Metabolic Aspects of Lentil–Fusarium Interactions
Abstract
:1. Introduction
2. Results
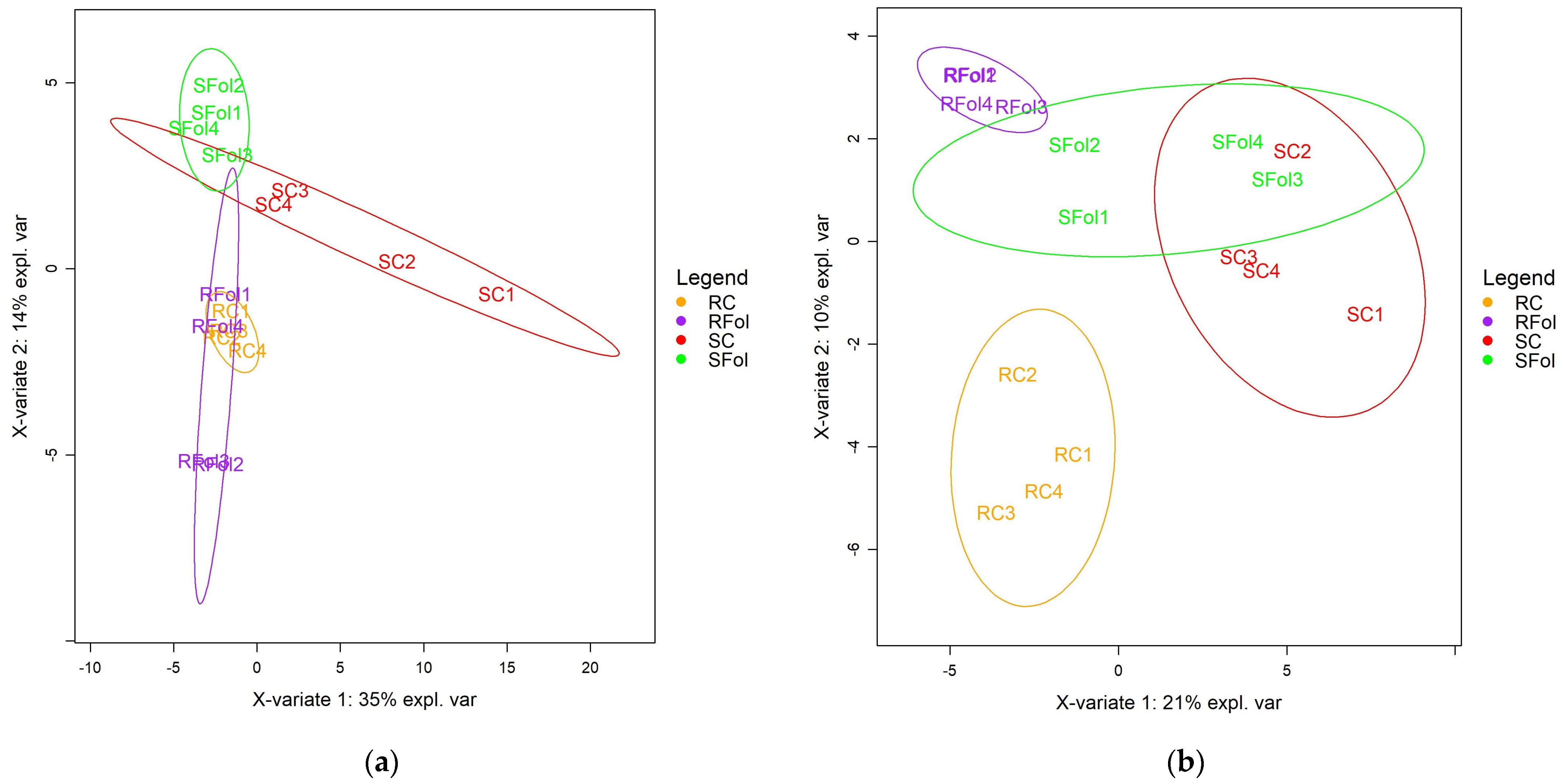
2.1. Comparative Metabolic Response of Contrasting Genotypes to Fol-Inoculation
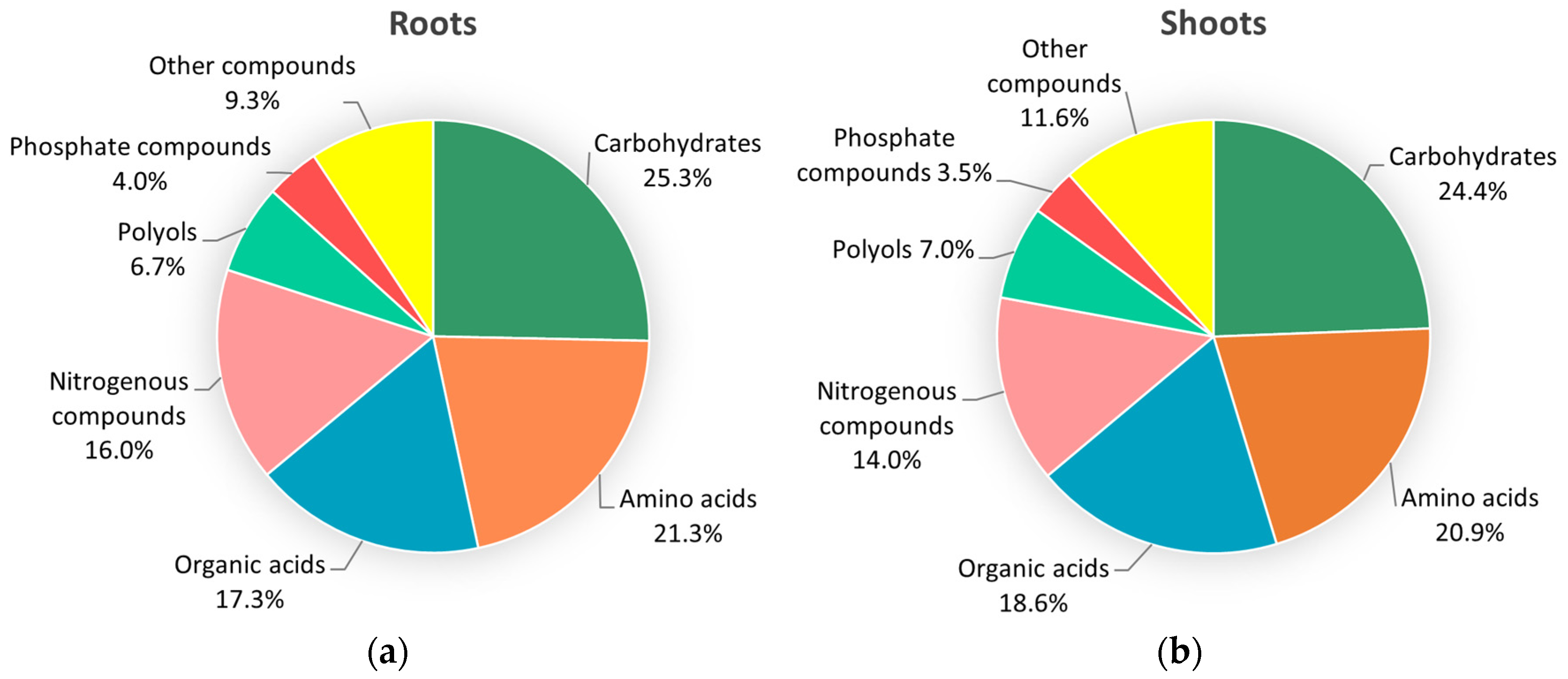
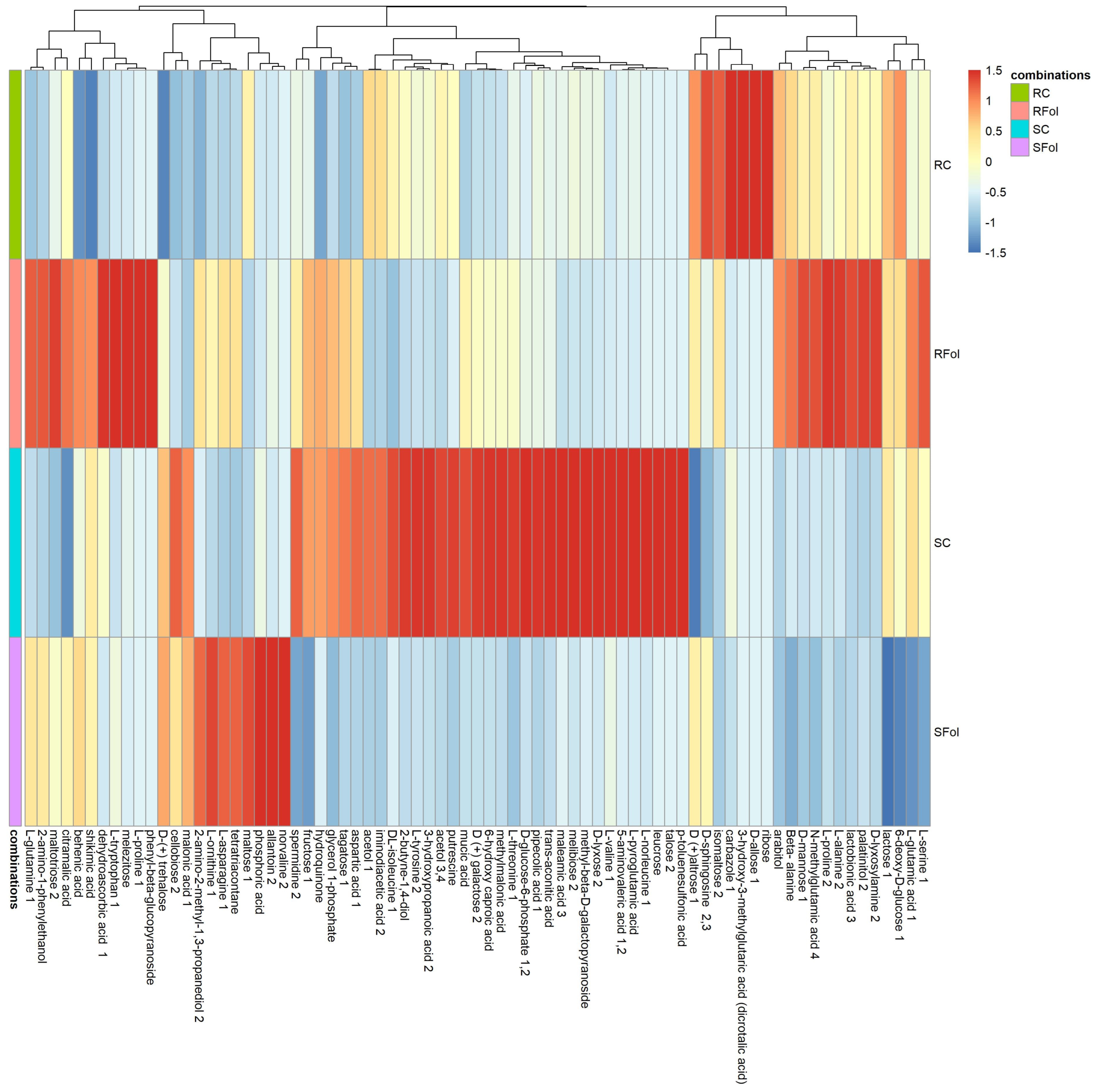
2.2. Global Fluctuations in the Metabolome of Contrasting Genotypes in Response to Fol-Inoculation
2.2.1. Carbohydrates
2.2.2. Amino Acids
2.2.3. Polyols
2.2.4. Organic Acids
2.2.5. Carboxylic Acids
2.2.6. Nitrogenous Compounds
2.2.7. Phosphate Compounds
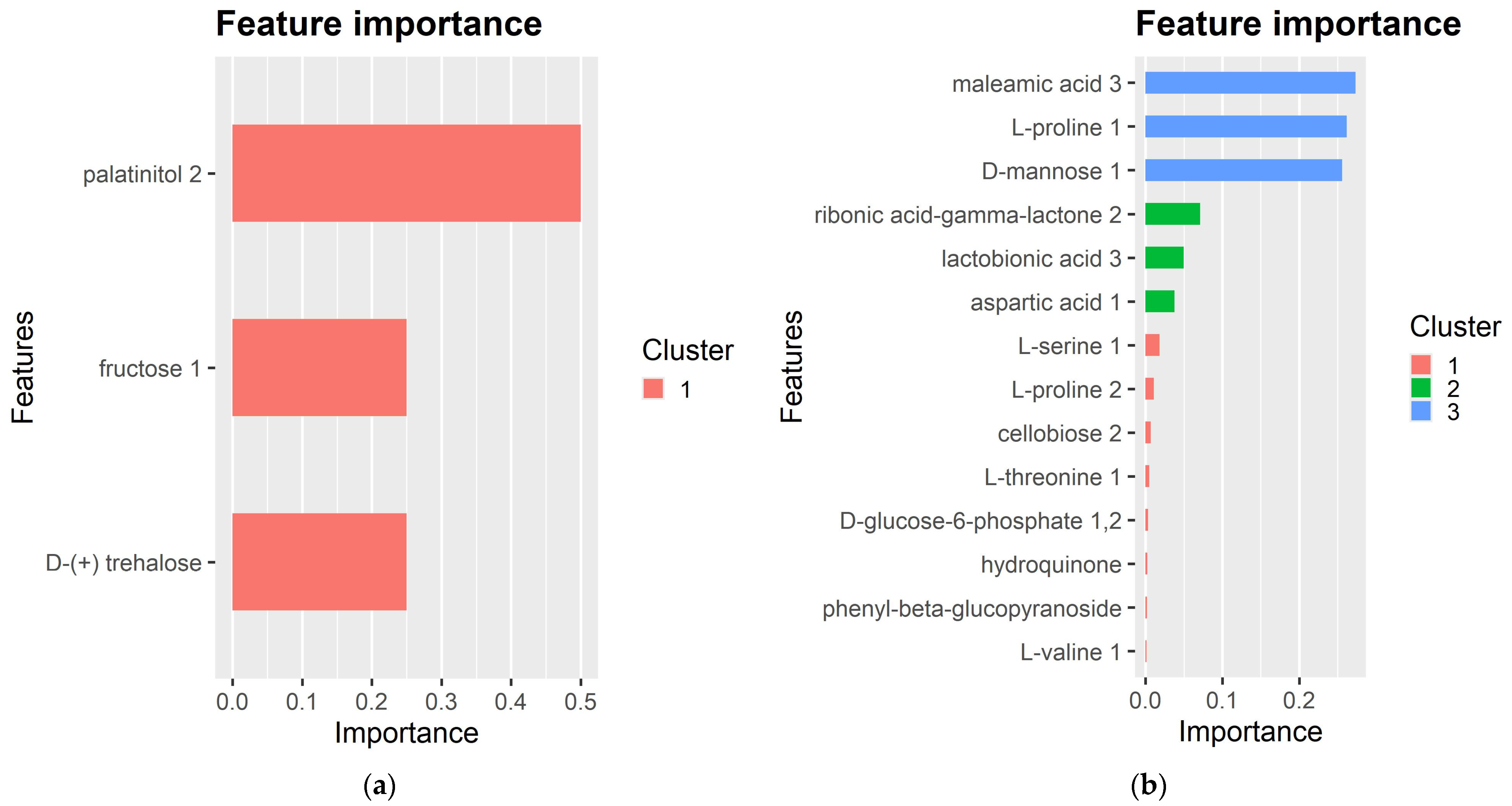
2.3. Exploring the Metabolic Response to Fol-Inoculation Based on an XGBoost Prediction Model
3. Discussion
4. Materials and Methods
4.1. Genetic Material
4.2. Fol-Inoculations and Experimental Design
4.3. Gas Chromatography–Mass Spectrometry (GC–MS)-Based Metabolomic Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ladizinsky, G. The origin of lentil and wild gene pool. Euphytica 1979, 28, 179–187. [Google Scholar] [CrossRef]
- Duran, Y.; Fratini, R.; Garcia, P.; De La Vega, M.P. An inter subspecific genetic map of Lens. Theor. Appl. Genet. 2004, 108, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, W.J.; Ramsay, M.D.; Makkouk, K.M.; Bretag, T.W.; Acikgoz, N.; Kumar, J.; Nutter, F.W. Foliar diseases of cool season food legumes and their control. In Linking Research and Marketing Opportunities for Pulses in the 21st Century. Current Plant Science and Biotechnology in Agriculture; Knight, R., Ed.; Springer: Dordrecht, The Netherlands, 2000; Volume 34, pp. 437–455. [Google Scholar] [CrossRef]
- Erskine, W.; Bayaa, B. Yield loss, incidence and inoculum density associated with vascular wilt of lentil. Phytopathol. Mediterr. 1996, 35, 24–32. [Google Scholar]
- Bayaa, B.; Erskine, W.; Singh, M. Screening lentil for resistance to Fusarium wilt: Methodology and sources of resistance. Euphytica 1997, 98, 69–74. [Google Scholar] [CrossRef]
- Tosi, L.; Cappelli, C. First Report of Fusarium oxysporum f. sp. lentis of lentil in Italy. Plant Dis. 2001, 85, 562. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, J.; Singh, S.; Ahmed, S.; Chaudhary, R.G.; Sarker, A. Vascular wilt disease of lentil: A review. J. Lentil Res. 2010, 4, 411–414. [Google Scholar] [CrossRef]
- Chen, W.; Sharma, H.C.; Muehlbauer, F.J. Compendium of Chickpea and Lentil Diseases and Pests; The American Phytopathological Society (APS): St. Paul, MN, USA, 2011; p. 164. [Google Scholar] [CrossRef]
- Tiwari, N.; Ahmed, S.; Sarker, A. Fusarium wilt: A killer disease of lentil. In Fusarium-Plant Diseases, Pathogen Diversity, Genetic Diversity, Resistance and Molecular Markers, Askun, T., Ed.; IntechOpen: Rijeka, Croatia, 2018; pp. 119–138. [Google Scholar] [CrossRef]
- Khare, M.N.; Bayaa, B.; Beniwal, S.P.S. Selection methods for disease resistance in lentil. In Breeding for Stress Tolerance in Cool-Season Food Legumes; Singh, K.B., Saxena, M.C., Eds.; Wiley: Chichester, UK, 1993; pp. 107–122. [Google Scholar]
- Bayaa, B.; Erskine, W.; Hamdi, A. Evaluation of a wild lentil collection for resistance to vascular wilt. Genet. Resour. Crop Evol. 1995, 42, 231–235. [Google Scholar] [CrossRef]
- Kraft, J.M.; Haware, M.P.; Halila, H.; Bayaa, B. Soilborne diseases and their control. In Linking Research and Marketing Opportunities for Pulses in the 21st Century. Current Plant Science and Biotechnology in Agriculture; Knight, R., Ed.; Springer: Dordrecht, The Netherlands, 2000; Volume 34, pp. 457–466. [Google Scholar] [CrossRef]
- Taylor, P.; Lindbeck, K.; Chen, W.; Ford, R. Lentil diseases. In Lentil: An Ancient Crop for Modern Times; Yadav, S.S., McNeil, D., Stevenson, P.C., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 291–313. [Google Scholar] [CrossRef]
- Mohammadi, N.; Pouralibaba, H.R.; Mohammadi Goltapeh, A.; Babaie Ahari, A.; Pakdaman Sararood, B. Advanced lentil lines screened for resistance to Fusarium oxysporum f.sp. lentis under greenhouse and field conditions. Phytoparasitica 2012, 40, 69–76. [Google Scholar] [CrossRef]
- Pouralibaba, H.R.; Rubiales, D.; Fondevilla, S. Identification of resistance to Fusarium oxysposbrorum f. sp. lentis in Spanish lentil germplasm. Eur. J. Plant Pathol. 2015, 143, 399–405. [Google Scholar] [CrossRef]
- Bayaa, B.; Erskine, W.; Abbas, A. Evaluating different methods for screening lentil germplasm for resistance to lentil wilt caused by F. oxysporum f. sp. lentis. Arab J. Plant Prot. 1994, 12, 83–91. [Google Scholar]
- Kraft, J.M.; Haware, M.P.; Jiménez-Diaz, R.M.; Bayaa, B.; Harrabi, M. Screening techniques and sources of resistance to root rots and wilts in cool season food legumes. Euphytica 1994, 73, 27–39. [Google Scholar] [CrossRef]
- Bayaa, B.; Erskine, W. Screening technique for resistance to vascular wilt in lentil. Arab J. Plant Prot. 1990, 8, 30–33. [Google Scholar]
- Eujayl, I.; Erskine, W.; Bayaa, B.; Baum, M.; Pehu, E. Fusarium vascular wilt in lentil: Inheritance and identification of DNA markers for resistance. Plant Breed. 2006, 177, 497–499. [Google Scholar] [CrossRef]
- Meena, J.K.; Singh, A.; Dikshit, H.K.; Mishra, G.P.; Aski, M.; Srinivasa, N.; Gupta, S.; Singh, D.; Tripathi, A. Screening of lentil (Lens culinaris Medikus sub sp. culinaris) germplasm against Fusarium wilt (Fusarium oxysporum f. sp. lentis). Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2533–2541. [Google Scholar]
- Riccioni, L.; Giambattista, G.D.; Valvassori, M.; Malta, R.; Porta-Puglia, A. Fungi associated with root rot of lentil landrace from Sicily. J. Plant Pathol. 2003, 85, 300. [Google Scholar]
- Infantino, A.; Kharrat, M.; Riccioni, L.; Coyne, C.J.; McPhee, K.E.; Grunwald, N.J. Screening techniques and sources of resistance to root diseases of food legumes. Euphytica 2006, 147, 201–221. [Google Scholar] [CrossRef]
- Kumar, Y.; Dholakia, B.B.; Panigrahi, P.; Kadoo, N.Y.; Giri, A.P.; Gupta, V.S. Metabolic profiling of chickpea-Fusarium interaction identifies differential modulation of disease resistance pathways. Phytochemistry 2015, 116, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Bohra, A.; Pandey, S.; Parida, S.K. Breeding, genetics, and genomics approaches for improving Fusarium wilt resistance in major grain legumes. Front. Genet. 2020, 11, 1001. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, A.M.; Araújo, S.d.S.; Rubiales, D.; Vaz Patto, M.C. Fusarium wilt management in legume crops. Agronomy 2020, 10, 1073. [Google Scholar] [CrossRef]
- Chen, F.; Ma, R.; Chen, X.-L. Advances of metabolomics in fungal pathogen-plant interactions. Metabolites 2019, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Muth, D.; Kachlicki, P.; Krajewski, P.; Przystalski, M.; Stobiecki, M. Differential metabolic response of narrow leafed lupine (Lupinus angustifolius) leaves to infection with Colletotrichum lupini. Metabolomics 2009, 5, 354–362. [Google Scholar] [CrossRef]
- Wojakowska, A.; Muth, D.; Narożna, D.; Mądrzak, C.; Stobiecki, M.; Kachlicki, P. Changes of phenolic secondary metabolite profiles in the reaction of narrow leaf lupin (Lupinus angustifolius) plants to infections with Colletotrichum lupini fungus or treatment with its toxin. Metabolomics 2013, 9, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Robison, F.M.; Turner, M.F.; Jahn, C.E.; Schwartz, H.F.; Prenni, J.E.; Brick, M.A.; Heuberger, A.L. Common bean varieties demonstrate differential physiological and metabolic responses to the pathogenic fungus Sclerotinia sclerotiorum. Plant Cell Environ. 2018, 41, 2141–2154. [Google Scholar] [CrossRef] [PubMed]
- Aliferis, K.A.; Faubert, D.; Jabaji, S. A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS ONE 2014, 9, e111930. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Westrick, N.M.; Jain, S.; Piotrowski, J.S.; Ranjan, M.; Kessens, R.; Stiegman, L.; Grau, C.R.; Conley, S.P.; Smith, D.L. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 2019, 17, 1567–1581. [Google Scholar] [CrossRef] [PubMed]
- Kumaraswamy, G.K.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Differential metabolic response of barley genotypes, varying in resistance, to trichothecene-producing and -nonproducing (tri5−) isolates of Fusarium graminearum. Plant Pathol. 2011, 61, 509–521. [Google Scholar] [CrossRef]
- Tomas, C.; Marta, V.; Zbynek, D.; Lukas, V.; Jaroslava, O.; Jana, H. Rapid LC-MS-based metabolomics method to study the Fusarium infection of barley. J. Sep. Sci. 2014, 37, 912–919. [Google Scholar] [CrossRef]
- Gunnaiah, R.; Kushalappa, A.C. Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Biochem. 2014, 83, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Scandiani, M.M.; Luque, A.G.; Razori, M.V.; Ciancio Casalini, L.; Aoki, T.; O’Donnell, K.; Cervigni, G.D.; Spampinato, C.P. Metabolic profiles of soybean roots during early stages of Fusarium tucumaniae infection. J. Exp. Bot. 2014, 66, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Zhang, L.; Panigrahi, P.; Dholakia, B.B.; Dewangan, V.; Chavan, S.G.; Kunjir, S.M.; Wu, X.; Li, N.; Rajmohanan, P.R. Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics. Plant Biotechnol. J. 2016, 14, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Rubiales, D.; Fondevilla, S.; Chen, W.; Gentzbittel, L.; Higgins, T.J.; Castillejo, M.A.; Singh, K.B.; Rispail, N. Achievements and challenges in legume breeding for pest and disease resistance. Crit. Rev. Plant Sci. 2014, 34, 195–236. [Google Scholar] [CrossRef]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for plant improvement: Status and prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed]
- Muscolo, A.; Junker, A.; Klukas, C.; Weigelt-Fischer, K.; Riewe, D.; Altmann, T. Phenotypic and metabolic responses to drought and salinity of four contrasting lentil accessions. J. Exp. Bot. 2015, 18, 5467–5480. [Google Scholar] [CrossRef]
- Skliros, D.; Kalloniati, C.; Karalias, G.; Skaracis, G.; Rennenberg, H.; Flemetakis, E. Global metabolomics analysis reveals distinctive tolerance mechanisms in different plant organs of lentil (Lens culinaris) upon salinity stress. Plant Soil 2018, 429, 451–468. [Google Scholar] [CrossRef]
- Foti, C.; Kalampokis, I.F.; Aliferis, K.A.; Pavli, O.I. Metabolic responses of two contrasting lentil genotypes to PEG-induced drought stress. Agronomy 2021, 11, 1190. [Google Scholar] [CrossRef]
- Bolouri Moghaddam, M.R.; Van den Ende, W. Sugars and plant innate immunity. J. Exp. Bot. 2012, 63, 3989–3998. [Google Scholar] [CrossRef] [PubMed]
- Scheideler, M.; Schlaich, N.L.; Fellenberg, K.; Beissbarth, T.; Hauser, N.C.; Vingron, M.; Slusarenko, A.J.; Hoheisel, J.D. Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem. 2002, 277, 10555–10561. [Google Scholar] [CrossRef]
- Heuberger, A.L.; Robison, F.M.; Lyons, S.M.A.; Broeckling, C.D.; Prenni, J.E. Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Front. Plant Sci. 2014, 5, 291. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, Q.; He, T.; Lan, J.; Ding, L.; Liu, T.; Wu, Q.; Pan, Y.; Chen, T. Transcriptomic and metabolomic changes triggered by Fusarium solani in common bean (Phaseolus vulgaris L.). Genes 2020, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Purvis, J.E.; Yomano, L.P.; Ingram, L.O. Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl. Environ. Microbiol. 2005, 71, 3761–3769. [Google Scholar] [CrossRef] [PubMed]
- Reignault, P.; Cogan, A.; Muchembled, J.; Sahraoui, A.L.H.; Durand, R.; Sancholle, M. Trehalose induces resistance to powdery mildew in wheat. New Phytol. 2001, 149, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, Y.; Dai, B.; Wang, B.; Zhang, H.; Zhu, Z.; Xu, Y.; Jiang, Y.; Zhang, G. Trehalose is an important mediator of Cap1p oxidative stress response in Candida albicans. Biol. Pharm. Bull. 2008, 31, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Louis, J.; Ayre, B.G.; Reese, J.C.; Shah, J. Trehalose phosphate synthase11-dependent trehalose metabolism promotes Arabidopsis thaliana defense against the phloem-feeding insect Myzus persicae. Plant J. 2011, 67, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.K.; Kim, J.K.; Ranwala, A.P.; Choi, Y.D.; Kochian, L.V.; Wu, R.J. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef] [PubMed]
- Keunen, E.; Peshev, D.; Vangronsveld, J.; van den Ende, W.; Cuypers, A. Plant sugars are crucial players in the oxidative challenge during abiotic stress: Extending the traditional concept. Plant Cell Environ. 2013, 36, 1242–1255. [Google Scholar] [CrossRef]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.A.; Li, S.; Lin, A.Z.; Boutrot, F.; Grossmann, G.; Zipfel, C.; Somerville, S.C. Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol. 2017, 173, 2383–2398. [Google Scholar] [CrossRef] [PubMed]
- Locci, F.; Benedetti, M.; Pontiggia, D.; Citterico, M.; Caprari, C.; Mattei, B.; Cervone, F.; De Lorenzo, G. An Arabidopsis berberine bridge enzyme-like protein specifically oxidizes cellulose oligomers and plays a role in immunity. Plant J. 2019, 98, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Barghahn, S.; Arnal, G.; Jain, N.; Petutschnig, E.; Brumer, S.; Lipka, V. Mixed linkage β-1,3/1,4-glucan oligosaccharides induce defense responses in Hordeum vulgare and Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 682439. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Motoyama, N.; Ikeda, A.; Wada, M.; Kamiya, K.; Hayafune, M.; Kaku, H.; Shibuya, N. Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 2012, 53, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Kurup, S.P.; Tarleton, R.L. Perpetual expression of PAMPs necessary for optimal immune control and clearance of a persistent pathogen. Nat. Commun. 2013, 4, 2616. [Google Scholar] [CrossRef] [PubMed]
- Wawra, S.; Fesel, P.; Widmer, H.; Timm, M.; Seibel, L.; Leson, L.; Kesseler, L.; Nostadt, R.; Hilbert, M.; Langen, G.; et al. The fungal-specific β-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat. Commun. 2016, 7, 13188. [Google Scholar] [CrossRef] [PubMed]
- Kesten, C.; Menna, A.; Sánchez-Rodríguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Liu, R.; Pang, J.H.; Rin, B.; Zhou, H.B.; Wang, G.; Wang, E.; Liu, J. Poaceae-specific cell wall-derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice. Nat. Commun. 2021, 12, 2178. [Google Scholar] [CrossRef] [PubMed]
- Zarattini, M.; Corso, M.; Kadowaki, M.A.; Monclaro, A.; Magri, S.; Milanese, I.; Jolivet, S.; Ortiz de Godoy, M.; Hermans, C.; Fagard, M.; et al. LPMO-oxidized cellulose oligosaccharides evoke immunity in Arabidopsis conferring resistance towards necrotrophic fungus B. cinerea. Commun. Biol. 2021, 4, 727. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Kong, M.; Qian, Y.; Gong, M.; Lv, G.; Song, J. Cellobiose elicits immunity in lettuce conferring resistance to Botrytis cinerea. J. Exp. Bot. 2023, 74, 1022–1038. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Arjona, F.M.; Vitale, S.; Voxeur, A.; Dora, S.; Müller, S.; Sancho-Andrés, G.; Montesinos, J.C.; Di Pietro, A.; Sánchez-Rodríguez, C. Impairment of the cellulose degradation machinery enhances Fusarium oxysporum virulence but limits its reproductive fitness. Sci. Adv. 2022, 8, abl9734. [Google Scholar] [CrossRef] [PubMed]
- Moormann, J.; Heinemann, B.; Hildebrandt, T.M. News about amino acid metabolism in plant–microbe interactions. Trends Biochem. Sci. 2022, 47, 839–850. [Google Scholar] [CrossRef]
- Zhu, L.; Zhou, Y.; Li, X.; Zhao, J.; Guo, N.; Xing, H. Metabolomics analysis of soybean hypocotyls in response to Phytophthora sojae infection. Front. Plant Sci. 2018, 9, 1530. [Google Scholar] [CrossRef] [PubMed]
- Tavernier, V.; Cadiou, S.; Pageau, K.; Laugé, R.; Reisdorf-Cren, M.; Langin, T.; Masclaux-Daubresse, C. The plant nitrogen mobilization promoted by Colletotrichum lindemuthianum in Phaseolus leaves depends on fungus pathogenicity. J. Exp. Bot. 2007, 58, 3351–3360. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.; Beckmann, M.; Zubair, H.; Enot, D.; Caracuel-Rios, Z.; Overy, D.; Snowdon, S.; Talbot, N.; Draper, J. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009, 59, 723–773. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Sawikowska, A.; Witaszak, N.; Waśkiewicz, A.; Kańczurzewska, M.; Kaczmarek, J.; Lalak-Kańczugowska, J. Metabolomic aspects of conservative and resistance-related elements of response to Fusarium culmorum in the grass family. Cells 2022, 11, 3213. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Sharma, S. Proline metabolism and its implications for plant-environment interaction. Arab. Book 2010, 8, e0140. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.E.; Savouré, A.; Szabados, L. Proline metabolism as regulatory hub. Trends Plant Sci. 2022, 27, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Fabro, G.; Kovács, I.; Pavet, V.; Szabados, L.; Alvarez, M.E. Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Int. Soc. Mol. Plant-Microbe Interact. 2004, 17, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, Y.S.; Cecchini, N.M.; Fabro, G.; Alvarez, M.E. Differential control and function of Arabidopsis ProDH1 and ProDH2 genes on infection with biotrophic and necrotrophic pathogens. Mol. Plant Pathol. 2017, 18, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Qamar, A.; Senthil-Kumar, M. Arabidopsis exhibits differential response in basal immunity and proline metabolism during defense against host and nonhost pathogen infection. Plant Physiol. Rep. 2019, 24, 496–506. [Google Scholar] [CrossRef]
- Yang, Z.; Rogers, L.M.; Song, Y.; Guo, W.; Kolattukudy, P.E. Homoserine and asparagine are host signals that trigger in planta expression of a pathogenesis gene in Nectria haematococca. Proc. Natl. Acad. Sci. USA 2005, 102, 4197–4202. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.S.; An, S.H.; Hwang, B.K. Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 2011, 67, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Peluffo, L.; Lia, V.; Troglia, C.; Maringolo, C.; Norma, P.; Escande, A.; Hopp, E.H.; Lytovchenko, A.; Fernie, A.R.; Heinz, R.; et al. Metabolic profiles of sunflower genotypes with contrasting response to Sclerotinia sclerotiorum infection. Phytochemistry 2010, 71, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.H.; Lee, S.H. The regulation of ornithine decarboxylase gene expression by sucrose and small upstream open reading frame in tomato (Lycopersicon esculentum Mill). Plant Cell Physiol. 2001, 42, 314–323. [Google Scholar] [CrossRef]
- Chang, K.S.; Lee, S.H.; Hwang, S.B.; Park, K.Y. Characterization and translational regulation of the arginine decarboxylase gene in carnation (Dianthus caryophyllus L.). Plant J. 2000, 24, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Hamzehzarghani, H.; Paranidharan, V.; Abu-Nada, Y.; Kushalappa, A.C.; Dion, Y.; Rioux, S.; Comeau, A.; Yaylayan, V.; Marshall, W.D. Metabolite profiling coupled with statistical analyses for potential high-throughput screening of quantitative resistance to Fusarium head blight in wheat. Can. J. Plant Pathol. 2008, 30, 24–36. [Google Scholar] [CrossRef]
- Paranidharan, V.; Abu-Nada, Y.; Hamzehzarghani, H.; Kushalappa, A.C.; Mamer, O.; Dion, Y.; Rioux, S.; Comeau, A.; Choiniere, L. Resistance-related metabolites in wheat against Fusarium graminearum and the virulence factor deoxynivalenol (DON). Botany 2008, 86, 1168–1179. [Google Scholar] [CrossRef]
- Bönnighausen, J.; Schauer, N.; Schäfer, W.; Bormann, J. Metabolic profiling of wheat rachis node infection by Fusarium graminearum-decoding deoxynivalenol-dependent susceptibility. New Phytol. 2019, 221, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Kinnersley, A.M.; Turano, F.J. Gamma aminobutyric acid (GABA) and plant responses to stress. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Williamson, J.D.; Jennings, D.B.; Guo, W.W.; Pharr, D.M.; Ehrenshaft, M. Sugar alcohols, salt stress, and fungal resistance: Polyols-multifunctional plant protection. J. Am. Soc. Hortic. Sci. 2002, 127, 467–473. [Google Scholar] [CrossRef]
- Kanayama, Y. Physiological roles of polyols in horticultural crops. J. Jpn. Soc. Hortic. Sci. 2009, 78, 158–168. [Google Scholar] [CrossRef]
- Abood, J.K.; Losel, D.M. Changes in carbohydrate composition of cucumber leaves during the development of powdery mildew infection. Plant Pathol. 2003, 52, 256–265. [Google Scholar] [CrossRef]
- Clark, A.J.; Blissett, K.J.; Oliver, R.P. Investigating the role of polyols in Cladosporium fulvum during growth under hyper-osmotic stress and in planta. Planta 2003, 216, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Jobic, C.; Boisson, A.; Gout, E.; Rascle, C.; Fevre, M.; Cotton, P.; Bligny, R. Metabolic processes and carbon nutrient exchanges between host and pathogen sustain the disease development during sunflower infection by Sclerotinia sclerotiorum. Planta 2007, 226, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Draper, J.; Rasmussen, S.; Zubair, H. Metabolite analysis and metabolomics in the study of biotrophic interactions between plants and microbes. Ann. Plant Rev. 2018, 43, 25–59. [Google Scholar] [CrossRef]
- Choudhary, A.K.; Kumar, S.; Patil, B.S.; Bhat, J.S.; Sharma, M.; Kemal, S.; Ontagodi, T.P.; Datta, S.; Patil, P.; Chaturvedi, S.K.; et al. Narrowing yield gaps through genetic improvement for Fusarium wilt resistance in three pulse crops of the semi-arid tropics. SABRAO J. Breed. Genet. 2013, 45, 341–370. [Google Scholar]
- Vlachostergios, D.N.; Roupakias, D.G. Screening under low plant density reinforces the identification of lentil plants with resistance to Fusarium wilt. Crop Sci. 2017, 57, 1285–1294. [Google Scholar] [CrossRef]
- Choudhary, A.K.; Kumar, S. Genetic improvement of lentil for Fusarium wilt resistance. In Scientific Lentil Production; Singh, A.K., Bhakta, N., Sangle, U.R., Manibhushan, Sundram, P.K., Kumar, S., Yasin, J.K., Eds.; Society for Upliftment of Rural Economy: Varanasi, India, 2018; pp. 91–108. [Google Scholar]
- Chaudhry, M.A.; Ilyas, M.B.; Ghazanfar, M.U. Sources of resistance from lentil international Fusarium wilt nursery 2006-7. Pak. J. Phytopathol. 2008, 20, 122–124. [Google Scholar]
- Foti, C. Evaluation and Improvement of Lentil’s Resistance against Biotic and Abiotic Stresses. Ph.D. Thesis, University of Thessaly, Volos, Greece, 2020. [Google Scholar]
- Patelou, M.; Infante, C.; Dardelle, F.; Randewig, D.; Kouri, A.D.; Udvardi, M.K.; Tsiplakou, E.; Mantecón, L.; Flemetakis, E. Transcriptomic and metabolomic adaptation of Nannochloropsis gaditana grown under different light regimes. Algal Res. 2020, 45, 101735. [Google Scholar] [CrossRef]
- Stein, S.E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 1999, 10, 770–781. [Google Scholar] [CrossRef]
- Fiehn, O.; Barupal, D.K.; Kind, T. Extending biochemical databases by metabolomic surveys. J. Biol. Chem. 2011, 286, 23637–23643. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. MixOmics: An R Package for omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Ramraj, S.; Nishant, U.; Sunil, R.; Shatadeep, B. Experimenting XGBoost algorithm for prediction and classification of different datasets. Int. J. Control Theory Appl. 2016, 9, 651–662. [Google Scholar]

| Ratio Fol/C | p | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Roots | Shoots | Root | Shoot | |||||||||
| Metabolite | RT | Fragment Ion m/z | ILL-590 | ILL-6031 | ILL-590 | ILL-6031 | G | T | G × T | G | T | G × T |
| Carbohydrates | ||||||||||||
| Cellobiose | 41.71 | 204 | 2.650 | 0.656 | 0.556 | 3.603 | 0.032 | 0.947 | 0.399 | 0.683 | 0.566 | 0.615 |
| Leucrose | 41.06 | 361 | # | 0.000 | 0.000 | 0.000 | 0.028 | 0.111 | 0.112 | 0.720 | 0.507 | 0.876 |
| D-trehalose | 41.05 | 361 | 2.999 | 1.063 | 0.814 | 0.692 | 0.021 | 0.354 | 0.402 | 0.455 | 0.865 | 0.876 |
| Talose | 27.13 | 319 | # | 0.000 | 0.000 | 0.000 | 0.005 | 0.005 | 0.010 | 0.720 | 0.507 | 0.876 |
| Amino acids | ||||||||||||
| L-proline 2 | 12.44 | 142 | 3.946 | 0.284 | 2.266 | 0.468 | 0.170 | 0.328 | 0.380 | 0.044 | 0.561 | 0.615 |
| L-asparagine | 20.86 | 188 | 11.250 | 33.646 | 23.109 | 5.416 | 0.547 | 0.111 | 0.399 | 0.088 | 0.010 | 0.221 |
| L-ornithine | 20.37 | 142 | ## | ## | ## | 0.642 | 0.437 | 0.029 | 0.399 | 0.720 | 0.845 | 0.615 |
| Polyols | ||||||||||||
| Palatinitol | 43.15 | 361 | 2.478 | 2.036 | 0.773 | 1.345 | 0.000 | 0.017 | 0.064 | 0.777 | 0.868 | 0.876 |
| Acetol * | 23.96/43.79 | 217 | 0.000 1 | 0.000 | # | 0.355 | 0.564 | 0.275 | 0.555 | 0.020 | 0.561 | 0.615 |
| 2-butyne-1,4-diol | 27.75 | 147 | 0.000 | 0.000 | # | 0.344 | 0.654 | 0.275 | 0.672 | 0.011 | 0.363 | 0.615 |
| Arabitol | 23.06 | 217 | 1.173 | 0.269 | 4.132 | 1.118 | 0.047 | 0.947 | 0.650 | 0.683 | 0.561 | 0.876 |
| Organic acids | ||||||||||||
| Maleamic acid | 11.94 | 244 | 0.715 | 0.386 | 0.499 | 0.840 | 0.032 | 0.019 | 0.080 | 0.192 | 0.445 | 0.615 |
| Fumaric acid | 13.71 | 245 | # | 0.000 | # | 0.002 | - | - | - | 0.002 | 0.002 | 0.002 |
| Inimodiacetic acid | 18.08 | 232 | 0.049 | 0.007 | 0.817 | 0.524 | 0.564 | 0.302 | 0.536 | 0.036 | 0.619 | 0.705 |
| Carboxylic acids | ||||||||||||
| 3-hydroxypropanoic acid | 15.58 | 219 | 0.000 | 0.000 | # | 0.000 | 0.547 | 0.275 | 0.509 | 0.004 | 0.003 | 0.004 |
| Nitrogenous compounds | ||||||||||||
| 2-amino-1-phenylethanol | 11.67 | 174 | 12.346 | 7.249 | 19.689 | 2.500 | 0.437 | 0.019 | 0.399 | 0.173 | 0.343 | 0.495 |
| Putrescine | 22.95 | 174 | 0.392 | 0.000 | 0.500 | 0.517 | 0.032 | 0.001 | 0.017 | 0.911 | 0.561 | 0.876 |
| Phosphate compounds | ||||||||||||
| Glycerol 1-phosphate | 23.87 | 357 | 1.983 | 0.360 | ## | 1.332 | 0.642 | 0.769 | 0.100 | 0.182 | 0.002 | 0.071 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foti, C.; Zambounis, A.; Bataka, E.P.; Kalloniati, C.; Panagiotaki, E.; Nakas, C.T.; Flemetakis, E.; Pavli, O.I. Metabolic Aspects of Lentil–Fusarium Interactions. Plants 2024, 13, 2005. https://doi.org/10.3390/plants13142005
Foti C, Zambounis A, Bataka EP, Kalloniati C, Panagiotaki E, Nakas CT, Flemetakis E, Pavli OI. Metabolic Aspects of Lentil–Fusarium Interactions. Plants. 2024; 13(14):2005. https://doi.org/10.3390/plants13142005
Chicago/Turabian StyleFoti, Chrysanthi, Antonios Zambounis, Evmorfia P. Bataka, Chrysanthi Kalloniati, Evangelia Panagiotaki, Christos T. Nakas, Emmanouil Flemetakis, and Ourania I. Pavli. 2024. "Metabolic Aspects of Lentil–Fusarium Interactions" Plants 13, no. 14: 2005. https://doi.org/10.3390/plants13142005
APA StyleFoti, C., Zambounis, A., Bataka, E. P., Kalloniati, C., Panagiotaki, E., Nakas, C. T., Flemetakis, E., & Pavli, O. I. (2024). Metabolic Aspects of Lentil–Fusarium Interactions. Plants, 13(14), 2005. https://doi.org/10.3390/plants13142005








