Xylem Sap Proteome Analysis Provides Insight into Root–Shoot Communication in Response to flg22
Abstract
:1. Introduction
2. Results
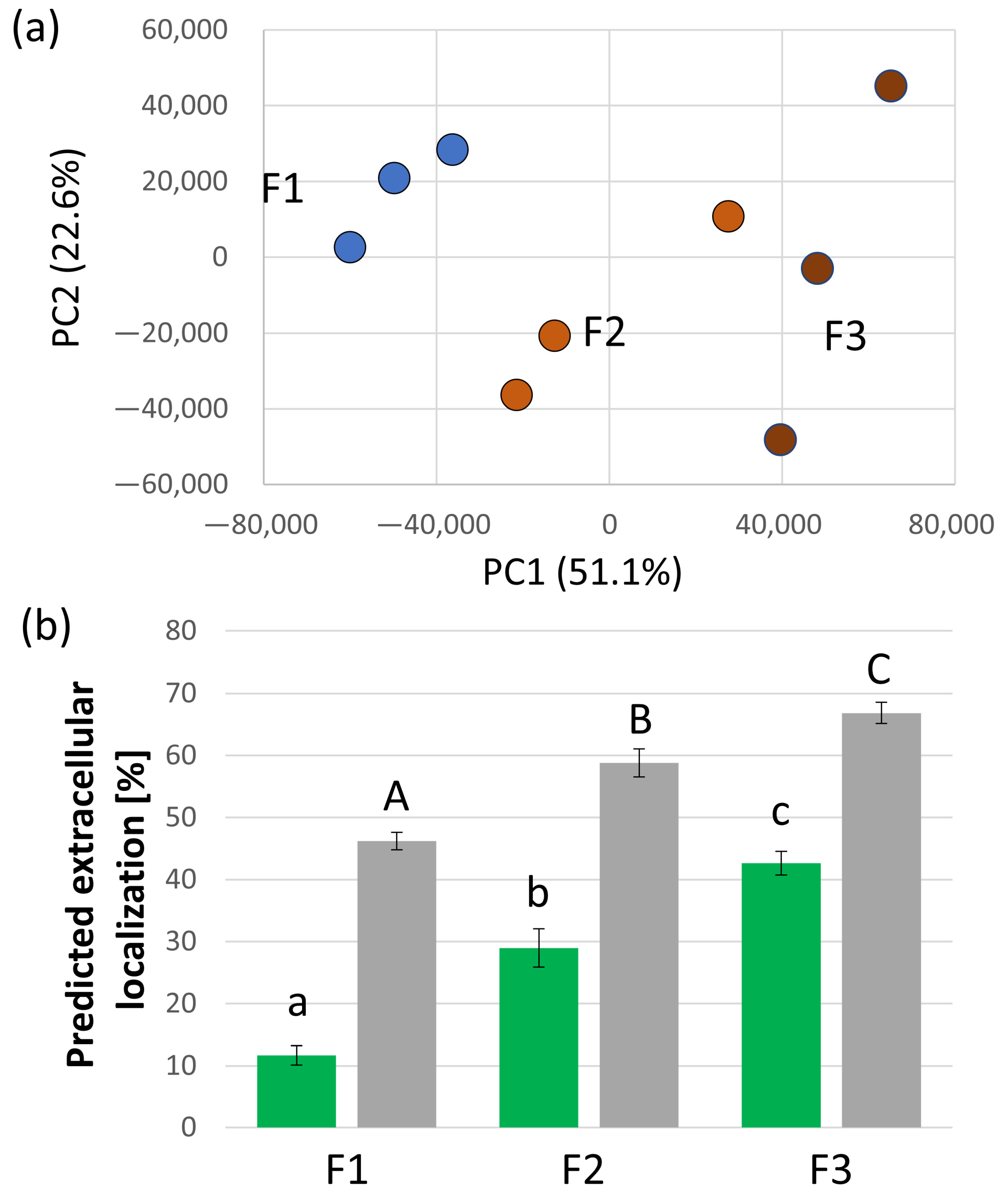
2.1. Majority of the Identified S. tuberosum Proteins in Xylem Sap Is Predicted to Be Extracellular
2.2. Functional Diversity of Putative Extracellular Xylem Sap Proteins in Solanum tuberosum
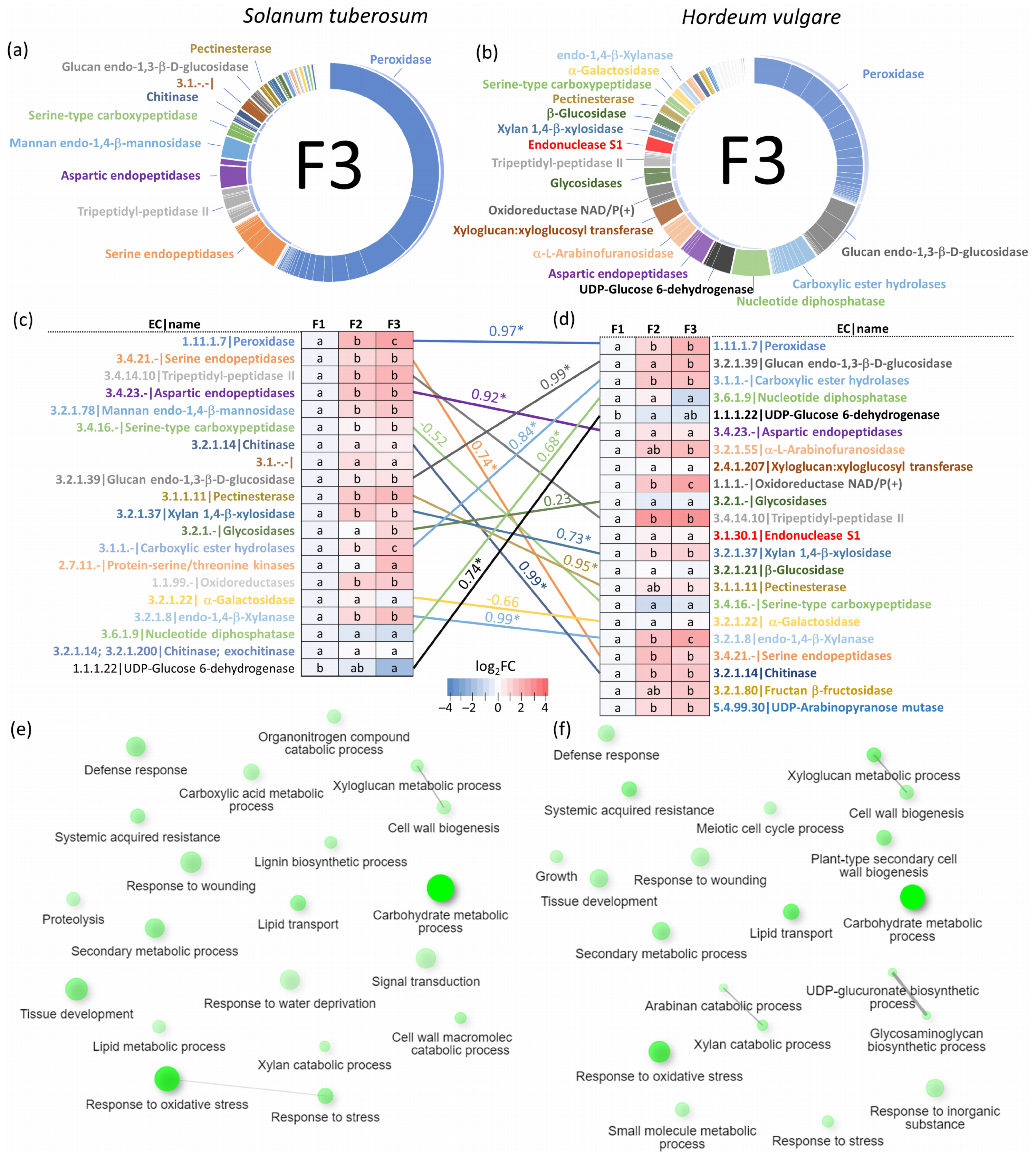
2.3. Solanum tuberosum Sap Proteome Response to flg22
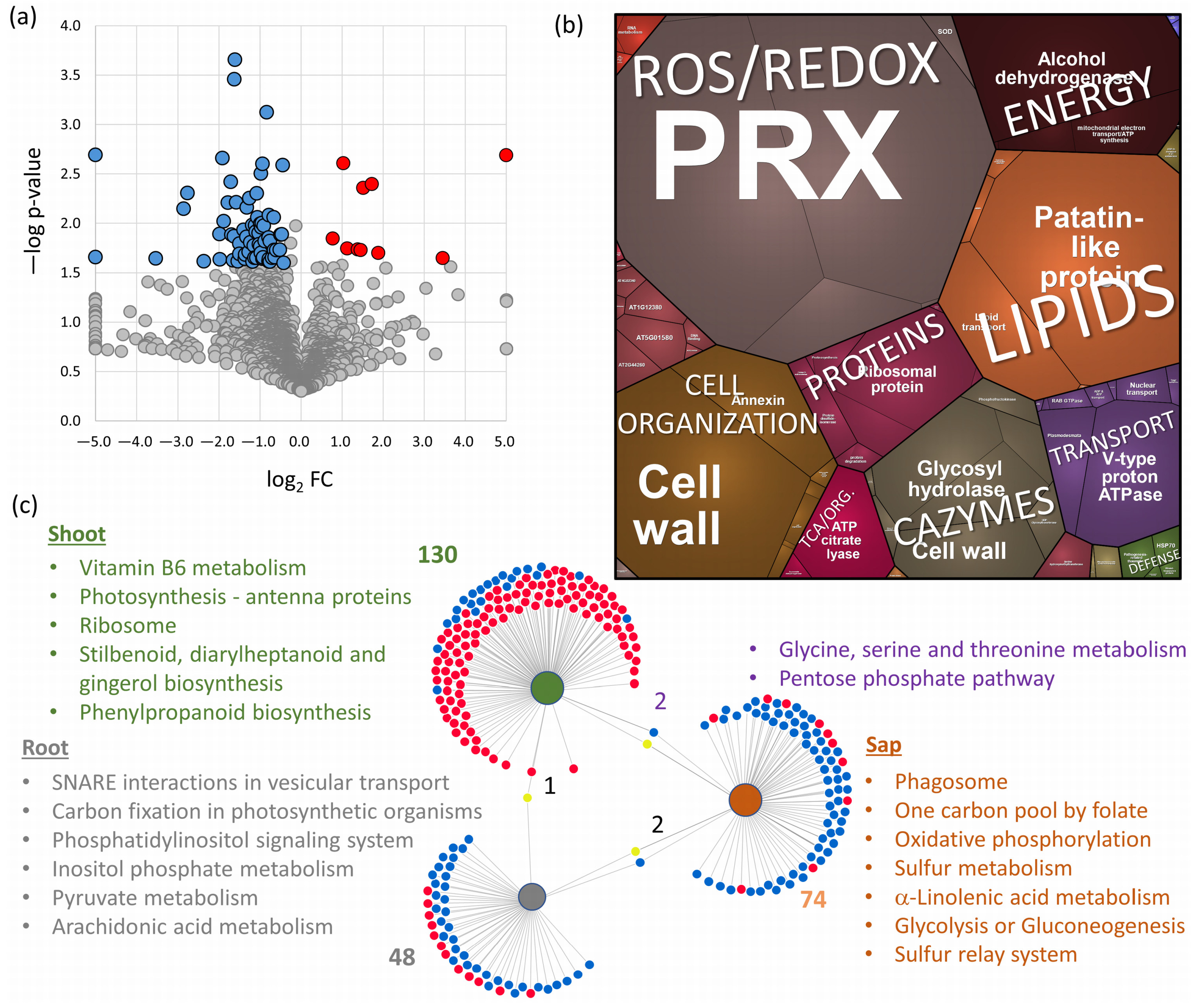
2.4. Root and Shoot Proteome Analyses Corroborate the Utility of Sap Proteome Profiling
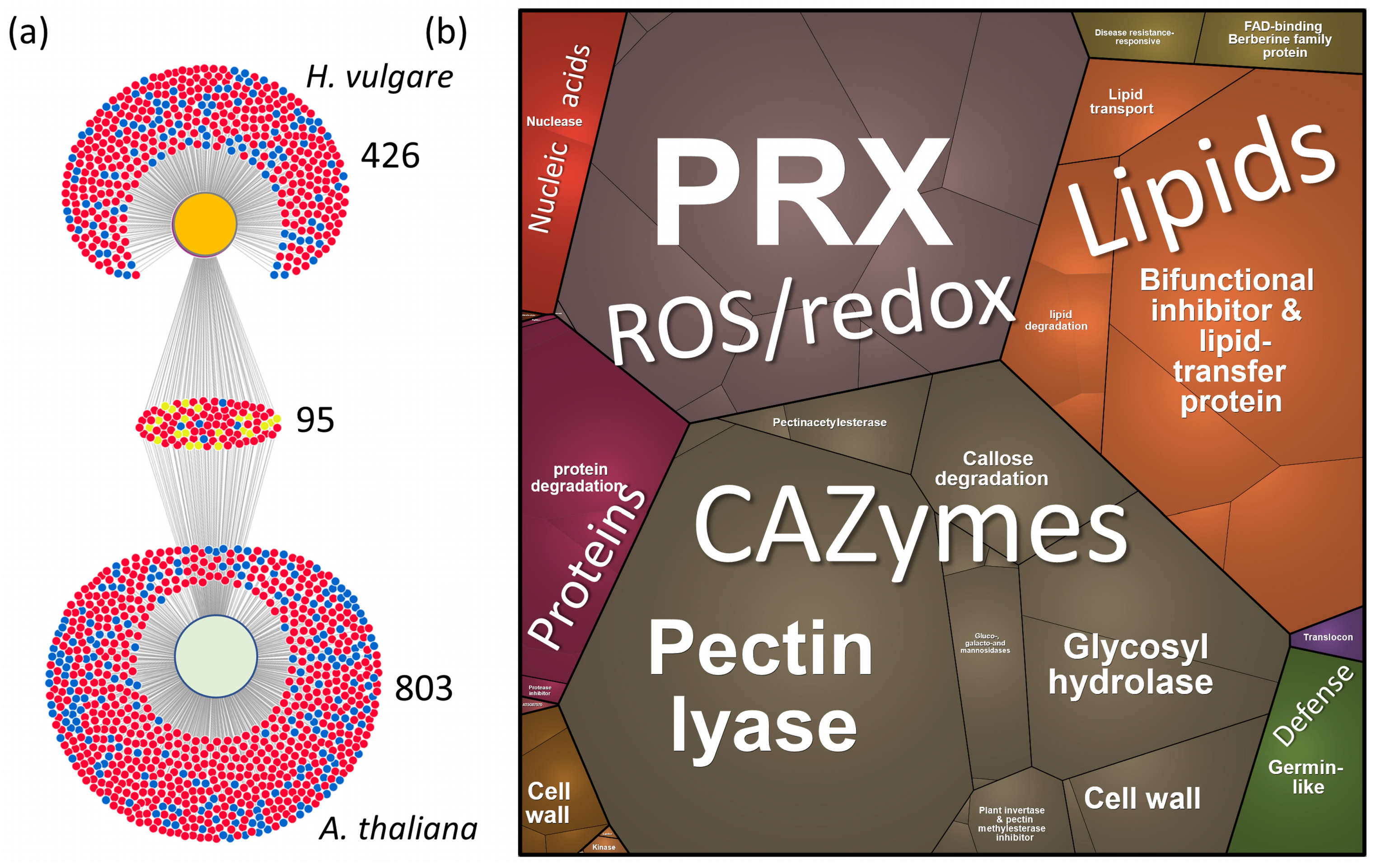
2.5. Xylem Sap Proteome Analysis of Hordeum Vulgare Revealed a Rich Protein Landscape and a Much Stronger Response to flg22
3. Discussion
3.1. Novel flg22 Response Proteins Identified by Xylem Sap Proteome Analysis
3.2. Response of Lipid Metabolism to flg22 Is Evolutionarily Conserved in H. vulgare and S. tuberosum
3.3. HSP70 Family Proteins in Response to flg22 Suggests Their Extracellular Role
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Elicitor Treatment
4.3. Proteome Analysis
4.4. Data Analysis and Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Koenig, A.M.; Hoffmann-Benning, S. The interplay of phloem-mobile signals in plant development and stress response. Biosci. Rep. 2020, 40, BSR20193329. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.; Helariutta, Y.; He, X.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The Plant Vascular System: Evolution, Development and Functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Taleski, M.; Jin, M.; Chapman, K.; Taylor, K.; Winning, C.; Frank, M.; Imin, N.; Djordjevic, M.A. CEP hormones at the nexus of nutrient acquisition and allocation, root development, and plant–microbe interactions. J. Exp. Bot. 2024, 75, 538–552. [Google Scholar] [CrossRef]
- Ladeynova, M.; Kuznetsova, D.; Mudrilov, M.; Vodeneev, V. Integration of Electrical Signals and Phytohormones in the Control of Systemic Response. Int. J. Mol. Sci. 2023, 24, 847. [Google Scholar] [CrossRef]
- Houmani, H.; Corpas, F.J. Can nutrients act as signals under abiotic stress? Plant Physiol. Biochem. 2024, 206, 108313. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-Q.; Morin, H.; Marcourt, L.; Yang, T.-H.; Wolfender, J.-L.; Farmer, E.E. Chloride, glutathiones, and insect-derived elicitors introduced into the xylem trigger electrical signaling. Plant Physiol. 2024, 194, 1091–1103. [Google Scholar] [CrossRef]
- Rüscher, D.; Vasina, V.V.; Knoblauch, J.; Bellin, L.; Pommerrenig, B.; Alseekh, S.; Fernie, A.R.; Neuhaus, H.E.; Knoblauch, M.; Sonnewald, U.; et al. Symplasmic phloem loading and subcellular transport in storage roots are key factors for carbon allocation in cassava. Plant Physiol. 2024, in press. [Google Scholar] [CrossRef]
- Akhiyarova, G.; Finkina, E.I.; Zhang, K.; Veselov, D.; Vafina, G.; Ovchinnikova, T.V.; Kudoyarova, G. The Long-Distance Transport of Some Plant Hormones and Possible Involvement of Lipid-Binding and Transfer Proteins in Hormonal Transport. Cells 2024, 13, 364. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; White, R.G.; Djordjevic, M.A.; Ruan, Y.-L.; Mathesius, U. Root-to-shoot signalling: Integration of diverse molecules, pathways and functions. Funct. Plant Biol. 2016, 43, 87. [Google Scholar] [CrossRef]
- De Schepper, V.; De Swaef, T.; Bauweraerts, I.; Steppe, K. Phloem transport: A review of mechanisms and controls. J. Exp. Bot. 2013, 64, 4839–4850. [Google Scholar] [CrossRef]
- Garg, V.; Kühn, C. What determines the composition of the phloem sap? Is there any selectivity filter for macromolecules entering the phloem sieve elements? Plant Physiol. Biochem. 2020, 151, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Tolstyko, E.A.; Lezzhov, A.A.; Morozov, S.Y.; Solovyev, A.G. Phloem transport of structured RNAs: A widening repertoire of trafficking signals and protein factors. Plant Sci. 2020, 299, 110602. [Google Scholar] [CrossRef]
- Hu, C.; Ham, B.; El-shabrawi, H.M.; Alexander, D.; Zhang, D.; Ryals, J.; Lucas, W.J. Proteomics and metabolomics analyses reveal the cucurbit sieve tube system as a complex metabolic space. Plant J. 2016, 87, 442–454. [Google Scholar] [CrossRef]
- Brodersen, C.R.; Roddy, A.B.; Wason, J.W.; McElrone, A.J. Functional Status of Xylem Through Time. Annu. Rev. Plant Biol. 2019, 70, 407–433. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Celma, J.; Ceballos-Laita, L.; Grusak, M.A.; Abadía, J.; López-Millán, A.-F. Plant fluid proteomics: Delving into the xylem sap, phloem sap and apoplastic fluid proteomes. BBA—Proteins Proteom. 2016, 1864, 991–1002. [Google Scholar] [CrossRef]
- Wheeldon, C.D.; Bennett, T. There and back again: An evolutionary perspective on long-distance coordination of plant growth and development. Semin. Cell Dev. Biol. 2021, 109, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. 2021, 105, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Seto, Y.; Yamaguchi, S. Strigolactone biosynthesis, transport and perception. Plant J. 2021, 105, 335–350. [Google Scholar] [CrossRef]
- Pérez-Pérez, J.G.; Puertolas, J.; Albacete, A.; Dodd, I.C. Alternation of wet and dry sides during partial rootzone drying irrigation enhances leaf ethylene evolution. Environ. Exp. Bot. 2020, 176, 104095. [Google Scholar] [CrossRef]
- Regnault, T.; Davière, J.-M.; Wild, M.; Sakvarelidze-Achard, L.; Heintz, D.; Carrera Bergua, E.; Lopez Diaz, I.; Gong, F.; Hedden, P.; Achard, P. The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat. Plants 2015, 1, 15073. [Google Scholar] [CrossRef]
- Thorpe, M.R.; Ferrieri, A.P.; Herth, M.M.; Ferrieri, R.A. 11C-imaging: Methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 2007, 226, 541. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-W.; Kaimoyo, E.; Kumar, D.; Mosher, S.; Klessig, D.F. Methyl Salicylate Is a Critical Mobile Signal for Plant Systemic Acquired Resistance. Science 2007, 318, 113–116. [Google Scholar] [CrossRef]
- Broussard, L.; Abadie, C.; Lalande, J.; Limami, A.M.; Lothier, J.; Tcherkez, G. Phloem Sap Composition: What Have We Learnt from Metabolomics? Int. J. Mol. Sci. 2023, 24, 6917. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Suzui, N.; Fujimaki, S.; Dohmae, N.; Yonekura-Sakakibara, K.; Fujiwara, T.; Hayashi, H.; Yamaya, T.; Sakakibara, H. Destination-Selective Long-Distance Movement of Phloem Proteins. Plant Cell 2005, 17, 1801–1814. [Google Scholar] [CrossRef]
- Djordjevic, M.A.; Oakes, M.; Li, D.X.; Hwang, C.H.; Hocart, C.H.; Gresshoff, P.M. The Glycine max Xylem Sap and Apoplast Proteome. J. Proteome Res. 2007, 6, 3771–3779. [Google Scholar] [CrossRef]
- Carella, P.; Merl-Pham, J.; Wilson, D.C.; Dey, S.; Hauck, S.M.; Vlot, C.; Cameron, R.K. Comparative Proteomics Analysis of Arabidopsis Phloem Exudates Collected During the Induction of Systemic Acquired Resistance. Plant Physiol. 2016, 171, 1495–1510. [Google Scholar] [CrossRef]
- Rep, M.; Dekker, H.L.; Vossen, J.H.; de Boer, A.D.; Houterman, P.M.; Speijer, D.; Back, J.W.; de Koster, C.G.; Cornelissen, B.J.C. Mass Spectrometric Identification of Isoforms of PR Proteins in Xylem Sap of Fungus-Infected Tomato. Plant Physiol. 2002, 130, 904–917. [Google Scholar] [CrossRef]
- Pu, Z.; Ino, Y.; Kimura, Y.; Tago, A.; Shimizu, M.; Natsume, S.; Sano, Y.; Fujimoto, R.; Kaneko, K.; Shea, D.J.; et al. Changes in the Proteome of Xylem Sap in Brassica oleracea in Response to Fusarium oxysporum Stress. Front. Plant Sci. 2016, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Abeysekara, N.S.; Bhattacharyya, M.K. Analyses of the Xylem Sap Proteomes Identified Candidate Fusarium virguliforme Proteinacious Toxins. PLoS ONE 2014, 9, e93667. [Google Scholar] [CrossRef]
- Floerl, S.; Druebert, C.; Majcherczyk, A.; Karlovsky, P.; Kües, U.; Polle, A. Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms. BMC Plant Biol. 2008, 8, 129. [Google Scholar] [CrossRef]
- Zheng, T.; Haider, M.S.; Zhang, K.; Jia, H.; Fang, J. Biological and functional properties of xylem sap extracted from grapevine (cv. Rosario bianco). Sci. Hortic. 2020, 272, 109563. [Google Scholar] [CrossRef]
- Notaguchi, M.; Okamoto, S. Dynamics of long-distance signaling via plant vascular tissues. Front. Plant Sci. 2015, 6, 161. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Buhtz, A.; Giavalisco, P. Analysis of xylem sap proteins from Brassica napus. BMC Plant Biol. 2005, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Buhtz, A.; Kolasa, A.; Arlt, K.; Walz, C.; Kehr, J. Xylem sap protein composition is conserved among different plant species. Planta 2004, 219, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Yadeta, K.A.J.; Thomma, B.P.H. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef] [PubMed]
- Jelenska, J.; Davern, S.M.; Standaert, R.F.; Mirzadeh, S.; Greenberg, J.T. Flagellin peptide flg22 gains access to long-distance trafficking in Arabidopsis via its receptor, FLS2. J. Exp. Bot. 2017, 68, 1769–1783. [Google Scholar] [CrossRef] [PubMed]
- Moroz, N.; Tanaka, K. FlgII-28 Is a Major Flagellin-Derived Defense Elicitor in Potato. Mol. Plant Microbe Interact. 2020, 33, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Colaianni, N.R.; Parys, K.; Lee, H.-S.; Conway, J.M.; Kim, N.H.; Edelbacher, N.; Mucyn, T.S.; Madalinski, M.; Law, T.F.; Jones, C.D.; et al. A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe 2021, 29, 635–649.e9. [Google Scholar] [CrossRef]
- Buscaill, P.; Chandrasekar, B.; Sanguankiattichai, N.; Kourelis, J.; Kaschani, F.; Thomas, E.L.; Morimoto, K.; Kaiser, M.; Preston, G.M.; Ichinose, Y.; et al. Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 2019, 364, eaav0748. [Google Scholar] [CrossRef]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef]
- Hooper, C.M.; Castleden, I.R.; Aryamanesh, N.; Jacoby, R.P.; Millar, A.H. Finding the Subcellular Location of Barley, Wheat, Rice and Maize Proteins: The Compendium of Crop Proteins with Annotated Locations (CropPAL). Plant Cell Physiol. 2016, 57, e9. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An Arabidopsis GPI-Anchor Plasmodesmal Neck Protein with Callose Binding Activity and Potential to Regulate Cell-to-Cell Trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Park, J.; Choi, G.; Kim, S.; Vo, K.T.X.; Jeon, J.; Kang, S.; Lee, Y. A rice gene encoding glycosyl hydrolase plays contrasting roles in immunity depending on the type of pathogens. Mol. Plant Pathol. 2022, 23, 400–416. [Google Scholar] [CrossRef] [PubMed]
- Schimoler-O’Rourke, R.; Richardson, M.; Selitrennikoff, C.P. Zeamatin Inhibits Trypsin and α-Amylase Activities. Appl. Environ. Microbiol. 2001, 67, 2365–2366. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Yu, L.; Chen, J.; Hu, L.; Cao, S.; Wang, Y. Characterization of the Fasciclin-like arabinogalactan gene family in Brassica napus and the negative regulatory role of BnFLA39 in response to clubroot disease stress. Ind. Crop Prod. 2023, 196, 116400. [Google Scholar] [CrossRef]
- Sun, L.; Dong, S.; Ge, Y.; Fonseca, J.P.; Robinson, Z.T.; Mysore, K.S.; Mehta, P. DiVenn: An Interactive and Integrated Web-Based Visualization Tool for Comparing Gene Lists. Front. Genet. 2019, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Berka, M.; Černý, M.; Novák, J.; Luklová, M.; Brzobohatý, B.; Saiz-Fernández, I. Cytokinin Deficiency Alters Leaf Proteome and Metabolome during Effector-Triggered Immunity in Arabidopsis thaliana Plants. Plants 2022, 11, 2123. [Google Scholar] [CrossRef] [PubMed]
- Narváez-Barragán, D.A.; Tovar-Herrera, O.E.; Guevara-García, A.; Serrano, M.; Martinez-Anaya, C. Mechanisms of plant cell wall surveillance in response to pathogens, cell wall-derived ligands and the effect of expansins to infection resistance or susceptibility. Front. Plant Sci. 2022, 13, 969343. [Google Scholar] [CrossRef]
- Raiola, A.; Lionetti, V.; Elmaghraby, I.; Immerzeel, P.; Mellerowicz, E.J.; Salvi, G.; Cervone, F.; Bellincampi, D. Pectin Methylesterase Is Induced in Arabidopsis upon Infection and Is Necessary for a Successful Colonization by Necrotrophic Pathogens. Mol. Plant Microbe Interact. 2011, 24, 432–440. [Google Scholar] [CrossRef]
- Zhou, X.; Gao, H.; Zhang, X.; Khashi u Rahman, M.; Mazzoleni, S.; Du, M.; Wu, F. Plant extracellular self-DNA inhibits growth and induces immunity via the jasmonate signaling pathway. Plant Physiol. 2023, 192, 2475–2491. [Google Scholar] [CrossRef] [PubMed]
- Pernis, M.; Salaj, T.; Bellová, J.; Danchenko, M.; Baráth, P.; Klubicová, K. Secretome analysis revealed that cell wall remodeling and starch catabolism underlie the early stages of somatic embryogenesis in Pinus nigra. Front. Plant Sci. 2023, 14, 1225424. [Google Scholar] [CrossRef] [PubMed]
- Batailler, B.; Lemaître, T.; Vilaine, F.; Sanchez, C.; Renard, D.; Cayla, T.; Beneteau, J.; Dinant, S. Soluble and filamentous proteins in Arabidopsis sieve elements. Plant Cell Environ. 2012, 35, 1258–1273. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.R.; Knauer, T.; Furch, A.C.U. Collection of Phloem Sap in Phytoplasma-Infected Plants. In Phytoplasmas. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; pp. 291–299. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, T.; Valencia, M.V.; Zhang, C.; Lv, Z. Unraveling the Roles of Vascular Proteins Using Proteomics. Molecules 2021, 26, 667. [Google Scholar] [CrossRef] [PubMed]
- Maricchiolo, E.; Panfili, E.; Pompa, A.; De Marchis, F.; Bellucci, M.; Pallotta, M.T. Unconventional Pathways of Protein Secretion: Mammals vs. Plants. Front. Cell Dev. Biol. 2022, 10, 895853. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.G.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Takai, R.; Isogai, A.; Takayama, S.; Che, F.-S. Analysis of Flagellin Perception Mediated by Flg22 Receptor OsFLS2 in Rice. Mol. Plant Microbe Interact. 2008, 21, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Balaceanu, A.; Rufian, J.S.; Segonzac, C.; Zhao, A.; Morcillo, R.J.L.; Macho, A.P. An immune receptor complex evolved in soybean to perceive a polymorphic bacterial flagellin. Nat. Commun. 2020, 11, 3763. [Google Scholar] [CrossRef] [PubMed]
- Hind, S.R.; Strickler, S.R.; Boyle, P.C.; Dunham, D.M.; Bao, Z.; O’Doherty, I.M.; Baccile, J.A.; Hoki, J.S.; Viox, E.G.; Clarke, C.R.; et al. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2016, 2, 16128. [Google Scholar] [CrossRef]
- Murakami, T.; Katsuragi, Y.; Hirai, H.; Wataya, K.; Kondo, M.; Che, F.-S. Distribution of flagellin CD2-1, flg22, and flgii-28 recognition systems in plant species and regulation of plant immune responses through these recognition systems. Biosci. Biotechnol. Biochem. 2022, 86, 490–501. [Google Scholar] [CrossRef]
- Sun, Y.; Qiao, Z.; Muchero, W.; Chen, J.-G. Lectin Receptor-like Kinases: The Sensor and Mediator at the Plant Cell Surface. Front. Plant Sci. 2020, 11, 596301. [Google Scholar] [CrossRef] [PubMed]
- Yip Delormel, T.; Boudsocq, M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 2019, 224, 585–604. [Google Scholar] [CrossRef] [PubMed]
- Lampl, N.; Alkan, N.; Davydov, O.; Fluhr, R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 2013, 74, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Tunc-Ozdemir, M.; Jones, A.M. BRL3 and AtRGS1 cooperate to fine tune growth inhibition and ROS activation. PLoS ONE 2017, 12, e0177400. [Google Scholar] [CrossRef] [PubMed]
- Matsui, S.; Noda, S.; Kuwata, K.; Nomoto, M.; Tada, Y.; Shinohara, H.; Matsubayashi, Y. Arabidopsis SBT5.2 and SBT1.7 subtilases mediate C-Terminal Cleavage of flg22 epitope from bacterial flagellin. Nat. Commun. 2024, 15, 3762. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ma, K.; Ji, G.; Pan, L.; Zhou, Q. Lipid transfer proteins involved in plant–pathogen interactions and their molecular mechanisms. Mol. Plant Pathol. 2022, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Berka, M.; Kopecká, R.; Berková, V.; Brzobohatý, B.; Černý, M. Regulation of heat shock proteins 70 and their role in plant immunity. J. Exp. Bot. 2022, 73, 1894–1909. [Google Scholar] [CrossRef] [PubMed]
- Lopez, V.; Cauvi, D.M.; Arispe, N.; De Maio, A. Bacterial Hsp70 (DnaK) and mammalian Hsp70 interact differently with lipid membranes. Cell Stress. Chaperones 2016, 21, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Dufková, H.; Berka, M.; Psota, V.; Brzobohatý, B.; Černý, M. Environmental impacts on barley grain composition and longevity. J. Exp. Bot. 2023, 74, 1609–1628. [Google Scholar] [CrossRef]
- Berková, V.; Berka, M.; Kameniarová, M.; Kopecká, R.; Kuzmenko, M.; Shejbalová, Š.; Abramov, D.; Čičmanec, P.; Frejlichová, L.; Jan, N.; et al. Salicylic Acid Treatment and Its Effect on Seed Yield and Seed Molecular Composition of Pisum sativum under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 5454. [Google Scholar] [CrossRef]
- Dorfer, V.; Pichler, P.; Stranzl, T.; Stadlmann, J.; Taus, T.; Winkler, S.; Mechtler, K. MS Amanda, a Universal Identification Algorithm Optimized for High Accuracy Tandem Mass Spectra. J. Proteome Res. 2014, 13, 3679–3684. [Google Scholar] [CrossRef] [PubMed]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Dufková, H.; Berka, M.; Greplová, M.; Shejbalová, Š.; Hampejsová, R.; Luklová, M.; Domkářová, J.; Novák, J.; Kopačka, V.; Brzobohatý, B.; et al. The Omics Hunt for Novel Molecular Markers of Resistance to Phytophthora infestans. Plants 2022, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Liebermeister, W.; Noor, E.; Flamholz, A.; Davidi, D.; Bernhardt, J.; Milo, R. Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. USA 2014, 111, 8488–8493. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopecká, R.; Černý, M. Xylem Sap Proteome Analysis Provides Insight into Root–Shoot Communication in Response to flg22. Plants 2024, 13, 1983. https://doi.org/10.3390/plants13141983
Kopecká R, Černý M. Xylem Sap Proteome Analysis Provides Insight into Root–Shoot Communication in Response to flg22. Plants. 2024; 13(14):1983. https://doi.org/10.3390/plants13141983
Chicago/Turabian StyleKopecká, Romana, and Martin Černý. 2024. "Xylem Sap Proteome Analysis Provides Insight into Root–Shoot Communication in Response to flg22" Plants 13, no. 14: 1983. https://doi.org/10.3390/plants13141983
APA StyleKopecká, R., & Černý, M. (2024). Xylem Sap Proteome Analysis Provides Insight into Root–Shoot Communication in Response to flg22. Plants, 13(14), 1983. https://doi.org/10.3390/plants13141983






