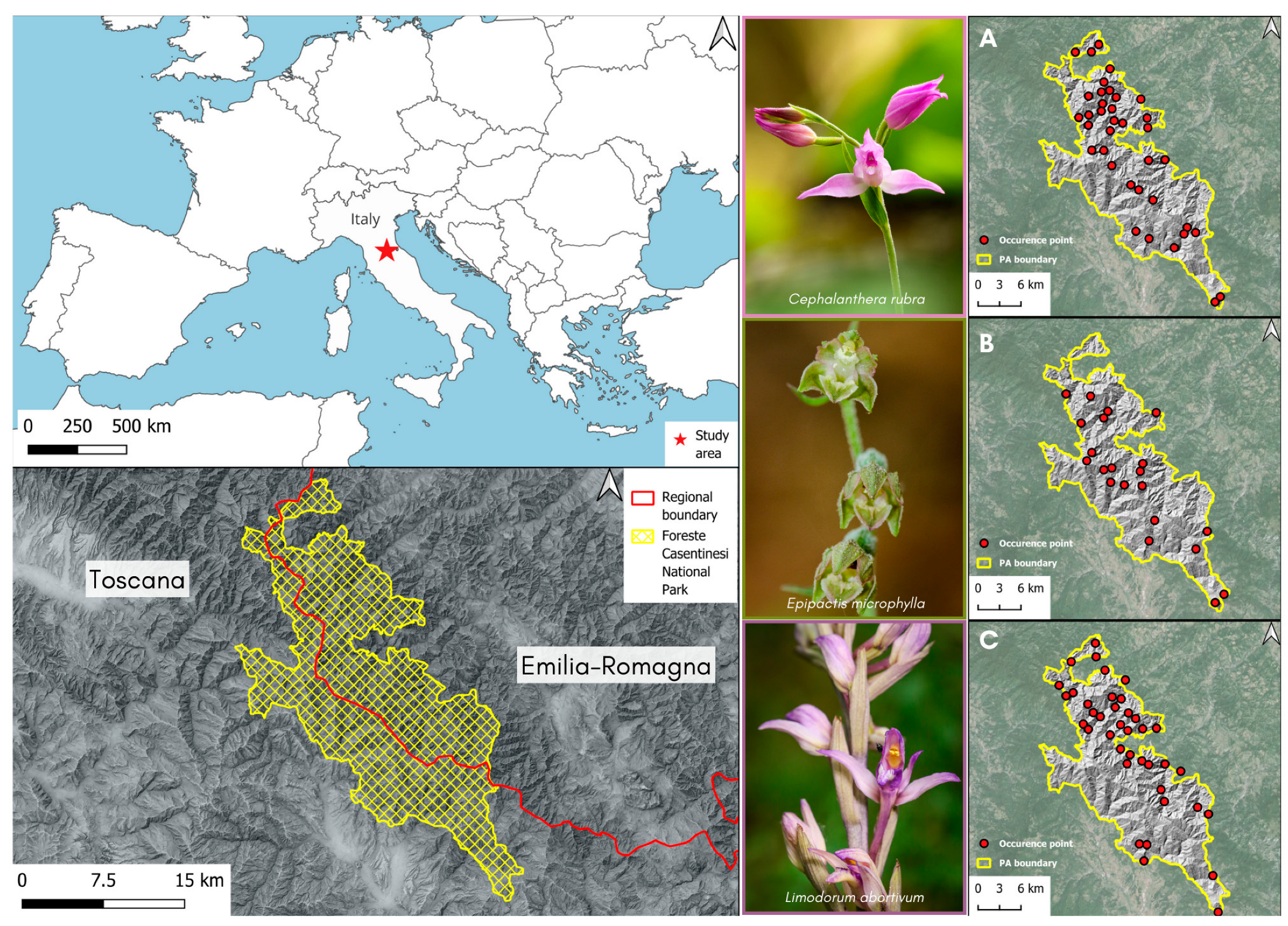
2.1. Species Occurrence and Environmental Variables
2.2. Model Optimization and Evaluation Results
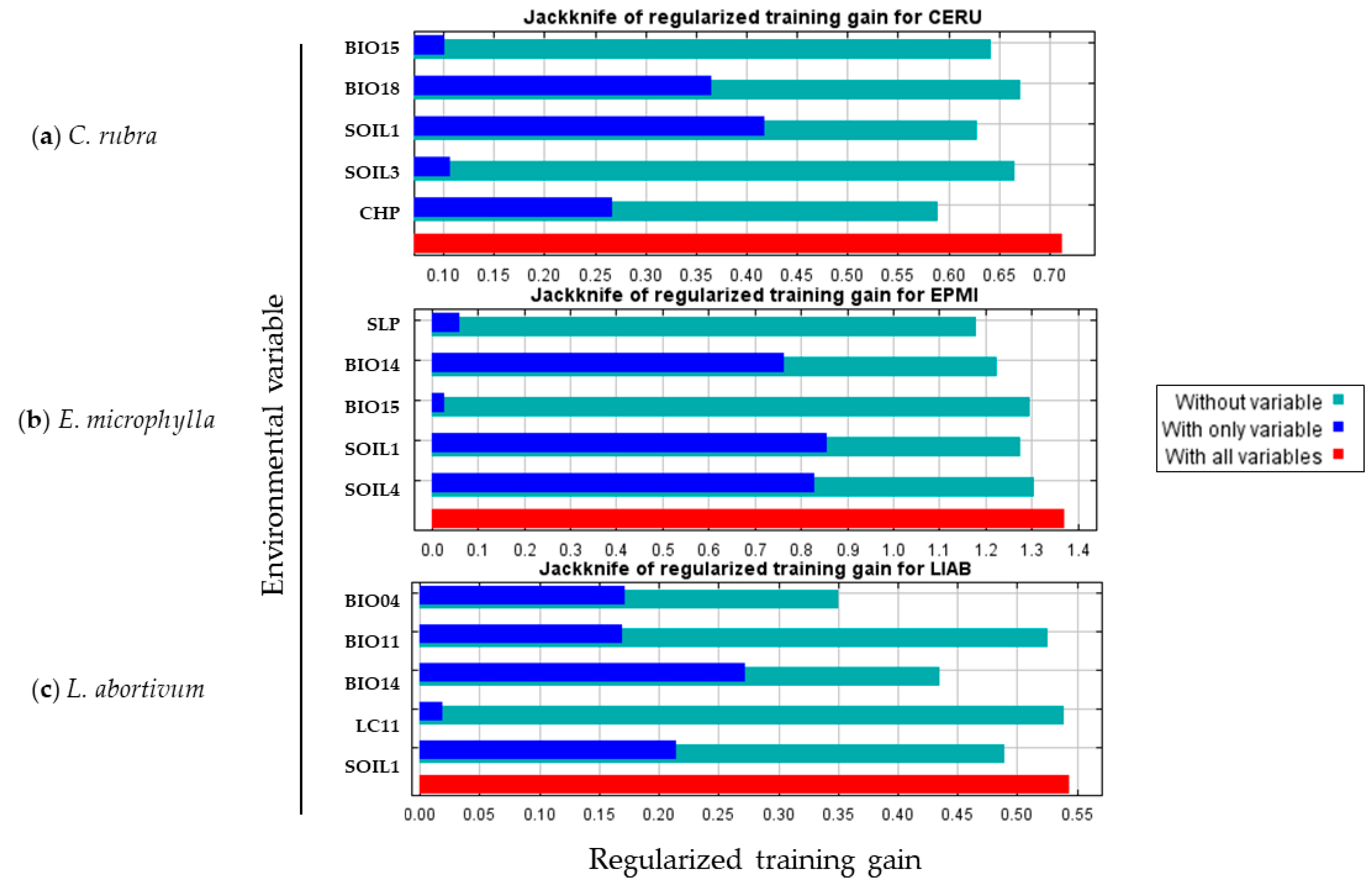
2.3. Contribution of Environmental Variables
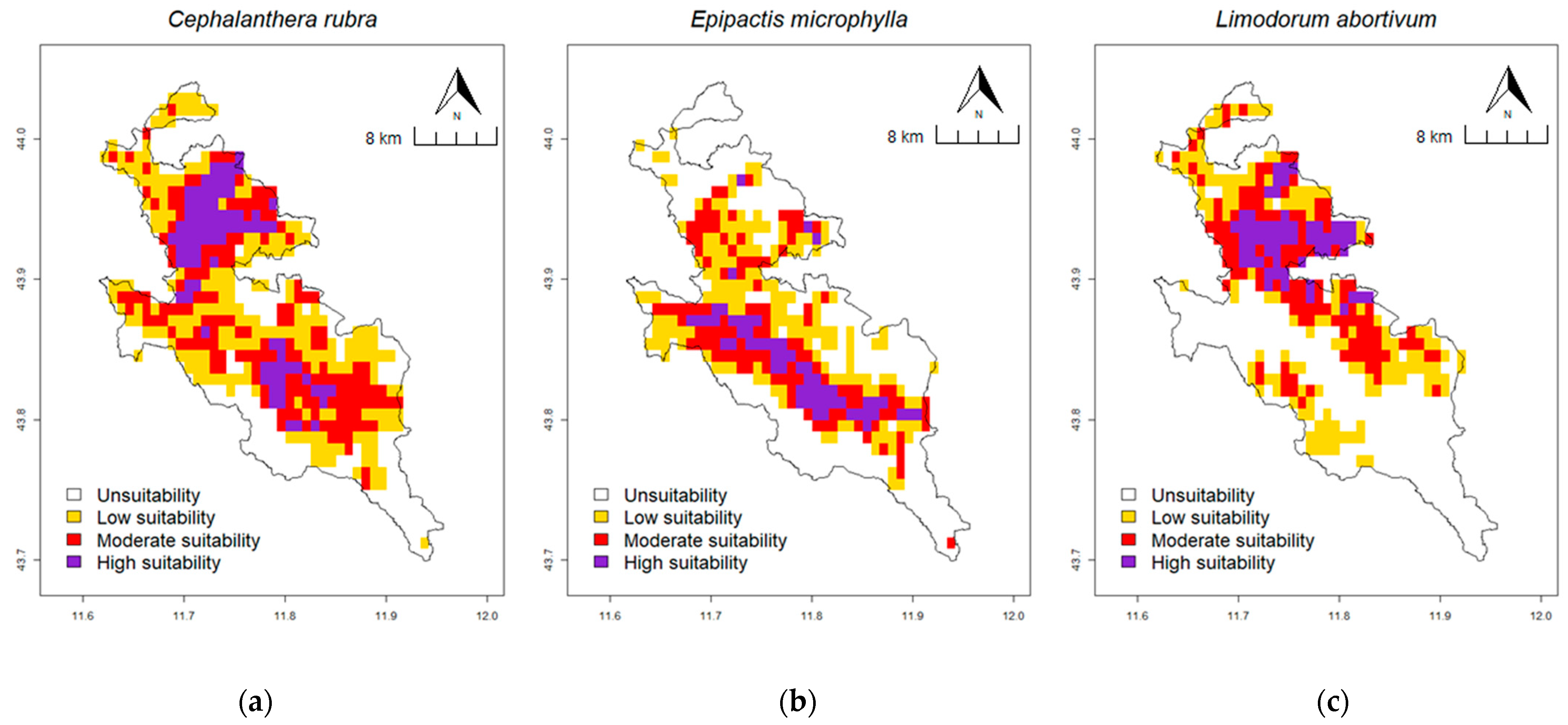
2.4. Prediction of Potential Suitable Habitat Distribution under Current Climate Conditions
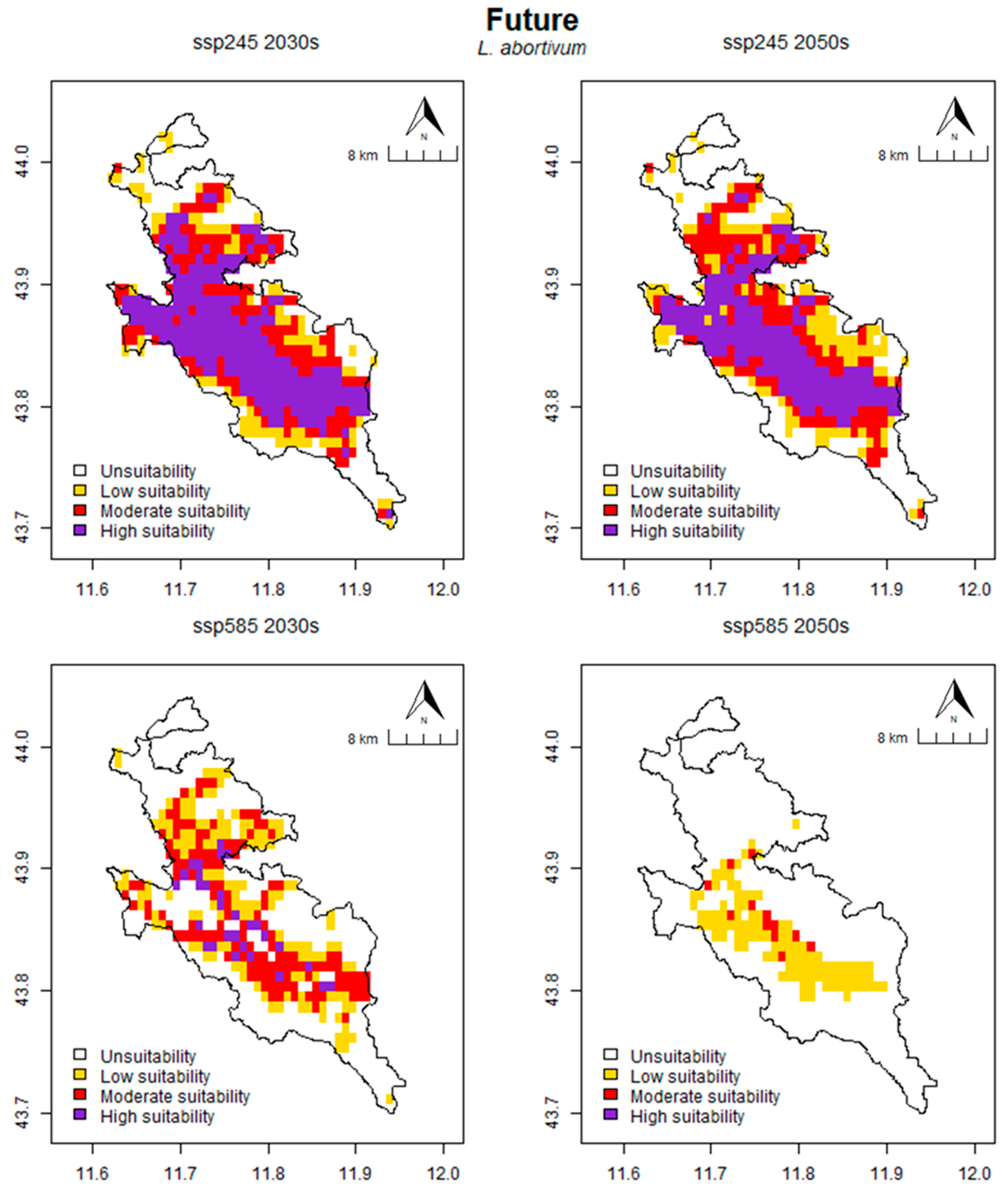
2.5. Prediction of Potential Suitable Habitat Distribution under Future Climate Conditions
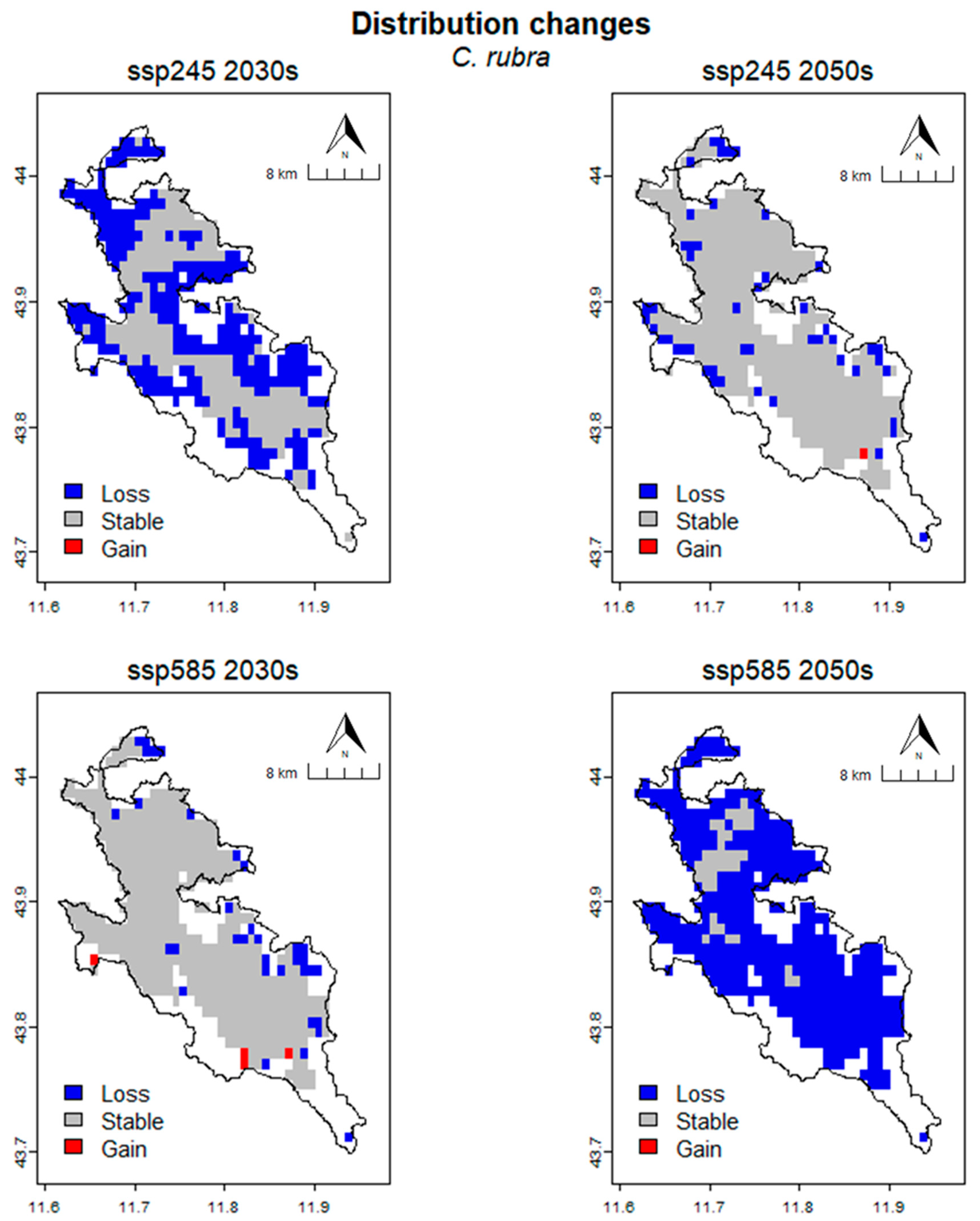
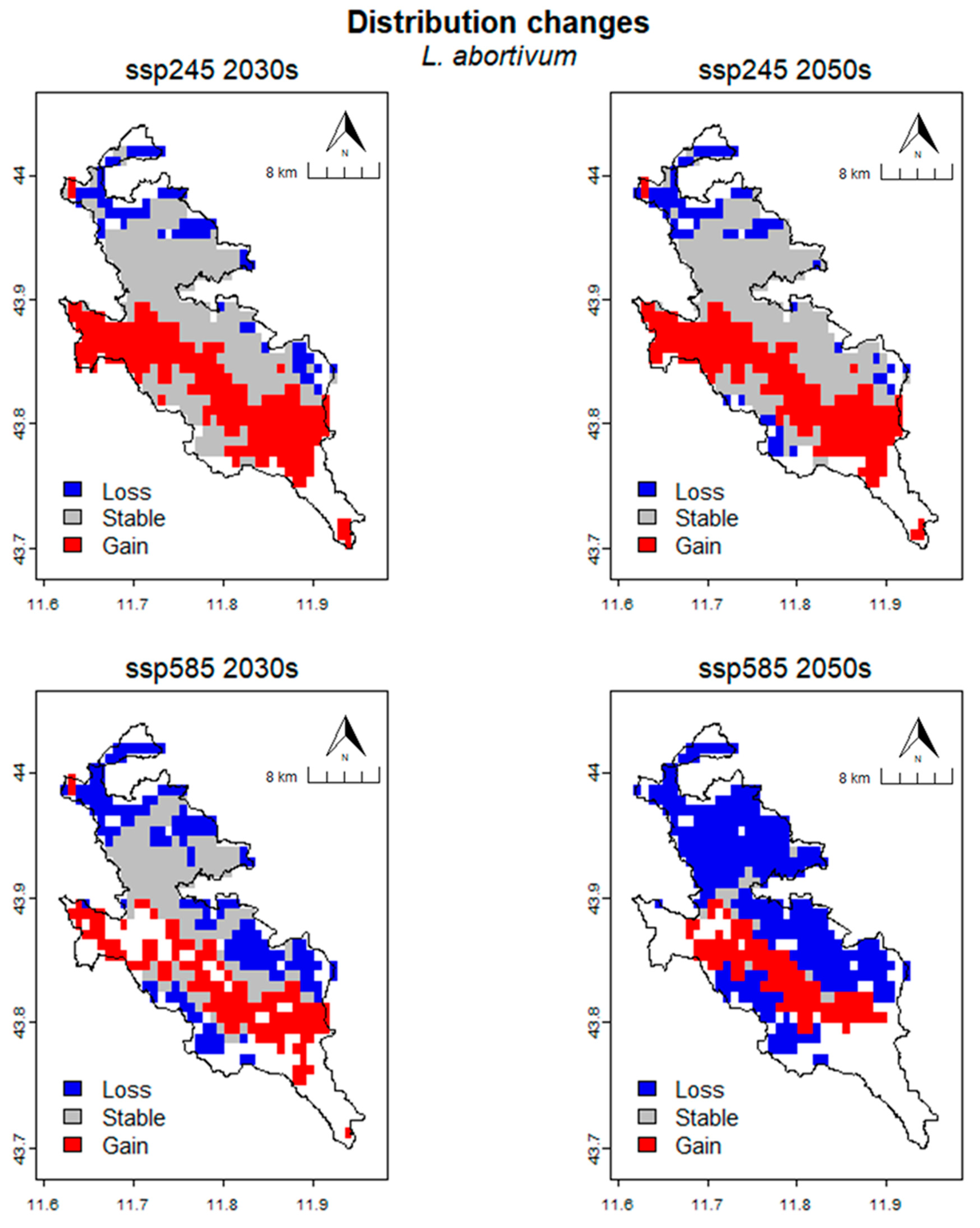
2.6. Distribution Changes under Future Climate Conditions
3. Discussion
4. Materials and Methods
4.1. Study Area
4.2. Species Occurrence Data
4.3. Environmental Variables
- Data matrixes creation: using the QGIS 3.32.3 software [116] we combined each species occurrence record with the spatial value of the corresponding attribute for the 46 variables, i.e., the cell value of the environmental variable in which the occurrence point falls. A matrix for all investigated species was obtained.
- EV screening and ranking: A preliminary modelling exercise was initiated utilising MaxEnt to identify the number and nature of environmental variables influencing the model. The initial model was constructed by applying the default ‘Auto features’ setting (default FC and RM settings) and then three replicate runs were initiated. The mean value of permutation importance for each of the variables included in the model in the three runs was then obtained [138]. The jackknife test [139] was applied to assess the significance of each environmental variable in elucidating the distribution of a species within the MaxEnt model. This enables the assessment of the impact of predictors on the model performance in terms of gain. Variables that lead to a reduction in gain when excluded are considered more important during the modelling process. The light blue bars indicate the impact on the model when the single variable is not included, the dark blue bars indicate the impact with only the variable included, and the red bar indicates the inclusion of all variables. Following, a ranking of the variables was established according to their permutation importance in the model. The use of permutation importance over percent contribution is preferable because it depends on the final model, not on the path used for each run, and this is better for correctly assessing the importance of each variable [140,141]. Furthermore, we decided to directly eliminate variables with a small contribution rate to the model, as this was deemed to be too low [141,142,143]. Variables with a contribution rate of less than 1% were removed [144,145].
- Multicollinearity test #1: Pearson correlation coefficient (r) between each pair of environmental variables was calculated using Past 4.04 software [136]. If |r| ≥ 0.8 [135,145,146,147,148] there is a correlation between the variables and one of the two must be excluded. Based on the permutation importance rank, in the screening and ranking phase, the variable with the greatest contribution for the model was retained while the second was discarded. This operation was performed for each pair of environmental variables.
- Multicollinearity test #2: to further reduce the multicollinearity between the environmental variables selected by the first multicollinearity test, Variance Inflation Factor (VIF), was calculated with the R package usdm [149]. A precautionary threshold was chosen at 5. Variables with VIF > 5 were excluded because they were strongly correlated with each other [84,135,150,151].
- Final environmental dataset: finally, variables that respect both multicollinearity conditions tests (|r| ≤ 0.8 and VIF < 5) were chosen as final predictors. From these, based on the ranking obtained in the screening and ranking phase, the top five variables contributing to the model were selected, sorted according to permutation importance values. These variables, five for each species, are used to build the final models.
4.4. Final Model Construction, Optimization and Evaluation
4.5. Habitat Suitability Analysis and Visualization
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chase, M.W.; Cameron, K.M.; Freudenstein, J.V.; Pridgeon, A.M.; Salazar, G.; van den Berg, C.; Schuiteman, A. An Updated Classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- WFO World Flora Online. Available online: http://www.worldfloraonline.org/ (accessed on 16 January 2024).
- Kühn, R.; Pedersen, H.; Cribb, P. Field Guide to the Orchids of Europe and the Mediterranean; Royal Botanic Gardens, Kew: Richmond, UK, 2019; pp. 1–430. [Google Scholar]
- Djordjević, V.; Tsiftsis, S. The Role of Ecological Factors in Distribution and Abundance of Terrestrial Orchids. In Orchids Phytochemistry, Biology and Horticulture: Fundamentals and Applications; Mérillon, J.-M., Kodja, H., Eds.; Reference Series in Phytochemistry; Springer International Publishing: Cham, Switzerland, 2022; pp. 3–72. ISBN 978-3-030-38392-3. [Google Scholar]
- Kirillova, I.A.; Dubrovskiy, Y.A.; Degteva, S.V.; Novakovskiy, A.B. Ecological and Habitat Ranges of Orchids in the Northernmost Regions of Their Distribution Areas: A Case Study from Ural Mountains, Russia. Plant Divers. 2023, 45, 211–218. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The Number of Known Plants Species in the World and Its Annual Increase. Phytotaxa 2016, 261, 201. [Google Scholar] [CrossRef]
- Hinsley, A.; de Boer, H.J.; Fay, M.F.; Gale, S.W.; Gardiner, L.M.; Gunasekara, R.S.; Kumar, P.; Masters, S.; Metusala, D.; Roberts, D.L.; et al. A Review of the Trade in Orchids and Its Implications for Conservation. Bot. J. Linn. Soc. 2018, 186, 435–455. [Google Scholar] [CrossRef]
- Vitt, P.; Taylor, A.; Rakosy, D.; Kreft, H.; Meyer, A.; Weigelt, P.; Knight, T.M. Global Conservation Prioritization for the Orchidaceae. Sci. Rep. 2023, 13, 6718. [Google Scholar] [CrossRef]
- GBIF GBIF Secretariat. Orchidaceae. GBIF Backbone Taxonomy. Checklist Dataset. Available online: https://www.gbif.org/species/7689 (accessed on 16 January 2024).
- POWO Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet. Available online: http://www.plantsoftheworldonline.org/ (accessed on 16 January 2024).
- Ya, J.-D.; Wang, W.-T.; Liu, Y.-L.; Jiang, H.; Han, Z.-D.; Zhang, T.; Huang, H.; Cai, J.; Li, D.-Z. Five New and Noteworthy Species of Epidendroideae (Orchidaceae) from Southwestern China Based on Morphological and Phylogenetic Evidence. PhytoKeys 2023, 235, 211–236. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, D.S.; Zabalgoitia, A.; Velázquez-Ríos, P.; Muñiz-Castro, M.Á.; Jiménez-Machorro, R.; Guerrero-Hernández, R.; Huerta-Galván, O. A New Species of Triphora (Orchidaceae: Triphoreae, Triphorinae) from Mexico. Phytotaxa 2023, 599, 89–99. [Google Scholar] [CrossRef]
- Moreno, J.S.; Galindo-Tarazona, R.; Alegría-Valencia, M.; Wilson, M.; Tróchez, A.Z. A New Species of Pleurothallis (Pleurothallidinae) from the Southwestern Andes of Colombia in the National Natural Park Farallones de Cali. Lankesteriana Int. J. Orchid. 2023, 23, 401–407. [Google Scholar] [CrossRef]
- Krahl, D.R.P.; Schmal, P.; Chiron, G.; da Silva, J.B.F.; Krahl, A.H.; Cantuária, P.d.C. Catasetum queirozii (Orchidaceae: Catasetinae): A New Species from the Brazilian Amazon. Acta Amaz. 2024, 54, e54bc23180. [Google Scholar] [CrossRef]
- Delforge, P. Orchidées d’Europe, d’Afrique du Nord et du Proche-Orient, 4th ed.; Delachaux & Nistlé: Paris, France, 2016. [Google Scholar]
- Bartolucci, F.; Peruzzi, L.; Galasso, G.; Alessandrini, A.; Ardenghi, N.M.G.; Bacchetta, G.; Banfi, E.; Barberis, G.; Bernardo, L.; Bouvet, D.; et al. A Second Update to the Checklist of the Vascular Flora Native to Italy. Plant Biosyst.—Int. J. Deal. Asp. Plant Biol. 2024, 158, 219–296. [Google Scholar] [CrossRef]
- Portal to the Flora of Italy Portal to the Flora of Italy. Available online: http://dryades.units.it/floritaly (accessed on 6 February 2024).
- Biagioli, M.; De Simoni, M.G.; Bogani, P.; Doro, D.; Klaver, J.M.I.; Puglisi, M. Orchidee d’Italia, 3rd ed.; GIROS—Gruppo Italiano per la Ricerca sule Orchidee Spontanee APS, Ed.; Il Castello: Cornaredo, Italy, 2024; ISBN 978-88-276-0447-2. [Google Scholar]
- Swarts, N.D.; Dixon, K.W. Terrestrial Orchid Conservation in the Age of Extinction. Ann. Bot. 2009, 104, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Wraith, J.; Pickering, C. A Continental Scale Analysis of Threats to Orchids. Biol. Conserv. 2019, 234, 7–17. [Google Scholar] [CrossRef]
- Kumar, J.; Katoch, D.; Thakur, A.; Pathania, A.; Anand, A.; Choudhary, K.; Shelja, S. A Comprehensive Review on Threats and Conservation Status of Orchids. J. Appl. Biol. Biotechnol. 2024, 12, 43–47. [Google Scholar] [CrossRef]
- Wraith, J.; Pickering, C. Tourism and Recreation a Global Threat to Orchids. Biodivers. Conserv. 2017, 26, 3407–3420. [Google Scholar] [CrossRef]
- Wraith, J.; Pickering, C. Quantifying Anthropogenic Threats to Orchids Using the IUCN Red List. Ambio 2018, 47, 307–317. [Google Scholar] [CrossRef]
- Liu, H.; Jacquemyn, H.; Chen, W.; Janssens, S.B.; He, X.; Yu, S.; Huang, Y. Niche Evolution and Historical Biogeography of Lady Slipper Orchids in North America and Eurasia. J. Biogeogr. 2021, 48, 2727–2741. [Google Scholar] [CrossRef]
- Ongaro, S.; Martellos, S.; Bacaro, G.; De Agostini, A.; Cogoni, A.; Cortis, P. Distributional Pattern of Sardinian Orchids under a Climate Change Scenario. Community Ecol. 2018, 19, 223–232. [Google Scholar] [CrossRef]
- Tsiftsis, S.; Djordjević, V. Modelling Sexually Deceptive Orchid Species Distributions under Future Climates: The Importance of Plant–Pollinator Interactions. Sci. Rep. 2020, 10, 10623. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Rawat, S. Modeling the Effect of Climate Change on the Distribution of Threatened Medicinal Orchid Satyrium nepalense D. Don in India. Environ. Sci. Pollut. Res. 2022, 29, 72431–72444. [Google Scholar] [CrossRef]
- Fay, M.F. Orchid Conservation: How Can We Meet the Challenges in the Twenty-First Century? Bot. Stud. 2018, 59, 16. [Google Scholar] [CrossRef]
- Gale, S.W.; Fischer, G.A.; Cribb, P.J.; Fay, M.F. Orchid Conservation: Bridging the Gap between Science and Practice. Bot. J. Linn. Soc. 2018, 186, 425–434. [Google Scholar] [CrossRef]
- Wraith, J.; Norman, P.; Pickering, C. Orchid Conservation and Research: An Analysis of Gaps and Priorities for Globally Red Listed Species. Ambio 2020, 49, 1601–1611. [Google Scholar] [CrossRef]
- Nicolè, F.; Brzosko, E.; Till-Bottraud, I. Population Viability Analysis of Cypripedium calceolus in a Protected Area: Longevity, Stability and Persistence. J. Ecol. 2005, 93, 716–726. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, J.; Corlett, R.T.; Fan, X.; Yu, D.; Yang, H.; Gao, J. Orchid conservation in the biodiversity hotspot of southwestern China. Conserv. Biol. 2015, 29, 1563–1572. [Google Scholar] [CrossRef]
- Khapugin, A.A.; Chugunov, G.G.; Vargot, E.V. Cypripedium calceolus Is Considered as One of the Flagship Plant Species of Nature Conservation. It Is legal Cypripedium calceolus (Orchidaceae) in Central Russia: A Case Study for Its Populations in Two Protected Areas in the Republic of Mordovia (Russia). Lankesteriana Int. J. Orchid. 2017, 17, 417–431. [Google Scholar] [CrossRef][Green Version]
- Estopinan, J.; Servajean, M.; Bonnet, P.; Joly, A.; Munoz, F. AI-Based Mapping of the Conservation Status of Orchid Assemblages at Global Scale. arXiv 2024. [Google Scholar]
- Lussu, M.; Ancillotto, L.; Labadessa, R.; Di Musciano, M.; Zannini, P.; Testolin, R.; Santi, F.; Dolci, D.; Conti, M.; Marignani, M.; et al. Prioritizing Conservation of Terrestrial Orchids: A Gap Analysis for Italy. Biol. Conserv. 2024, 289, 110385. [Google Scholar] [CrossRef]
- Gama, M.; Crespo, D.; Dolbeth, M.; Anastácio, P. Predicting Global Habitat Suitability for Corbicula fluminea Using Species Distribution Models: The Importance of Different Environmental Datasets. Ecol. Model. 2016, 319, 163–169. [Google Scholar] [CrossRef]
- Lee, C.M.; Lee, D.-S.; Kwon, T.-S.; Athar, M.; Park, Y.-S. Predicting the Global Distribution of Solenopsis geminata (Hymenoptera: Formicidae) under Climate Change Using the MaxEnt Model. Insects 2021, 12, 229. [Google Scholar] [CrossRef]
- Marage, D.; Garraud, L.; Rameau, J.-C. The Influence of Management History on Spatial Prediction of Eryngium spinalba, an Endangered Endemic Species. Appl. Veg. Sci. 2008, 11, 139–148. [Google Scholar] [CrossRef]
- Gogol-Prokurat, M. Predicting Habitat Suitability for Rare Plants at Local Spatial Scales Using a Species Distribution Model. Ecol. Appl. 2011, 21, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum Entropy Modeling of Species Geographic Distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Elith, J.H.; Graham, C.P.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel Methods Improve Prediction of Species’ Distributions from Occurrence Data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N.E.; Elith, J.; Graham, C.H.; Phillips, S.; Peterson, A.T. What Matters for Predicting the Occurrences of Trees: Techniques, Data, or Species’ Characteristics? Ecol. Monogr. 2007, 77, 615–630. [Google Scholar] [CrossRef]
- Wisz, M.S.; Hijmans, R.J.; Li, J.; Peterson, A.T.; Graham, C.H.; Guisan, A.; Group, N.P.S.D.W. Effects of Sample Size on the Performance of Species Distribution Models. Divers. Distrib. 2008, 14, 763–773. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J. The Contribution of Species Distribution Modelling to Conservation Prioritization. In Spatial Conservation Prioritization: Quantitative Methods and Computational Tools; Moilanen, A., Wilson, K.A., Possingham, H.P., Eds.; Oxford University Press: Oxford, UK, 2009; ISBN 978-0-19-954776-0. [Google Scholar]
- Deb, C.R.; Jamir, N.S.; Kikon, Z.P. Distribution Prediction Model of a Rare Orchid Species (Vanda bicolor Griff.) Using Small Sample Size. Am. J. Plant Sci. 2017, 8, 1388–1398. [Google Scholar]
- Zhao, Q.; Li, H.; Chen, C.; Fan, S.; Wei, J.; Cai, B.; Zhang, H. Potential Global Distribution of Paracoccus marginatus, under Climate Change Conditions, Using MaxEnt. Insects 2024, 15, 98. [Google Scholar] [CrossRef]
- Ferraz, K.M.P.M.D.B.; Ferraz, S.F.D.B.; Paula, R.C.D.; Beisiegel, B.; Breitenmoser, C. Species Distribution Modeling for Conservation Purposes. Nat. Conserv. 2012, 10, 214–220. [Google Scholar] [CrossRef]
- Schwartz, M.W. Using Niche Models with Climate Projections to Inform Conservation Management Decisions. Biol. Conserv. 2012, 155, 149–156. [Google Scholar] [CrossRef]
- Wang, H.-H.; Wonkka, C.L.; Treglia, M.L.; Grant, W.E.; Smeins, F.E.; Rogers, W.E. Species Distribution Modelling for Conservation of an Endangered Endemic Orchid. AoB Plants 2015, 7, plv039. [Google Scholar] [CrossRef]
- Engler, J.O.; Stiels, D.; Schidelko, K.; Strubbe, D.; Quillfeldt, P.; Brambilla, M. Avian SDMs: Current State, Challenges, and Opportunities. J. Avian Biol. 2017, 48, 1483–1504. [Google Scholar] [CrossRef]
- Frans, V.F.; Augé, A.A.; Fyfe, J.; Zhang, Y.; McNally, N.; Edelhoff, H.; Balkenhol, N.; Engler, J.O. Integrated SDM Database: Enhancing the Relevance and Utility of Species Distribution Models in Conservation Management. Methods Ecol. Evol. 2022, 13, 243–261. [Google Scholar] [CrossRef]
- Wang, X.-M.; Tang, Y.; Peng, X.-F.; Wang, J.; Zhang, S.-Q.; Feng, Y.; Peng, P.-H. Identifying Priorities under Highly Heterogeneous Environments through Species Distribution Models to Facilitate Orchid Conservation. Biodivers. Conserv. 2024, 33, 647–665. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Brys, R.; Hermy, M.; Willems, J.H. Does Nectar Reward Affect Rarity and Extinction Probabilities of Orchid Species? An Assessment Using Historical Records from Belgium and the Netherlands. Biol. Conserv. 2005, 121, 257–263. [Google Scholar] [CrossRef]
- Štípková, Z.; Tsiftsis, S.; Kindlmann, P. Pollination Mechanisms Are Driving Orchid Distribution in Space. Sci. Rep. 2020, 10, 850. [Google Scholar] [CrossRef] [PubMed]
- Wotavová, K.; Balounová, Z.; Kindlmann, P. Factors Affecting Persistence of Terrestrial Orchids in Wet Meadows and Implications for Their Conservation in a Changing Agricultural Landscape. Biol. Conserv. 2004, 118, 271–279. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Brys, R.; Adriaens, D.; Honnay, O.; Roldán-Ruiz, I. Effects of Population Size and Forest Management on Genetic Diversity and Structure of the Tuberous Orchid Orchis Mascula. Conserv. Genet. 2009, 10, 161–168. [Google Scholar] [CrossRef]
- Hurskainen, S.; Jäkäläniemi, A.; Ramula, S.; Tuomi, J. Tree Removal as a Management Strategy for the Lady’s Slipper Orchid, a Flagship Species for Herb-Rich Forest Conservation. For. Ecol. Manag. 2017, 406, 12–18. [Google Scholar] [CrossRef]
- Williams, J.L.; Jacquemyn, H.; Ochocki, B.M.; Brys, R.; Miller, T.E.X. Life History Evolution under Climate Change and Its Influence on the Population Dynamics of a Long-Lived Plant. J. Ecol. 2015, 103, 798–808. [Google Scholar] [CrossRef]
- van der Meer, S.; Jacquemyn, H.; Carey, P.D.; Jongejans, E. Recent Range Expansion of a Terrestrial Orchid Corresponds with Climate-Driven Variation in Its Population Dynamics. Oecologia 2016, 181, 435–448. [Google Scholar] [CrossRef]
- Kolanowska, M.; Jakubska-Busse, A. Is the Lady’s-Slipper Orchid (Cypripedium calceolus) Likely to Shortly Become Extinct in Europe?—Insights Based on Ecological Niche Modelling. PLoS ONE 2020, 15, e0228420. [Google Scholar] [CrossRef] [PubMed]
- Geppert, C.; Perazza, G.; Wilson, R.J.; Bertolli, A.; Prosser, F.; Melchiori, G.; Marini, L. Consistent Population Declines but Idiosyncratic Range Shifts in Alpine Orchids under Global Change. Nat. Commun. 2020, 11, 5835. [Google Scholar] [CrossRef]
- Selosse, M.A.; Weiß, M.; Jany, J.L.; Tillier, A. Communities and Populations of Sebacinoid Basidiomycetes Associated with the Achlorophyllous Orchid Neottia nidus-avis (L.) L.C.M. Rich. and Neighbouring Tree Ectomycorrhizae. Mol. Ecol. 2002, 11, 1831–1844. [Google Scholar] [CrossRef]
- Selosse, M.A.; Faccio, A.; Scappaticci, G.; Bonfante, P. Chlorophyllous and Achlorophyllous Specimens of Epipactis microphylla (Neottieae, Orchidaceae) Are Associated with Ectomycorrhizal Septomycetes, Including Truffles. Microb. Ecol. 2004, 47, 416–426. [Google Scholar] [CrossRef]
- Liebel, H.T.; Gebauer, G. Stable Isotope Signatures Confirm Carbon and Nitrogen Gain through Ectomycorrhizas in the Ghost Orchid Epipogium Aphyllum Swartz*. Plant Biol. 2011, 13, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Bellino, A.; Alfani, A.; Selosse, M.-A.; Guerrieri, R.; Borghetti, M.; Baldantoni, D. Nutritional Regulation in Mixotrophic Plants: New Insights from Limodorum abortivum. Oecologia 2014, 175, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.; Yokoya, K.; Kendon, J.P.; Sarasan, V. Diversity of Root-Associated Culturable Fungi of Cephalanthera rubra (Orchidaceae) in Relation to Soil Characteristics. PeerJ 2020, 8, e8695. [Google Scholar] [CrossRef]
- Pica, A.; Laghi, P. Atlante delle Orchidee del Parco Nazionale delle Foreste Casentinesi, Monte Falterona e Campigna. Guida alle Specie e Chiavi di Riconoscimento; Parco Nazionale delle Foreste Casentinesi, Monte Falterona e Campigna: Premiato Stabilimento Tipografico dei Comuni: Santa Sofia, Italy, 2023; ISBN 978-88-95719-04-7. [Google Scholar]
- Biondi, E.; Blasi, C.; Burrascano, S.; Casavecchia, S.; Copiz, R.; Del Vico, E.; Galdenzi, D.; Gigante, D.; Lasen, C.; Spampinato, G.; et al. Manuale Italiano di Interpretazione degli Habitat della Direttiva 92/43/CEE. 2009. Società Botanica Italiana. Ministero dell’Ambiente e della Tutela del Territorio e del Mare, D.P.N. Available online: http://vnr.unipg.it/habitat/ (accessed on 16 January 2024).
- Guisan, A.; Thuiller, W.; Zimmermann, N.E. Habitat Suitability and Distribution Models: With Applications in R; Cambridge University Press: Cambridge, UK, 2017; ISBN 978-0-521-76513-8. [Google Scholar]
- Djordjević, V.; Tsiftsis, S.; Lakušić, D.; Jovanović, S.; Stevanović, V. Factors Affecting the Distribution and Abundance of Orchids in Grasslands and Herbaceous Wetlands. Syst. Biodivers. 2016, 14, 355–370. [Google Scholar] [CrossRef]
- Tsiftsis, S.; Štípková, Z.; Kindlmann, P. Role of Way of Life, Latitude, Elevation and Climate on the Richness and Distribution of Orchid Species. Biodivers. Conserv. 2019, 28, 75–96. [Google Scholar] [CrossRef]
- Štípková, Z.; Kindlmann, P. Factors Determining the Distribution of Orchids—A Review with Examples from the Czech Republic. Eur. J. Environ. Sci. 2021, 11, 21–30. [Google Scholar] [CrossRef]
- Djordjević, V.; Tsiftsis, S.; Lakušić, D.; Jovanović, S.; Jakovljević, K.; Stevanović, V. Patterns of Distribution, Abundance and Composition of Forest Terrestrial Orchids. Biodivers. Conserv. 2020, 29, 4111–4134. [Google Scholar] [CrossRef]
- Štípková, Z.; Romportl, D.; Černocká, V.; Kindlmann, P. Factors Associated with the Distributions of Orchids in the Jeseníky Mountains, Czech Republic. Eur. J. Environ. Sci. 2017, 7, 135–145. [Google Scholar] [CrossRef]
- Štípková, Z.; Romportl, D.; Kindlmann, P. Which Environmental Factors Drive Distribution of Orchids? A Case Study from South Bohemia, Czech Republic. In Orchids Phytochemistry, Biology and Horticulture; Merillon, J.-M., Kodja, H., Eds.; Reference Series in Phytochemistry; Springer International Publishing: Cham, Swizerland, 2020; pp. 1–33. ISBN 978-3-030-11257-8. [Google Scholar]
- Evans, A.; Janssens, S.; Jacquemyn, H. Impact of Climate Change on the Distribution of Four Closely Related Orchis (Orchidaceae) Species. Diversity 2020, 12, 312. [Google Scholar] [CrossRef]
- Kolanowska, M. The Future of a Montane Orchid Species and the Impact of Climate Change on the Distribution of Its Pollinators and Magnet Species. Glob. Ecol. Conserv. 2021, 32, e01939. [Google Scholar] [CrossRef]
- Evans, A.; Jacquemyn, H. Range Size and Niche Breadth as Predictors of Climate-Induced Habitat Change in Epipactis (Orchidaceae). Front. Ecol. Evol. 2022, 10, 894616. [Google Scholar] [CrossRef]
- Kolanowska, M.; Nowak, S.; Rewicz, A. Will Greenland Be the Last Refuge for the Continental European Small-White Orchid? Niche Modeling of Future Distribution of Pseudorchis albida. Front. Environ. Sci. 2022, 10, 912428. [Google Scholar] [CrossRef]
- Bohl, C.L.; Kass, J.M.; Anderson, R.P. A New Null Model Approach to Quantify Performance and Significance for Ecological Niche Models of Species Distributions. J. Biogeogr. 2019, 46, 1101–1111. [Google Scholar] [CrossRef]
- Namyatova, A.A. Climatic Niche Comparison between Closely Related Trans-Palearctic Species of the Genus Orthocephalus (Insecta: Heteroptera: Miridae: Orthotylinae). PeerJ 2020, 8, e10517. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lu, X.; Li, K.; Wang, C.; Guna, A.; Zhang, J. Prediction of Potential Geographical Distribution Patterns of Actinidia arguta under Different Climate Scenarios. Sustainability 2021, 13, 3526. [Google Scholar] [CrossRef]
- Laface, V.L.A.; Musarella, C.M.; Tavilla, G.; Sorgonà, A.; Cano-Ortiz, A.; Quinto Canas, R.; Spampinato, G. Current and Potential Future Distribution of Endemic Salvia ceratophylloides Ard. (Lamiaceae). Land 2023, 12, 247. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, L.; Zhou, X.; Chen, R.; Zhao, G.; Zhang, F. Prediction of the Potentially Suitable Areas of Leonurus japonicus in China Based on Future Climate Change Using the Optimized MaxEnt Model. Ecol. Evol. 2023, 13, e10597. [Google Scholar] [CrossRef]
- Kolanowska, M.; Rewicz, A.; Baranow, P. Ecological Niche Modeling of the Pantropical Orchid Polystachya concreta (Orchidaceae) and Its Response to Climate Change. Sci. Rep. 2020, 10, 14801. [Google Scholar] [CrossRef]
- Tsiftsis, S.; Djordjević, V.; Tsiripidis, I. Neottia Cordata (Orchidaceae) at Its Southernmost Distribution Border in Europe: Threat Status and Effectiveness of Natura 2000 Network for Its Conservation. J. Nat. Conserv. 2019, 48, 27–35. [Google Scholar] [CrossRef]
- Fekete, R.; Vincze, O.; Nagy, J.; Löki, V.; Süveges, K.; Bódis, J.; Malkócs, T.; Lovas-kiss, Á.; Molnár V., A. North-Facing Roadside Slopes: Anthropogenic Climate Microrefugia for Orchids. Glob. Ecol. Conserv. 2023, 47, e02642. [Google Scholar] [CrossRef]
- Zhang, Z.; Yan, Y.; Tian, Y.; Li, J.; He, J.-S.; Tang, Z. Distribution and Conservation of Orchid Species Richness in China. Biol. Conserv. 2015, 181, 64–72. [Google Scholar] [CrossRef]
- Reina-Rodríguez, G.A.; Rubiano Mejía, J.E.; Castro Llanos, F.A.; Soriano, I. Orchids Distribution and Bioclimatic Niches as a Strategy to Climate Change in Areas of Tropical Dry Forest in Colombia. Lankesteriana 2017, 17, 17–47. [Google Scholar] [CrossRef][Green Version]
- Yudaputra, A.; Munawaroh, E.; Usmadi, D.; Purnomo, D.W.; Astuti, I.P.; Puspitaningtyas, D.M.; Handayani, T.; Garvita, R.V.; Aprilianti, P.; Wawangningrum, H.; et al. Vulnerability of Lowland and Upland Orchids in Their Spatially Response to Climate Change and Land Cover Change. Ecol. Inform. 2024, 80, 102534. [Google Scholar] [CrossRef]
- Anibaba, Q.A.; Dyderski, M.K.; Jagodziński, A.M. Predicted Range Shifts of Invasive Giant Hogweed (Heracleum Mantegazzianum) in Europe. Sci. Total Environ. 2022, 825, 154053. [Google Scholar] [CrossRef]
- Zhang, F.-X.; Yang, L.-H.; Wang, C.-J.; Zhang, C.-H.; Wan, J.-Z. Distribution and Conservation of Plants in the Northeastern Qinghai–Tibet Plateau under Climate Change. Diversity 2022, 14, 956. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.; Yang, Y.; Wang, T.; Wu, C.; Zhang, X. Prediction of Historical, Current, and Future Configuration of Tibetan Medicinal Herb Gymnadenia orchidis Based on the Optimized MaxEnt in the Qinghai–Tibet Plateau. Plants 2024, 13, 645. [Google Scholar] [CrossRef]
- Lenoir, J.; Gégout, J.C.; Marquet, P.A.; de Ruffray, P.; Brisse, H. A Significant Upward Shift in Plant Species Optimum Elevation During the 20th Century. Science 2008, 320, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Parmesan, C.; Hanley, M.E. Plants and Climate Change: Complexities and Surprises. Ann. Bot. 2015, 116, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Gebrewahid, Y.; Abrehe, S.; Meresa, E.; Eyasu, G.; Abay, K.; Gebreab, G.; Kidanemariam, K.; Adissu, G.; Abreha, G.; Darcha, G. Current and Future Predicting Potential Areas of Oxytenanthera abyssinica (A. Richard) Using MaxEnt Model under Climate Change in Northern Ethiopia. Ecol. Process. 2020, 9, 6. [Google Scholar] [CrossRef]
- Charitonidou, M.; Kougioumoutzis, K.; Halley, J.M. An Orchid in Retrograde: Climate-Driven Range Shift Patterns of Ophrys helenae in Greece. Plants 2021, 10, 470. [Google Scholar] [CrossRef] [PubMed]
- Canturk, U.; Kulaç, Ş. The Effects of Climate Change Scenarios on Tilia Ssp. in Turkey. Environ. Monit. Assess. 2021, 193, 771. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Shi, C.; Wen, G.; Lü, Z.; Ye, L.; Huang, Q.; Zhang, G. Potential Impacts of Climate Change on the Distribution of the Relict Plant Shaniodendron subaequale. Heliyon 2023, 9, e14402. [Google Scholar] [CrossRef] [PubMed]
- Kolanowska, M.; Kras, M.; Lipińska, M.; Mystkowska, K.; Szlachetko, D.L.; Naczk, A.M. Global Warming Not so Harmful for All Plants-Response of Holomycotrophic Orchid Species for the Future Climate Change OPEN. Sci. Rep. 2017, 7, 12704. [Google Scholar] [CrossRef]
- Kolanowska, M. Loss of Fungal Symbionts and Changes in Pollinator Availability Caused by Climate Change Will Affect the Distribution and Survival Chances of Myco-Heterotrophic Orchid Species. Sci. Rep. 2023, 13, 6848. [Google Scholar] [CrossRef]
- Jakubska-Busse, A.; Pielech, R.; Szczęśniak, E. The Extinction of Terrestrial Orchids in Europe: Does Disappearance of Cephalanthera Rich., 1817 (Orchidaceae, Neottieae) Species Show Pattern Consistent with the Elevation Gradient? Life Sci. J. 2014, 11, 140–144. [Google Scholar]
- Evans, A.; Jacquemyn, H. Impact of Mating System on Range Size and Niche Breadth in Epipactis (Orchidaceae). Ann. Bot. 2020, 126, 1203–1214. [Google Scholar] [CrossRef]
- Guisan, A.; Tingley, R.; Baumgartner, J.B.; Naujokaitis-Lewis, I.; Sutcliffe, P.R.; Tulloch, A.I.T.; Regan, T.J.; Brotons, L.; McDonald-Madden, E.; Mantyka-Pringle, C.; et al. Predicting Species Distributions for Conservation Decisions. Ecol. Lett. 2013, 16, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Duflot, R.; Avon, C.; Roche, P.; Bergès, L. Combining Habitat Suitability Models and Spatial Graphs for More Effective Landscape Conservation Planning: An Applied Methodological Framework and a Species Case Study. J. Nat. Conserv. 2018, 46, 38–47. [Google Scholar] [CrossRef]
- Rondinini, C.; Di Marco, M.; Chiozza, F.; Santulli, G.; Baisero, D.; Visconti, P.; Hoffmann, M.; Schipper, J.; Stuart, S.N.; Tognelli, M.F.; et al. Global Habitat Suitability Models of Terrestrial Mammals. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 2633–2641. [Google Scholar] [CrossRef] [PubMed]
- Lyet, A.; Thuiller, W.; Cheylan, M.; Besnard, A. Fine-Scale Regional Distribution Modelling of Rare and Threatened Species: Bridging GIS Tools and Conservation in Practice. Divers. Distrib. 2013, 19, 651–663. [Google Scholar] [CrossRef]
- Araújo, M.B.; Alagador, D.; Cabeza, M.; Nogués-Bravo, D.; Thuiller, W. Climate Change Threatens European Conservation Areas. Ecol. Lett. 2011, 14, 484–492. [Google Scholar] [CrossRef] [PubMed]
- D’Amen, M.; Bombi, P.; Pearman, P.B.; Schmatz, D.R.; Zimmermann, N.E.; Bologna, M.A. Will Climate Change Reduce the Efficacy of Protected Areas for Amphibian Conservation in Italy? Biol. Conserv. 2011, 144, 989–997. [Google Scholar] [CrossRef]
- Signorello, G.; Prato, C.; Marzo, A.; Ientile, R.; Cucuzza, G.; Sciandrello, S.; Martínez-López, J.; Balbi, S.; Villa, F. Are Protected Areas Covering Important Biodiversity Sites? An Assessment of the Nature Protection Network in Sicily (Italy). Land Use Policy 2018, 78, 593–602. [Google Scholar] [CrossRef]
- Sebastiani, A.; Fares, S. Spatial Prioritization of Ecosystem Services for Land Conservation: The Case Study of Central Italy. Forests 2023, 14, 145. [Google Scholar] [CrossRef]
- Friedrichs, M.; Hermoso, V.; Bremerich, V.; Langhans, S.D. Evaluation of Habitat Protection under the European Natura 2000 Conservation Network—The Example for Germany. PLoS ONE 2018, 13, e0208264. [Google Scholar] [CrossRef]
- Alba-Patiño, D.; Martínez-Hernández, F.; Mota Poveda, J.F. Determination of Sites of Special Importance for the Conservation of Threatened Orchid Species in Colombia. Mediterr. Bot. 2021, 42, e67589. [Google Scholar] [CrossRef]
- Geoportale Nazionale. Available online: http://www.pcn.minambiente.it/viewer/ (accessed on 30 May 2024).
- Pesaresi, S.; Galdenzi, D.; Biondi, E.; Casavecchia, S. Bioclimate of Italy: Application of the Worldwide Bioclimatic Classification System. J. Maps 2014, 10, 538–553. [Google Scholar] [CrossRef]
- QGIS.org QGIS Geographic Information System. QGIS Association. Available online: https://www.qgis.org/it/site/ (accessed on 20 December 2023).
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.r-project.org/ (accessed on 20 December 2023).
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2023; Available online: http://www.rstudio.com/ (accessed on 20 December 2023).
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. spThin: An R Package for Spatial Thinning of Species Occurrence Records for Use in Ecological Niche Models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- Aidoo, O.F.; Souza, P.G.C.; da Silva, R.S.; Santana, P.A.; Picanço, M.C.; Kyerematen, R.; Sètamou, M.; Ekesi, S.; Borgemeister, C. Climate-Induced Range Shifts of Invasive Species (Diaphorina citri Kuwayama). Pest Manag. Sci. 2022, 78, 2534–2549. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Bertola, L.V.; Hoskin, C.J. Species Distribution Modelling of the Endangered Mahogany Glider (Petaurus gracilis) Reveals Key Areas for Targeted Survey and Conservation. Austral Ecol. 2023, 48, 289–312. [Google Scholar] [CrossRef]
- Boria, R.A.; Olson, L.E.; Goodman, S.M.; Anderson, R.P. Spatial Filtering to Reduce Sampling Bias Can Improve the Performance of Ecological Niche Models. Ecol. Model. 2014, 275, 73–77. [Google Scholar] [CrossRef]
- Kramer-Schadt, S.; Niedballa, J.; Pilgrim, J.D.; Schröder, B.; Lindenborn, J.; Reinfelder, V.; Stillfried, M.; Heckmann, I.; Scharf, A.K.; Augeri, D.M.; et al. The Importance of Correcting for Sampling Bias in MaxEnt Species Distribution Models. Divers. Distrib. 2013, 19, 1366–1379. [Google Scholar] [CrossRef]
- Fourcade, Y.; Engler, J.O.; Rödder, D.; Secondi, J. Mapping Species Distributions with MAXENT Using a Geographically Biased Sample of Presence Data: A Performance Assessment of Methods for Correcting Sampling Bias. PLoS ONE 2014, 9, e97122. [Google Scholar] [CrossRef] [PubMed]
- Mod, H.K.; Scherrer, D.; Luoto, M.; Guisan, A. What We Use Is Not What We Know: Environmental Predictors in Plant Distribution Models. J. Veg. Sci. 2016, 27, 1308–1322. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-Km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Tuanmu, M.-N.; Jetz, W. A Global 1-Km Consensus Land-Cover Product for Biodiversity and Ecosystem Modelling. Glob. Ecol. Biogeogr. 2014, 23, 1031–1045. [Google Scholar] [CrossRef]
- Venter, O.; Sanderson, E.W.; Magrach, A.; Allan, J.R.; Beher, J.; Jones, K.R.; Possingham, H.P.; Laurance, W.F.; Wood, P.; Fekete, B.M.; et al. Sixteen Years of Change in the Global Terrestrial Human Footprint and Implications for Biodiversity Conservation. Nat. Commun. 2016, 7, 12558. [Google Scholar] [CrossRef] [PubMed]
- Venter, O.; Sanderson, E.W.; Magrach, A.; Allan, J.R.; Beher, J.; Jones, K.R.; Levy, M.A.; Watson, J.E. Last of the Wild Project, Version 3 (LWP-3): 2009 Human Footprint, 2018 Release. 2018. Palisades, New York: NASA Socioeconomic Data and Applications Center (SEDAC). Available online: https://doi.org/10.7927/H46T0JQ4 (accessed on 16 January 2024).
- Poggio, L.; de Sousa, L.M.; Batjes, N.H.; Heuvelink, G.B.M.; Kempen, B.; Ribeiro, E.; Rossiter, D. SoilGrids 2.0: Producing Soil Information for the Globe with Quantified Spatial Uncertainty. SOIL 2021, 7, 217–240. [Google Scholar] [CrossRef]
- Turek, M.E.; Poggio, L.; Batjes, N.H.; Armindo, R.A.; de Jong van Lier, Q.; de Sousa, L.; Heuvelink, G.B.M. Global Mapping of Volumetric Water Retention at 100, 330 and 15 000 Cm Suction Using the WoSIS Database. Int. Soil Water Conserv. Res. 2023, 11, 225–239. [Google Scholar] [CrossRef]
- Júnior, P.D.M.; Nóbrega, C.C. Evaluating Collinearity Effects on Species Distribution Models: An Approach Based on Virtual Species Simulation. PLoS ONE 2018, 13, e0202403. [Google Scholar] [CrossRef]
- Sillero, N.; Barbosa, A.M. Common Mistakes in Ecological Niche Models. Int. J. Geogr. Inf. Sci. 2021, 35, 213–226. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A Review of Methods to Deal with It and a Simulation Study Evaluating Their Performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Shrestha, N. Detecting Multicollinearity in Regression Analysis. Am. J. Appl. Math. Stat. 2020, 8, 39–42. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4(1): 9pp. Available online: https://www.nhm.uio.no/english/research/resources/past/index.html (accessed on 22 December 2023).
- Wang, Y.; Zhao, R.; Zhou, X.; Zhang, X.; Zhao, G.; Zhang, F. Prediction of Potential Distribution Areas and Priority Protected Areas of Agastache rugosa Based on Maxent Model and Marxan Model. Front. Plant Sci. 2023, 14, 1200796. [Google Scholar] [CrossRef]
- Schnase, J.L.; Carroll, M.L.; Gill, R.L.; Tamkin, G.S.; Li, J.; Strong, S.L.; Maxwell, T.P.; Aronne, M.E.; Spradlin, C.S. Toward a Monte Carlo Approach to Selecting Climate Variables in MaxEnt. PLoS ONE 2021, 16, e0237208. [Google Scholar] [CrossRef]
- Miller, R.G. The Jackknife—A Review. Biometrika 1974, 61, 1–15. [Google Scholar] [CrossRef]
- Songer, M.; Delion, M.; Biggs, A.; Huang, Q. Modeling Impacts of Climate Change on Giant Panda Habitat. Int. J. Ecol. 2012, 2012, e108752. [Google Scholar] [CrossRef]
- Wei, B.; Wang, R.; Hou, K.; Wang, X.; Wu, W. Predicting the Current and Future Cultivation Regions of Carthamus Tinctorius L. Using MaxEnt Model under Climate Change in China. Glob. Ecol. Conserv. 2018, 16, e00477. [Google Scholar] [CrossRef]
- Wang, R.; Li, Q.; He, S.; Liu, Y.; Wang, M.; Jiang, G. Modeling and Mapping the Current and Future Distribution of Pseudomonas syringae Pv. Actinidiae under Climate Change in China. PLoS ONE 2018, 13, e0192153. [Google Scholar] [CrossRef] [PubMed]
- Worthington, T.A.; Zhang, T.; Logue, D.R.; Mittelstet, A.R.; Brewer, S.K. Landscape and Flow Metrics Affecting the Distribution of a Federally-Threatened Fish: Improving Management, Model Fit, and Model Transferability. Ecol. Model. 2016, 342, 1–18. [Google Scholar] [CrossRef]
- Ab Lah, N.Z.; Yusop, Z.; Hashim, M.; Mohd Salim, J.; Numata, S. Predicting the Habitat Suitability of Melaleuca cajuputi Based on the MaxEnt Species Distribution Model. Forests 2021, 12, 1449. [Google Scholar] [CrossRef]
- Deng, X.; Xu, D.; Liao, W.; Wang, R.; Zhuo, Z. Predicting the Distributions of Scleroderma guani (Hymenoptera: Bethylidae) under Climate Change in China. Ecol. Evol. 2022, 12, e9410. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhuo, Z.; Wang, R.; Ye, M.; Pu, B. Modeling the Distribution of Zanthoxylum armatum in China with MaxEnt Modeling. Glob. Ecol. Conserv. 2019, 19, e00691. [Google Scholar] [CrossRef]
- Zhao, Y.; Deng, X.; Xiang, W.; Chen, L.; Ouyang, S. Predicting Potential Suitable Habitats of Chinese Fir under Current and Future Climatic Scenarios Based on Maxent Model. Ecol. Inform. 2021, 64, 101393. [Google Scholar] [CrossRef]
- Chen, K.; Wang, B.; Chen, C.; Zhou, G. MaxEnt Modeling to Predict the Current and Future Distribution of Pomatosace filicula under Climate Change Scenarios on the Qinghai–Tibet Plateau. Plants 2022, 11, 670. [Google Scholar] [CrossRef]
- Naimi, B.; Hamm, N.A.S.; Groen, T.A.; Skidmore, A.K.; Toxopeus, A.G. Where Is Positional Uncertainty a Problem for Species Distribution Modelling? Ecography 2014, 37, 191–203. [Google Scholar] [CrossRef]
- Fois, M.; Cuena-Lombraña, A.; Fenu, G.; Bacchetta, G. Using Species Distribution Models at Local Scale to Guide the Search of Poorly Known Species: Review, Methodological Issues and Future Directions. Ecol. Model. 2018, 385, 124–132. [Google Scholar] [CrossRef]
- Tesfamariam, B.G.; Gessesse, B.; Melgani, F. MaxEnt-Based Modeling of Suitable Habitat for Rehabilitation of Podocarpus Forest at Landscape-Scale. Environ. Syst. Res. 2022, 11, 4. [Google Scholar] [CrossRef]
- Intergovernmental Panel on Climate Change (IPCC) (Ed.) Future Global Climate: Scenario-Based Projections and Near-Term Information. In Climate Change 2021—The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2023; pp. 553–672. ISBN 978-1-00-915788-9. [Google Scholar]
- Lovato, T.; Peano, D.; Butenschön, M. CMCC CMCC-ESM2 Model Output Prepared for CMIP6 ScenarioMIP. 2021. Version 20231230. Earth System Grid Federation. Available online: https://doi.org/10.22033/ESGF/CMIP6.13168 (accessed on 30 December 2023).
- Lovato, T.; Peano, D.; Butenschön, M.; Materia, S.; Iovino, D.; Scoccimarro, E.; Fogli, P.G.; Cherchi, A.; Bellucci, A.; Gualdi, S.; et al. CMIP6 Simulations With the CMCC Earth System Model (CMCC-ESM2). J. Adv. Model. Earth Syst. 2022, 14, e2021MS002814. [Google Scholar] [CrossRef]
- Kriegler, E.; O’Neill, B.C.; Hallegatte, S.; Kram, T.; Lempert, R.J.; Moss, R.H.; Wilbanks, T. The Need for and Use of Socio-Economic Scenarios for Climate Change Analysis: A New Approach Based on Shared Socio-Economic Pathways. Glob. Environ. Chang. 2012, 22, 807–822. [Google Scholar] [CrossRef]
- O’Neill, B.C.; Kriegler, E.; Riahi, K.; Ebi, K.L.; Hallegatte, S.; Carter, T.R.; Mathur, R.; van Vuuren, D.P. A New Scenario Framework for Climate Change Research: The Concept of Shared Socioeconomic Pathways. Clim. Chang. 2014, 122, 387–400. [Google Scholar] [CrossRef]
- O’Neill, B.C.; Kriegler, E.; Ebi, K.L.; Kemp-Benedict, E.; Riahi, K.; Rothman, D.S.; van Ruijven, B.J.; van Vuuren, D.P.; Birkmann, J.; Kok, K.; et al. The Roads Ahead: Narratives for Shared Socioeconomic Pathways Describing World Futures in the 21st Century. Glob. Environ. Chang. 2017, 42, 169–180. [Google Scholar] [CrossRef]
- Popp, A.; Calvin, K.; Fujimori, S.; Havlik, P.; Humpenöder, F.; Stehfest, E.; Bodirsky, B.L.; Dietrich, J.P.; Doelmann, J.C.; Gusti, M.; et al. Land-Use Futures in the Shared Socio-Economic Pathways. Glob. Environ. Chang. 2017, 42, 331–345. [Google Scholar] [CrossRef]
- Riahi, K.; van Vuuren, D.P.; Kriegler, E.; Edmonds, J.; O’Neill, B.C.; Fujimori, S.; Bauer, N.; Calvin, K.; Dellink, R.; Fricko, O.; et al. The Shared Socioeconomic Pathways and Their Energy, Land Use, and Greenhouse Gas Emissions Implications: An Overview. Glob. Environ. Chang. 2017, 42, 153–168. [Google Scholar] [CrossRef]
- Meinshausen, M.; Nicholls, Z.R.J.; Lewis, J.; Gidden, M.J.; Vogel, E.; Freund, M.; Beyerle, U.; Gessner, C.; Nauels, A.; Bauer, N.; et al. The Shared Socio-Economic Pathway (SSP) Greenhouse Gas Concentrations and Their Extensions to 2500. Geosci. Model Dev. 2020, 13, 3571–3605. [Google Scholar] [CrossRef]
- Kolanowska, M. Future Distribution of the Epiphytic Leafless Orchid (Dendrophylax lindenii), Its Pollinators and Phorophytes Evaluated Using Niche Modelling and Three Different Climate Change Projections. Sci. Rep. 2023, 13, 15242. [Google Scholar] [CrossRef] [PubMed]
- Intergovernmental Panel on Climate Change (IPCC) (Ed.) Summary for Policymakers. In Climate Change 2021—The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2023; pp. 3–32. ISBN 978-1-00-915788-9. [Google Scholar]
- Phillips, S.J.; Dudík, M. Modeling of Species Distributions with Maxent: New Extensions and a Comprehensive Evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Schapire, R.E. [Internet] Maxent Software for Modeling Species Niches and Distributions (Version 3.4.4). 2023. Available online: http://biodiversityinformatics.amnh.org/open_source/maxent/ (accessed on 2 November 2023).
- Shcheglovitova, M.; Anderson, R.P. Estimating Optimal Complexity for Ecological Niche Models: A Jackknife Approach for Species with Small Sample Sizes. Ecol. Model. 2013, 269, 9–17. [Google Scholar] [CrossRef]
- Radosavljevic, A.; Anderson, R.P. Making Better Maxent Models of Species Distributions: Complexity, Overfitting and Evaluation. J. Biogeogr. 2014, 41, 629–643. [Google Scholar] [CrossRef]
- Morales, N.S.; Fernández, I.C.; Baca-González, V. MaxEnt’s Parameter Configuration and Small Samples: Are We Paying Attention to Recommendations? A Systematic Review. PeerJ 2017, 5, e3093. [Google Scholar] [CrossRef] [PubMed]
- Muscarella, R.; Galante, P.J.; Soley-Guardia, M.; Boria, R.A.; Kass, J.M.; Uriarte, M.; Anderson, R.P. ENMeval: An R Package for Conducting Spatially Independent Evaluations and Estimating Optimal Model Complexity for Maxent Ecological Niche Models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- Kass, J.M.; Muscarella, R.; Galante, P.J.; Bohl, C.L.; Pinilla-Buitrago, G.E.; Boria, R.A.; Soley-Guardia, M.; Anderson, R.P. ENMeval 2.0: Redesigned for Customizable and Reproducible Modeling of Species’ Niches and Distributions. Methods Ecol. Evol. 2021, 12, 1602–1608. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A Statistical Explanation of MaxEnt for Ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; Chen, S.-T.; Gao, Y.; Yang, L.; Yu, H. Prediction of Global Potential Suitable Habitats of Nicotiana alata Link et Otto Based on MaxEnt Model. Sci. Rep. 2023, 13, 4851. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.L.; Seifert, S.N. Ecological Niche Modeling in Maxent: The Importance of Model Complexity and the Performance of Model Selection Criteria. Ecol. Appl. Publ. Ecol. Soc. Am. 2011, 21, 335–342. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A. A Practical Guide to MaxEnt for Modeling Species’ Distributions: What It Does, and Why Inputs and Settings Matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- West, A.M.; Kumar, S.; Brown, C.S.; Stohlgren, T.J.; Bromberg, J. Field Validation of an Invasive Species Maxent Model. Ecol. Inform. 2016, 36, 126–134. [Google Scholar] [CrossRef]
- Tang, C.Q.; Matsui, T.; Ohashi, H.; Dong, Y.-F.; Momohara, A.; Herrando-Moraira, S.; Qian, S.; Yang, Y.; Ohsawa, M.; Luu, H.T.; et al. Identifying Long-Term Stable Refugia for Relict Plant Species in East Asia. Nat. Commun. 2018, 9, 4488. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.S.; Kumar, L.; Shabani, F.; Picanço, M.C. Risk of Spread of Tomato Yellow Leaf Curl Virus (TYLCV) in Tomato Crops under Various Climate Change Scenarios. Agric. Syst. 2019, 173, 524–535. [Google Scholar] [CrossRef]
- Zhao, Q.; Mi, Z.; Lu, C.; Zhang, X.; Chen, L.; Wang, S.; Niu, J.; Wang, Z. Predicting Potential Distribution of Ziziphus spinosa (Bunge) H.H. Hu Ex F.H. Chen in China under Climate Change Scenarios. Ecol. Evol. 2022, 12, e8629. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Berry, P.M.; Dawson, T.P.; Pearson, R.G. Selecting Thresholds of Occurrence in the Prediction of Species Distributions. Ecography 2005, 28, 385–393. [Google Scholar] [CrossRef]
- Liu, C.; White, M.; Newell, G. Selecting Thresholds for the Prediction of Species Occurrence with Presence-Only Data. J. Biogeogr. 2013, 40, 778–789. [Google Scholar] [CrossRef]
- Liu, C.; Newell, G.; White, M. On the Selection of Thresholds for Predicting Species Occurrence with Presence-only Data. Ecol. Evol. 2015, 6, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, M.; Li, C.; Liu, Z. Optimized Maxent Model Predictions of Climate Change Impacts on the Suitable Distribution of Cunninghamia lanceolata in China. Forests 2020, 11, 302. [Google Scholar] [CrossRef]
- Khan, A.M.; Li, Q.; Saqib, Z.; Khan, N.; Habib, T.; Khalid, N.; Majeed, M.; Tariq, A. MaxEnt Modelling and Impact of Climate Change on Habitat Suitability Variations of Economically Important Chilgoza Pine (Pinus gerardiana Wall.) in South Asia. Forests 2022, 13, 715. [Google Scholar] [CrossRef]
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araújo, M.B. BIOMOD—A Platform for Ensemble Forecasting of Species Distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- Thuiller, W.; Georges, D.; Gueguen, M.; Engler, R.; Breiner, F.; Lafourcade, B.; Patin, R. Biomod2: Ensemble Platform for Species Distribution Modeling. R Package Version 4.2-5. Available online: https://biomodhub.github.io/biomod2/authors.html#citation (accessed on 4 January 2024).
- Zhang, Q.; Shen, X.; Jiang, X.; Fan, T.; Liang, X.; Yan, W. MaxEnt Modeling for Predicting Suitable Habitat for Endangered Tree Keteleeria davidiana (Pinaceae) in China. Forests 2023, 14, 394. [Google Scholar] [CrossRef]



| ID | Code | Variable Description | Unit |
|---|---|---|---|
| 1 | BIO1 | Annual Mean Temperature | °C |
| 2 | BIO2 | Mean Diurnal Range (Mean of monthly (max temp—min temp)) | °C |
| 3 | BIO3 | Isothermality (BIO2/BIO7) (×100) | % |
| 4 | BIO4 | Temperature Seasonality (standard deviation ×100) 3 | % |
| 5 | BIO5 | Max Temperature of Warmest Month | °C |
| 6 | BIO6 | Min Temperature of Coldest Month | °C |
| 7 | BIO7 | Temperature Annual Range (BIO5-BIO6) | °C |
| 8 | BIO8 | Mean Temperature of Wettest Quarter | °C |
| 9 | BIO9 | Mean Temperature of Driest Quarter | °C |
| 10 | BIO10 | Mean Temperature of Warmest Quarter | °C |
| 11 | BIO11 | Mean Temperature of Coldest Quarter 3 | °C |
| 12 | BIO12 | Annual Precipitation | mm |
| 13 | BIO13 | Precipitation of Wettest Month | mm |
| 14 | BIO14 | Precipitation of Driest Month 2,3 | mm |
| 15 | BIO15 | Precipitation Seasonality (Coefficient of Variation) 1,2 | % |
| 16 | BIO16 | Precipitation of Wettest Quarter | mm |
| 17 | BIO17 | Precipitation of Driest Quarter | mm |
| 18 | BIO18 | Precipitation of Warmest Quarter 1 | mm |
| 19 | BIO19 | Precipitation of Coldest Quarter | mm |
| 20 | LC01 | Evergreen/Deciduous Needleleaf Trees | % |
| 21 | LC02 | Evergreen Broadleaf Trees | % |
| 22 | LC03 | Deciduous Broadleaf Trees | % |
| 23 | LC04 | Mixed/Other Trees | % |
| 24 | LC05 | Shrubs | % |
| 25 | LC06 | Herbaceous Vegetation | % |
| 26 | LC07 | Cultivated and Managed Vegetation | % |
| 27 | LC08 | Regularly Flooded Vegetation | % |
| 28 | LC09 | Urban/Built-up | % |
| 29 | LC10 | Snow/Ice | % |
| 30 | LC11 | Barren 3 | % |
| 31 | LC12 | Open Water | % |
| 32 | SOIL1 | Bulk density of the fine earth fraction 1,2,3 | cg/cm3 |
| 33 | SOIL2 | Cation Exchange Capacity of the soil | mmol(c)/kg |
| 34 | SOIL3 | Volumetric fraction of coarse fragments (>2 mm) 1 | cm3/dm3 (vol‰) |
| 35 | SOIL4 | Proportion of clay particles (<0.002 mm) in the fine earth fraction 2 | g/kg |
| 36 | SOIL5 | Total nitrogen (N) | cg/kg |
| 37 | SOIL6 | Soil pH | pHx10 |
| 38 | SOIL7 | Proportion of sand particles (>0.05 mm) in the fine earth fraction | g/kg |
| 39 | SOIL8 | Proportion of silt particles (≥0.002 mm and ≤0.05 mm) in the fine earth fraction | g/kg |
| 40 | SOIL9 | Soil organic carbon content in the fine earth fraction | dg/kg |
| 41 | SOIL10 | Organic carbon density | hg/m3 |
| 42 | SOIL11 | Organic carbon stocks | t/ha |
| 43 | SLP | Slope 2 | degree |
| 44 | ELV | Elevation | meter |
| 45 | ASP | Aspect | degree |
| 46 | CHP | Cumulative human pressure on the environment 1 |
| Species | Environmental Variables (EV) | Permutation Importance (%) | Percent Contribution (%) |
|---|---|---|---|
| Cephalanthera rubra | SOIL1 BIO18 BIO15 CHP SOIL3 | 35.5 25.2 18.5 11.4 9.5 | 34 24.7 8.1 27.4 5.8 |
| Epipactis microphylla | SOIL1 BIO14 SOIL4 SLOPE BIO15 | 46.6 19 17.6 12.5 10.4 | 45 18.3 21.4 8.6 6.7 |
| Limodorum abortivum | BIO04 BIO14 SOIL1 BIO11 LC11 | 39.5 35.2 17.7 6.7 1 | 30.2 32.5 28.9 7.4 1 |
| Time | Scenario | Unit | Tot. | Unsuitability | Low Suitability | Moderate Suitability | High Suitability |
|---|---|---|---|---|---|---|---|
| Present | km2 | 315.28 | 97.92 | 144.44 | 113.89 | 56.94 | |
| % | 76.30 | 23.70 | 34.96 | 27.56 | 13.78 | ||
| 2030s | SSP245 | km2 | 140.28 | 272.92 | 85.42 | 46.53 | 8.33 |
| % | 33.95 | 66.05 | 20.67 | 11.26 | 2.02 | ||
| % inc./dec. | −55.51 | +178.72 | −40.87 | −59.15 | −85.37 | ||
| SSP585 | km2 | 294.44 | 118.75 | 166.67 | 81.25 | 46.53 | |
| % | 71.26 | 28.74 | 40.34 | 19.66 | 11.26 | ||
| % inc./dec. | −6.61 | +21.28 | +15.38 | −28.66 | −18.29 | ||
| 2050s | SSP245 | km2 | 281.94 | 131.25 | 166.67 | 80.56 | 34.72 |
| % | 68.24 | 31.76 | 40.34 | 19.50 | 8.40 | ||
| % inc./dec. | −10.57 | +34.04 | +15.38 | −29.27 | −39.02 | ||
| SSP585 | km2 | 27.08 | 386.11 | 25.69 | 1.39 | 0.00 | |
| % | 6.55 | 93.45 | 6.22 | 0.34 | 0.00 | ||
| % inc./dec. | −91.41 | +294.33 | −82.21 | −98.78 | −100.00 |
| Time | Scenario | Unit | Tot. | Unsuitability | Low Suitability | Moderate Suitability | High Suitability |
|---|---|---|---|---|---|---|---|
| Present | km2 | 225.69 | 187.50 | 107.64 | 79.17 | 38.89 | |
| % | 54.62 | 45.38 | 26.05 | 19.16 | 9.41 | ||
| 2030s | SSP245 | km2 | 144.44 | 268.75 | 97.92 | 37.50 | 9.03 |
| % | 34.96 | 65.04 | 23.70 | 9.08 | 2.18 | ||
| % inc./dec. | −36.00 | +43.33 | −9.03 | −52.63 | −76.79 | ||
| SSP585 | km2 | 198.61 | 214.58 | 115.97 | 64.58 | 18.06 | |
| % | 48.07 | 51.93 | 28.07 | 15.63 | 4.37 | ||
| % inc./dec. | −12.00 | +14.44 | +7.74 | −18.42 | −53.57 | ||
| 2050s | SSP245 | km2 | 195.14 | 218.06 | 120.83 | 51.39 | 22.92 |
| % | 47.23 | 52.77 | 29.24 | 12.44 | 5.55 | ||
| % inc./dec. | −13.54 | +16.30 | +12.26 | −35.09 | −41.07 | ||
| SSP585 | km2 | 11.81 | 401.39 | 11.81 | 0.00 | 0.00 | |
| % | 2.86 | 97.14 | 2.86 | 0.00 | 0.00 | ||
| % inc./dec. | −94.77 | +114.07 | −89.03 | −100.00 | −100.00 |
| Time | Scenario | Unit | Tot. | Unsuitability | Low Suitability | Moderate Suitability | High Suitability |
|---|---|---|---|---|---|---|---|
| Present | km2 | 217.36 | 195.83 | 93.75 | 82.64 | 40.97 | |
| % | 52.61 | 47.39 | 22.69 | 20.00 | 9.92 | ||
| 2030s | SSP245 | km2 | 320.83 | 92.36 | 59.72 | 95.14 | 165.97 |
| % | 77.65 | 22.35 | 14.45 | 23.03 | 40.17 | ||
| % inc./dec. | +47.60 | −52.84 | −36.30 | +15.13 | +305.08 | ||
| SSP585 | km2 | 202.78 | 210.42 | 91.67 | 90.97 | 20.14 | |
| % | 49.08 | 50.92 | 22.18 | 22.02 | 4.87 | ||
| % inc./dec. | −6.71 | +7.45 | −2.22 | +10.08 | −50.85 | ||
| 2050s | SSP245 | km2 | 309.03 | 104.17 | 75.69 | 100.69 | 132.64 |
| % | 74.79 | 25.21 | 18.32 | 24.37 | 32.10 | ||
| % inc./dec. | +42.17 | −46.81 | −19.26 | +21.85 | +223.73 | ||
| SSP585 | km2 | 79.17 | 334.03 | 70.83 | 8.33 | 0.00 | |
| % | 19.16 | 80.84 | 17.14 | 2.02 | 0.00 | ||
| % inc./dec. | −63.58 | +70.57 | −24.44 | −89.92 | −100.00 |
| Time | Scenario | Units | CRS | Loss | Stable | Gain | %Loss | %Gain | SRC |
|---|---|---|---|---|---|---|---|---|---|
| Present | cells | 454 | |||||||
| km2 | 315.28 | ||||||||
| 2030s | SSP245 | cells | 252 | 202 | 0 | 55.51 | 0.00 | −55.51 | |
| km2 | 175.00 | 140.28 | 0.00 | ||||||
| SSP585 | cells | 34 | 420 | 4 | 7.49 | 0.88 | −6.61 | ||
| km2 | 23.61 | 291.67 | 2.78 | ||||||
| 2050s | SSP245 | cells | 49 | 405 | 1 | 10.79 | 0.22 | −10.57 | |
| km2 | 34.03 | 281.25 | 0.69 | ||||||
| SSP585 | cells | 415 | 39 | 0 | 91.41 | 0.00 | −91.41 | ||
| km2 | 288.19 | 27.08 | 0.00 |
| Time | Scenario | Units | CRS | Loss | Stable | Gain | % Loss | % Gain | SRC |
|---|---|---|---|---|---|---|---|---|---|
| Present | cells | 325 | |||||||
| km2 | 225.69 | ||||||||
| 2030s | SSP245 | cells | 117 | 208 | 0 | 36.00 | 0.00 | −36.00 | |
| km2 | 81.25 | 144.44 | 0.00 | ||||||
| SSP585 | cells | 39 | 286 | 0 | 12.00 | 0.00 | −12.00 | ||
| km2 | 27.08 | 198.61 | 0.00 | ||||||
| 2050s | SSP245 | cells | 44 | 281 | 0 | 13.54 | 0.00 | −13.54 | |
| km2 | 30.56 | 195.14 | 0.00 | ||||||
| SSP585 | cells | 308 | 17 | 0 | 94.77 | 0.00 | −94.77 | ||
| km2 | 213.89 | 11.81 | 0.00 |
| Time | Scenario | Units | CRS | Loss | Stable | Gain | % Loss | % Gain | SRC |
|---|---|---|---|---|---|---|---|---|---|
| Present | cells | 313 | |||||||
| km2 | 217.36 | ||||||||
| 2030s | SSP245 | cells | 60 | 253 | 209 | 19.17 | 66.77 | 47.60 | |
| km2 | 41.67 | 175.69 | 145.14 | ||||||
| SSP585 | cells | 152 | 161 | 131 | 48.56 | 41.85 | −6.71 | ||
| km2 | 105.56 | 111.81 | 90.97 | ||||||
| 2050s | SSP245 | cells | 64 | 249 | 196 | 20.45 | 62.62 | 42.17 | |
| km2 | 44.44 | 172.92 | 136.11 | ||||||
| SSP585 | cells | 297 | 16 | 98 | 94.89 | 31.31 | −63.58 | ||
| km2 | 206.25 | 11.11 | 68.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pica, A.; Vela, D.; Magrini, S. Forest Orchids under Future Climate Scenarios: Habitat Suitability Modelling to Inform Conservation Strategies. Plants 2024, 13, 1810. https://doi.org/10.3390/plants13131810
Pica A, Vela D, Magrini S. Forest Orchids under Future Climate Scenarios: Habitat Suitability Modelling to Inform Conservation Strategies. Plants. 2024; 13(13):1810. https://doi.org/10.3390/plants13131810
Chicago/Turabian StylePica, Antonio, Daniele Vela, and Sara Magrini. 2024. "Forest Orchids under Future Climate Scenarios: Habitat Suitability Modelling to Inform Conservation Strategies" Plants 13, no. 13: 1810. https://doi.org/10.3390/plants13131810
APA StylePica, A., Vela, D., & Magrini, S. (2024). Forest Orchids under Future Climate Scenarios: Habitat Suitability Modelling to Inform Conservation Strategies. Plants, 13(13), 1810. https://doi.org/10.3390/plants13131810









