Abstract
The DOF (DNA-binding with one finger) transcription factors are exclusive to plants and play crucial roles in plant growth, development, and environmental adaptation. Although extensive research has been conducted on the Dof gene family in Arabidopsis, maize, and Solanum, investigations concerning the role of this gene family in Liriodendron remain unreported, leaving its biological function largely unknown. In this study, we performed a comprehensive genome-wide identification of the Dof gene family based on the Liriodendron genome, resulting in the discovery of a total of 17 LcDof gene members. Based on the results of phylogenetic analysis, the 17 LcDof proteins were classified into eight subfamilies. The motif analysis revealed the diverse nature of motifs within the D1 subfamily, which includes a distinct type of Dof transcription factor known as CDF (Cycling Dof Factor). We further characterized the chromosomal distribution, gene structure, conserved protein motifs, and cis-elements in the promoter regions. Additionally, utilizing transcriptome data from Liriodendron hybrids and conducting RT-qPCR experiments, we investigated the expression patterns of LhDofs under various abiotic stresses such as drought, cold, and heat stress. Notably, we found that several LhDofs, particularly LhDof4 and LhDof6, were significantly upregulated in response to abiotic stress. Furthermore, we cloned LhDof4 and LhDof6 genes and found that its encoding protein was mainly located in the nucleus by transient transformation in Liriodendron hybrids protoplast. Subsequently, we used LhDof6-overexpressing Liriodendron hybrid seedlings. We found that overexpression of LhDof6 enhanced the cold tolerance of the plants, increasing their survival rate at −20 °C. This result was further validated by changes in physiological indicators.
1. Introduction
The presence of abiotic stresses, such as high salinity, drought, extreme temperatures, and poor soil fertility, poses significant environmental challenges that critically impede plant growth, development, and overall productivity []. Throughout the process of evolution, plants have demonstrated their ability to gradually colonize a wide range of terrestrial environments, including those characterized by harsh and extreme conditions. This remarkable adaptability has been facilitated by the development of sophisticated molecular and physiological mechanisms that enable plants to regulate their growth in response to resource availability and prevailing environmental conditions. The aforementioned adaptations have played a pivotal role in enabling plants to flourish in various ecosystems, ranging from arid deserts to saline marshes, thereby showcasing their remarkable resilience and adaptability in surmounting environmental constraints [,,,,].
Numerous transcription factors (TFs) involved in the regulation of gene expression and signaling pathways related to abiotic stress have been identified, encompassing a diverse range of members from large gene families such as bHLH, HD-ZIP, WRKY, MYB, bZIP, DOF, and NAC [,,,,].
The Plant-specific DNA Binding with One Finger (DOF) proteins are a group of transcription factors (TFs) characterized by a conserved 50-amino-acid DNA-binding domain, typically located in their N-terminal region and connected to a basic region []. The conserved DOF domain is a distinct zinc finger domain, characterized by a C2–C2 finger structure. It specifically binds to cis-regulatory DNA elements featuring the core 5′-T/AAAG-3′ motif, which is found in the promoter regions of target genes [,]. Recent studies have revealed that, despite its initial identification as a DNA-binding domain, the DOF domain may possess a plethora of functions, including nuclear localization, interaction with other transcription factors and intercellular trafficking [,]. Previous studies have corroborated its functional role in plant growth and development, including flowering control [], maturation, seed development [], and germination [,]. In particular, mutant dag1 (which encodes a Dof transcription factor in Arabidopsis) seeds are induced to germinate by significantly red light fluence rates []; the COG1 gene (which encodes a Dof protein in Arabidopsis) functions as a negative regulator in phytochrome signaling pathways []. Additionally, compelling evidence suggests that CYCLING DOF FACTORS (CDFs), a class of Dof-type transcriptional repressors, have been experimentally proven to directly suppress the expression of CONSTANS (CO). CDFs possess the ability to inhibit the expression of photoperiodic genes, thereby influencing the perception of day length and ultimately impacting the floral transition in Arabidopsis []. More significantly, Dof transcription factors play a crucial role in plant phytohormone and stress responses. For instance, TDDF1, which encodes a Dof protein in tomato, enhances tolerance to drought, salt, various hormonal stresses, as well as resistance to late blight []. The salt and osmotic stress tolerance is enhanced by ThZFP1 and ThDof1.4 through the elevation of proline levels and improvement in ROS scavenging capability []. Therefore, the Dof gene family plays an essential role in the life cycle of plants.
The Liriodendron hybrids display significant heterosis, encompassing not only distinct foliar morphology and exotic floral characteristics but also notable adaptive capabilities and growth advantages. Historically, the Liriodendron hybrids have shown sensitivity to low-temperature stress, which has posed a considerable challenge. The study and functional validation of the Dof gene family may provide a potential avenue for identifying candidate genes that can be utilized for genetic improvement and the development of cold tolerance in Liriodendron hybrids. Furthermore, proposing further application of genetic engineering techniques for developing novel cold-resistant Liriodendron hybrids aims to extend these new varieties’ reach to facilitate ecosystem services across a broader geographical area.
2. Results
2.1. Identification and Protein Sequence Characterization of LcDofs
The Dof gene has been previously confirmed to be broadly involved in plant growth and development. Recent reports have also identified its significant role in plant responses to abiotic stress. The Dof gene in Liriodendron chinense (L. chinense) has been the subject of initial exploration and analysis. The L. chinense genome contains a total of 17 Dof genes, accounting for 0.048% of the overall gene count in the genome (Table 1).

Table 1.
LcDof genes and their related information. (Len: protein length; MW: molecular weight; PI: isoelectric point; AI: aliphatic index; II: Instability index).
The Dof transcription factor family in L. chinense is relatively small compared to the majority of species. Based on their chromosomal locations, these genes have been designated as LhDof1 to LhDof17 (Table 1). To investigate the genetic diversity within this family, we conducted a comprehensive analysis of the fundamental physicochemical characteristics of these 17 Dof proteins, encompassing protein sequence length, molecular weight (MW), isoelectric point (pI), and other pertinent properties. The analysis of the physicochemical properties of the LcDof gene family revealed that the Dof proteins exhibit a length range of 160 to 635 amino acids, with molecular weights varying from 17.01 to 71.60 kDa. The isoelectric point (pI) of LcDof7 (Lchi21078) was observed to be the lowest at 5.70, while the highest pI value of 9.56 was noted for LcDof5 (Lchi18955), with an average pI of 7.76. The analysis indicates that the majority of Dof proteins exhibit a weakly basic nature. Most LcDof proteins are characterized by high levels of instability, with the highest instability index score reaching 85.81. Among this group, only LcDof11 (Lchi08966) is considered stable, as it exhibits an instability index value below 40. The LcDof proteins are susceptible to denaturation or degradation, leading to potential alterations in their physicochemical properties and biological functions. This implies that LcDof proteins exhibit a high sensitivity towards changes in the external environment. The physicochemical properties of Dof proteins in L. chinense display variations, suggesting diverse regulatory roles in plant growth and development under different conditions. Therefore, it is crucial to conduct a comprehensive investigation into their classification and phylogenetic traits.
2.2. LcDofs Contain More Abundant Conserved Motifs and a More Homogeneous Gene Structure
A multiple sequence alignment of 17 LcDof amino acid sequences from L. chinense was performed using the ClustalX software (V2.1). The conserved domains were then analyzed. It was found that the N-terminal Dof domain of the L. chinense Dof protein contains a typical C2–C2 zinc finger protein structure, comprising 52 amino acid residues (Figure 1). The LcDof protein domain remains intact and exhibits a remarkable level of uniformity, comprising a solitary zinc finger protein composed of four cysteines. This observation signifies the highly conserved and complete nature of the Dof domain in L. chinense. The LcDof gene family is classified into 6 subgroups based on multiple sequence alignment and phylogenetic analysis, with each subgroup exhibiting nearly identical motif structure and distribution of LcDof proteins (Figure 2). Moreover, genes within the same subgroup share analogous intron-exon structures and gene lengths (Figure 2). All LcDof proteins contain Motif1 (Figure 2), which is consistent with the findings of previous studies. Motif1 is the conserved Dof motif of LcDof. Our findings indicate that the C-terminal structures of four LcDof proteins (Lchi19181, Lchi21078, Lchi14330, Lchi02891) within a specific subgroup are characterized by the presence of diverse types of motifs. This observation suggests that the LcDof proteins in this subgroup may play a role in a multitude of biological processes. The presence of other motifs, such as Motif 4, 6, and 9, is limited to only a single or a few phylogeographic subgroups. Interestingly, in certain categories, no additional conserved sequence motifs were identified besides the Dof motif, implying that the functions of these sequences may be unpredictable. A comprehensive analysis of these sequences may prove crucial in elucidating the functional diversity of the Dof family. Through gene structure analysis, it was observed that the LcDof genes exhibit a distribution of 1 (2 genes), 2 (11 genes), and 3 (4 genes) exons, respectively, suggesting that the predominant form of LcDof genes consists of two exons.

Figure 1.
The Dof concerved region in LcDofs, alignment of multiple protein sequences in LcDofs.
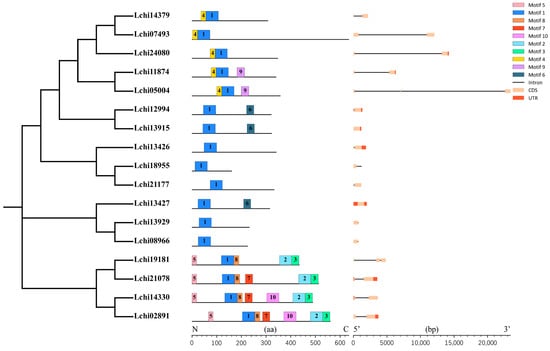
Figure 2.
Analysis of conserved motif elements and gene structures of LcDofs. Figure was made using the MEME program and TBtools.
2.3. Phylogenetic Structure of the LcDof Gene Family
In order to elucidate the molecular evolution and phylogenetic relationships of L. chinense Dof proteins, an unrooted phylogenetic tree was constructed containing 17 LcDofs and their homologs in Arabidopsis thaliana, Amborella trichopoda, Zea mays, and Oryza sativa. A multiple sequence alignment of Dof gene family members was performed using MAFFT software (v7.487) (https://mafft.cbrc.jp/alignment/software, accessed on 25 January 2022) with default parameters. The phylogenetic tree was constructed using MAGE 7.0, employing the neighbor-joining method with a bootstrap value of 1000 to analyze the evolution of the Dof gene in L. chinense (Figure 3). The results of multiple sequence alignment and phylogenetic tree clustering indicate that the Dof proteins from Liriodendron chinense, Arabidopsis thaliana, Amborella trichopoda, Zea mays, and Oryza sativa can be classified into eight distinct categories: Class A, B1, B2, C1, C2, C3, D1, and D2 (Figure 3). Additionally, L. chinense and A. trichopoda exhibit a closer phylogenetic relationship.
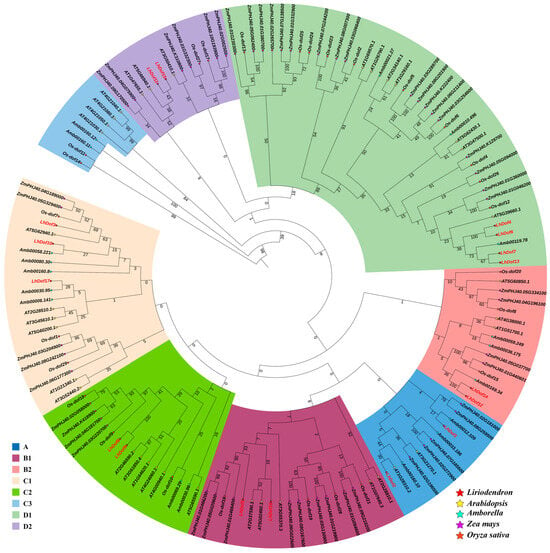
Figure 3.
Phylogenetic trees of Dof genes. Different colors represent different subclasses, and the red font represent L. chinense. Numbers on branches indicate percent reliability of bootstrap values based on 1000 replicates.
2.4. Analysis of the Whole-Genome Duplication Events in the LcDof Family
In order to gain insight into the chromosomal distribution of the LcDof family, we used Tbtools software along with the genomic localization information of the LcDof family members to display their chromosomal distribution (Figure 4). The distribution of LcDof family members across the chromosomes is not uniform, with Dof genes found only on chromosomes 1, 2, 4, 7, 8, 11, 12, 13, 15, 16, and 18 in L. chinense. Among them, chromosomes 1, 2, 4, 13, 16, and 18 each contain two Dof genes, and the genes are closely arranged on chromosomes 1, 13, and 16. The data indicates that each chromosome contains a maximum of two Dof genes, suggesting that LcDof genes are typically spaced at considerable distances and rarely clustered on the same chromosome. The majority of LcDof genes are located at the chromosomal ends, with fewer near the centromeric regions. The concentrated distribution of LcDof genes at the chromosomal ends in L. chinense may be related to the more open chromatin structure found at these locations, which could facilitate active gene expression. Meanwhile, the chromosomal center (near the centromeres) typically has a more compact chromatin structure, which may restrict gene expression [,].
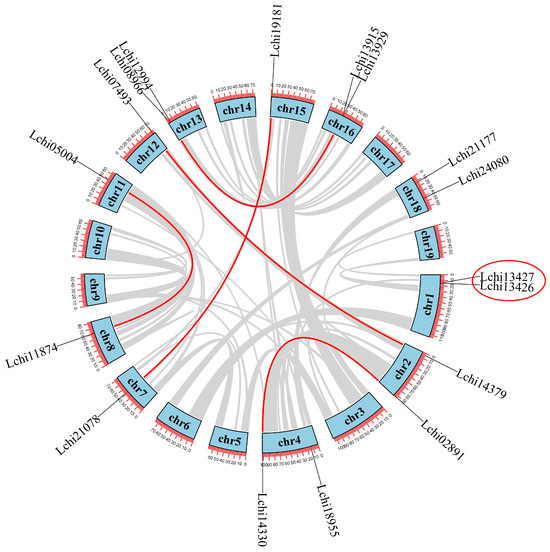
Figure 4.
Genome-wide synteny analysis of Dof gene family among L. chinense. Distribution and duplication of the LcDof gene in L. chinense. The circles are chromosomal genes, and the names are displayed outside the circles.
We have studied and mapped gene duplication events during the evolutionary process of L. chinense, which may also affect changes in the number and distribution of LcDof genes within the genome (Figure 4). It is known that duplication events occur regionally through a process known as tandem duplication, whereby gene sequences of less than 200 kb are copied in close proximity to the original gene. However, they can also occur over a broader range through segmental duplication, where larger fragments are duplicated. The former is typically attributed to DNA replication errors, whereas the latter may be attributed to polyploidy events resulting from chromosomal rearrangements [,]. There are six duplication events within the LcDof genes of L. chinense (Lchi21087–Lchi19181, Lchi05004–Lchi11874, Lchi14330–Lchi02891, Lchi07493–Lchi14379, Lchi08966–Lchi13929), of which only one pair (Lchi13427–Lchi13426) is due to tandem duplication, while the rest are caused by segmental duplication. The analysis of the expansion of the LcDof gene family has revealed that the majority of gene duplication events are the result of segmental duplication, which represents a crucial mechanism for gene expansion in plants. The presence of these duplication events reveals significant expansion strategies within the gene family during the evolutionary process and also indicates the dynamic changes and adaptive evolution of plant genomes. The aforementioned duplication pattern, in conjunction with the whole-genome duplication (WGD) event experienced by L. chinense, offers crucial insights into the expansion and adaptation of the LcDof gene family throughout plant evolution. Following whole-genome duplication events, some genes may be lost due to functional redundancy, while genes with essential functions or those providing adaptive advantages are often retained.
2.5. The Analysis of Cis-Element Regulation of Promoters Revealed That LcDofs Regulated Many Bioactive Processes
We used the PlantCARE website to analyze the 5′ upstream promoter regions (2000 bp) of the LcDof genes to predict all cis-acting elements. A variety of cis-acting elements were identified, and the promoter regions of all LcDof genes were found to contain a substantial number of them. We classified these cis-acting elements into four categories: growth and development-related elements, light-responsive elements, abiotic stress-responsive elements, and plant hormone-responsive elements (Figure 5). The promoters of LcDof genes were found to be rich in light-responsive and abiotic stress-responsive elements, which suggests that LcDof genes are primarily involved in biological processes related to light response and abiotic stress response. In summary, the distribution of cis-acting elements indicates that LcDofs are involved in light response, hormone response, stress response, and plant growth and development.
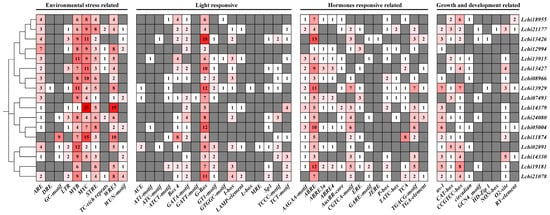
Figure 5.
Results of LcDofs gene promoter predictor elements from the PlantCare database are depicted. The abscissa axis displays the types of elements. Red squares indicate a higher predicted presence of the element in the LcDofs gene promoter, while gray and white squares signify that the element is either absent in the gene promoter or present in lesser numbers.
The presence of MYB and STRE environmental response factors in all LcDof genes indicates that these two cis-acting elements are crucial for LcDof genes to perceive environmental alterations. However, some cis-acting elements are specific to certain LcDofs. For example, only LcDof15 (Lchi13929) in Class D2 contains the DRE element, which suggests that Class D2 LcDofs may have differentiated to include proteins that are responsive to dehydration. The LcDof family members contain LTR functional elements, suggesting that some members may be specifically involved in cold response. While not all members within each class contain LTR elements, at least one member in each class is capable of responding to cold, indicating that the LcDof family exhibits some sensitivity to low temperatures.
G-Box, a crucial cis-regulatory element in light response, is abundant in all members of the LcDof family, playing a significant role in mediating light-dependent gene expression. The G-Box is commonly found in the promoters of many light-responsive genes, where it binds to light-activated transcription factors to promote transcriptional activation of these genes in response to light stimuli. Other light-responsive elements appear to be irregularly distributed in the promoters of each member, with LcDof4 (Lchi02891) containing more light-responsive elements than other members, indicating differences in light response capabilities within the family. As LcDofs contain abundant ABRE elements, they may be actively involved in the abscisic acid response of plants. The distribution and quantity of ABRE3a and ABRE4 elements are evenly spread among specific LcDof members. This pattern suggests that these genes likely play a consistent and significant role in plant’s response to ABA signaling. The specific distribution of ABRE3a and ABRE4 elements may ensure the coordinated expression of these genes throughout the plant, effectively regulating the physiological state of the plant to adapt to environmental changes [,]. While ABRE elements are widely distributed in certain LcDof members, this does not imply that all members have identical functions in all physiological processes or stress responses. The different ABRE types may act as mediators for the binding of specific transcription factors or co-activators, which in turn result in subtle functional differences that enable plants to adapt their response to abscisic acid in a precise manner [,]. The results suggest that these LcDof proteins may also be involved in different abscisic acid-mediated regulatory networks. The majority of the growth and development regulatory elements of LcDofs are concentrated in the meristematic and differentiation functions of tissues, suggesting that these genes play a pivotal role in tissue differentiation and development.
The cis-regulatory element analysis reveals that the functional elements in the promoter regions of LcDof genes are both abundant and comprehensive, indicating that they may function independently or simultaneously to regulate growth, development, and abiotic stress response. Accordingly, further investigation is required to elucidate the expression profiles of LcDofs under growth conditions and abiotic stress.
2.6. LcDof Gene Families of Class D1 under Abiotic Stress Has a Strong Reaction
To study the expression pattern of Liriodendron hybrid Dof genes under different stress conditions, we analyzed their expression profiles in the transcriptome data of leaves under low temperature, PEG6000-simulated drought, and high-temperature stress conditions (Figure 6). The results indicate that LhDof genes in the D1 class are actively expressed, and both LhDof4 (Lchi02891) and LhDof6 (Lchi14330) exhibit strong responses to cold and drought stress, with similar expression patterns (Figure 6A,B). Under low-temperature treatment, the expression levels of LhDof4 and LhDof6 were continuously upregulated from 0 h (CK) to 1 day, peaking at 12 h, and then downregulated from 1 day to 3 days (Figure 6A). Under PEG6000-simulated drought treatment, the expression levels of LhDof4 and LhDof6 were continuously upregulated from 0 h (CK) to 1 h, reaching their highest expression at 1 h, gradually downregulated from 1 h to 12 h, and then slowly upregulated from 12 h to 3 days, finally maintaining normal expression levels (Figure 6B).
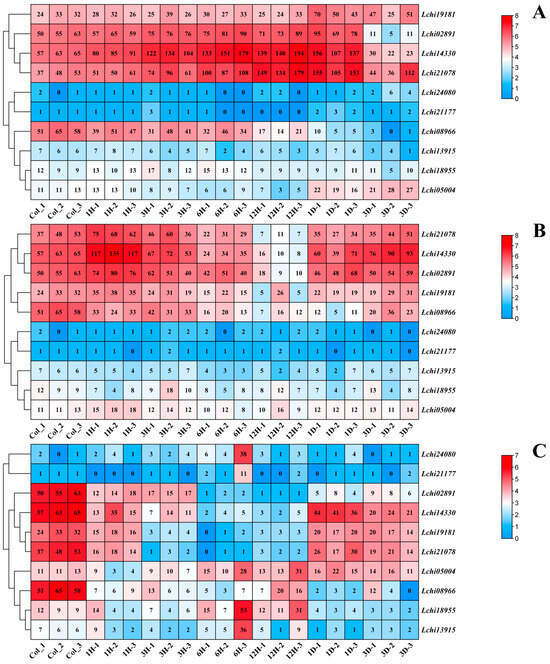
Figure 6.
Transcriptional expression patterns of Dof genes in L. hybrid under (A) cold, (B) drought, and (C) heat stress are depicted. The LhDofs were subjected to three different stress factors: cold, drought, and heat stress. The designations Heat_0h, Heat_1h, Heat_3h, Heat_6h, Heat_12h, Heat_1d, and Heat_3d represent three biological replicates for each time point (0 h, 1 h, 3 h, 6 h, 12 h, 1 day, 3 days). Transcript abundance levels are represented using the log2 (FPKM + 1) transformation. The values on the right panel of the heatmap indicate the expression level.
The expression levels of LhDof4 and LhDof6 were significantly downregulated under high-temperature treatment. However, they exhibited high responsiveness to low-temperature and drought stress, displaying similar expression patterns across all three stress conditions (Figure 6C).
The D1 class of the Dof family encompasses a specific category of Dof genes, designated as CDF genes, which typically demonstrate robust responses to abiotic stressors. We have confirmed this in Liriodendron hybrid as well. The CDF transcription factors LhDof4 and LhDof6 from the D1 class may play a positive regulatory role under low-temperature and drought stress. The sharply downregulated expression of these two factors under high temperature stress suggests that LhDof4 and LhDof6 have a negative regulatory role in response to high temperatures.
Two of the LcDof genes were not expressed under the two stresses, and the expressions of five LcDof genes were insignificant. In summary, only the class D1 CDF transcription factors exhibited a robust response to low-temperature, drought, and high-temperature stress, indicating that the LcDof gene family has evolved class D1 CDF transcription factors that are specifically responsive to diverse abiotic stresses.
This is consistent with recent studies on CDFs. For instance, CDF transcription factors can induce the expression of stress-response genes. The A. thaliana AtCDF3 has been shown to regulate the expression of multiple abiotic stress-response genes in plants that respond to extreme temperatures, drought, and osmotic stress []. The expression of two CDF genes was observed to undergo significant alterations in response to drought and elevated temperatures, with notable changes occurring within the first hour. This suggests that the LhCDF genes exhibit a high degree of sensitivity to these two stressors. In summary, the pair of CDF genes (LhDof4 and LhDof6) may co-regulate and respond to different abiotic stresses.
2.7. qRT-PCR Validation of LcCDFs under Low Temperature and Drought Stress
To verify the accuracy of the abiotic stress transcriptome data for LcCDFs (LhDof4 and LhDof6), we conducted qRT-PCR experiments on Liriodendron hybrids under cold and drought stress (Figure 7).
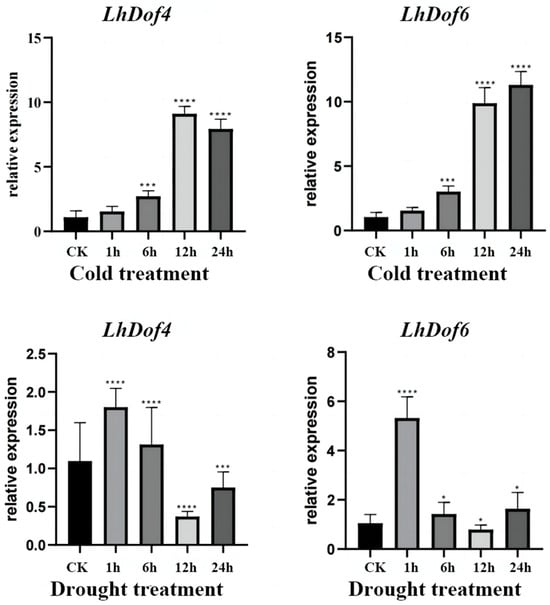
Figure 7.
The expression of LhDof4 and LhDof6 in cold and drought-treated samples was analyzed by qRT-PCR. Vertical bars represent standard deviation, with 18S serving as the internal reference gene, and each sample was repeated three times. * on the bar chart indicates that the difference between the two control groups reached a significant level, that is, p < 0.05, *** means p < 0.001, **** means the difference between the two control groups reached a very significant level, that is, p < 0.0001.
Under drought stress, the expression of LhDof4 and LhDof6 initially increases, then decreases, and subsequently increases again. The expression levels of both genes drop to their lowest at 12 h, below the control (CK) level, and then increase at 24 h to levels similar to those observed at 1 h. After 1 h, the expression levels significantly increased, then gradually decreased, and finally increased again to return to normal levels. This pattern is consistent with the transcriptome data results under drought conditions. Under low-temperature stress, the expression of LhDof4 and LhDof6 gradually increased over time, peaked at 12 h, maintained a high level until 1 day, and then gradually decreased, aligning with the transcriptome data results.
Under low-temperature stress, the expression levels of LhDof4 and LhDof6 gradually increased over time, peaking at 12 h. They maintained a high level until 24 h, after which they gradually decreased. This pattern aligns with the transcriptome data results.
2.8. LhDof4 and LhDof6 Are Localized to the Cell Nucleus
We conducted a subcellular localization prediction analysis for all LcDof genes, including LcDof4 and LcDof6, using an online tool. The results of this analysis are presented in Table 2. Most of the LcDof genes are predicted to localize to the nucleus, which is consistent with their roles as transcription factors. However, LcDof10 and LcDof11 are predicted to localize in the chloroplast, while LcDof13 is localized in the mitochondria. All CDFs are shown to be localized in the nucleus, likely due to the presence of one or more nuclear localization sequences in the N-terminal structure of CDF proteins. The LcDof genes also play regulatory roles in the chloroplasts or mitochondria of plant cells, not just in the nucleus.

Table 2.
Subcellular localization prediction results for LcDof genes.
We then used the cloned LhDof4 and LhDof6 sequences to construct fusion fluorescent protein vectors, p35S:LhDof4-GFP and p35S:LhDof6-GFP, with the unmodified pJlT166 vector serving as a control. The results showed that in protoplast cells transformed with p35S:GFP, the GFP green fluorescent protein was localized in the nucleus and also distributed in the cell membrane and cytoplasm. In contrast, in protoplast cells transformed with p35S:LhDof4-GFP and p35S:LhDof6-GFP, the LhDof4-GFP and LhDof6-GFP fusion proteins were primarily localized in the nucleus (Figure 8).
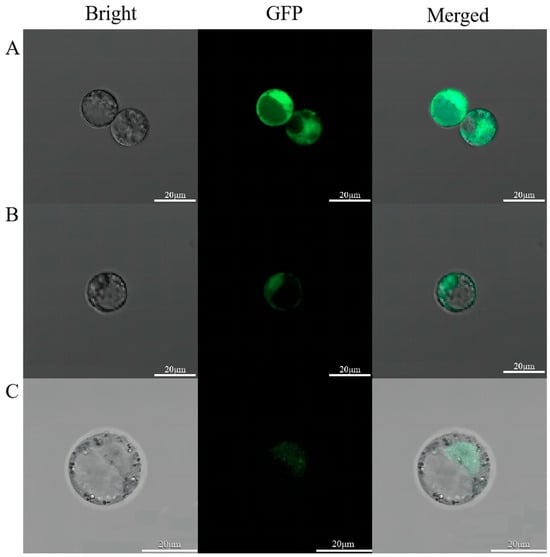
Figure 8.
Subcellular localization of LhDof4 and LhDof6. The first panel shows the bright field. The second panel shows green fluorescence (GFP). The third panel is a merged image of the green fluorescence and bright field. (A) p35S:GFP, (B) p35S:LhDof4-GFP; (C) p35S:LhDof6-GFP. Bar = 20 μm.
2.9. Overexpression of LhDof6 Improved the Cold Tolerance of Liriodendron Hybrid
Given that our expression analysis indicates that the LhCDFs gene may play a role in plant response to cold stress, we further investigated its molecular function in this context. We analyzed the response of LhDof6 overexpression plants to abiotic stress and characterized their phenotypes. We constructed overexpression vector (Figure S1). Using qRT-PCR, we selected two overexpression lines (Figure 9) and obtained LhDof6-OE plants through somatic embryogenesis (SE) technology of Liriodendron hybrid. We then compared the freezing stress tolerance of LhDof6-OE transgenic plants grown in 1/2 MS medium to that of the wild type. The results indicate that plants overexpressing LhDof6 exhibit enhanced cold tolerance.
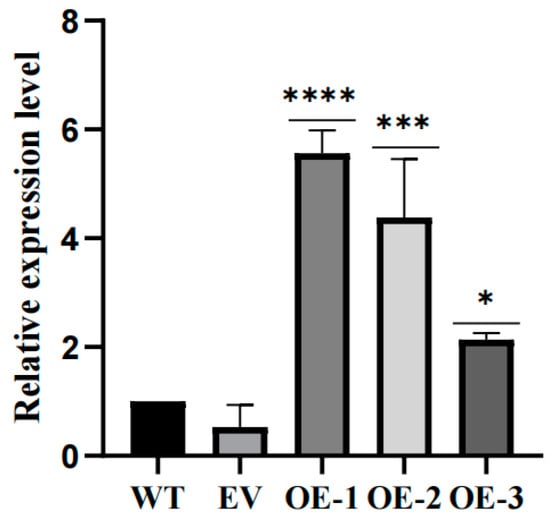
Figure 9.
The Relative expression of LhDof6 in wild-type, no-load and transgenic plants was determined by using 18S rRNA as the reference gene. * on the bar chart indicates that the difference between the two control groups reached a significant level, that is, p < 0.05, *** means p < 0.001, **** means the difference between the two control groups reached a very significant level, that is, p < 0.0001.
In each group, 28 similarly grown Liriodendron hybrid plants were selected and acclimated at 4 °C (16 h light and 8 h dark) for 7 days. They were then subjected to −20 °C for 20 min. After the treatment, the plants were transferred to a greenhouse at 22 °C for a 7-day recovery period. Photos were taken on the first and seventh days of recovery, and survival rates were calculated on the seventh day. The results showed that the leaves of both the overexpression group and the control group exhibited wilting on the first day of recovery (Figure 10). After 7 days, the leaves of the control group did not recover from wilting; most of them began to wither and die. In contrast, the overexpression group showed significant alleviation of leaf wilting, with leaves resuming spreading and exhibiting a low degree of mortality. Survival rate statistics indicated that the overexpression lines had a survival rate of 68–71%, compared to only 25% in the wild type (Figure 10).
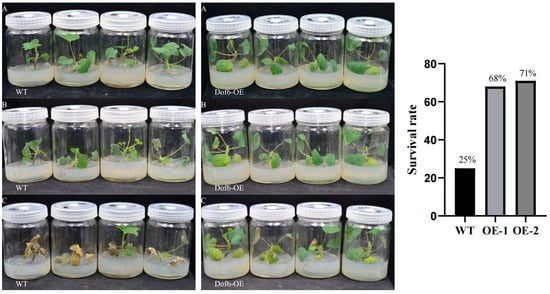
Figure 10.
LhDof6-OE seedling freeze stress treatment and survival rate statistics. (A) Seedlings grown under greenhouse conditions for six months; (B) After −20 °C treatment for 30 min followed by 1 d of recovery cultivation; (C) After −20 °C treatment for 30 min followed by 7 d of recovery cultivation; Survival rate statistics are displayed on the right.
The occurrence of low temperature stress in plants is widely recognized to induce numerous physiological and metabolic rearrangements, which are mediated by known determinants. Therefore, we conducted measurements on a range of physiological indicators to further elucidate the role of LhDof6 under freezing stress, as depicted in (Figure 11). After exposure to freezing stress, the MDA content exhibited an increase in both plant types; however, the rise was more pronounced in the wild type, indicating that low temperatures can induce damage to plant cell membranes to some extent, with the wild type experiencing more severe impairment (Figure 11A). Overexpression of LhDof6 leads to a modest increase in plant proline levels, while under freezing stress conditions, the overexpression lines exhibit significantly enhanced proline accumulation compared to the wild type. This accumulation of proline plays a crucial role in safeguarding plant cells (Figure 11C).
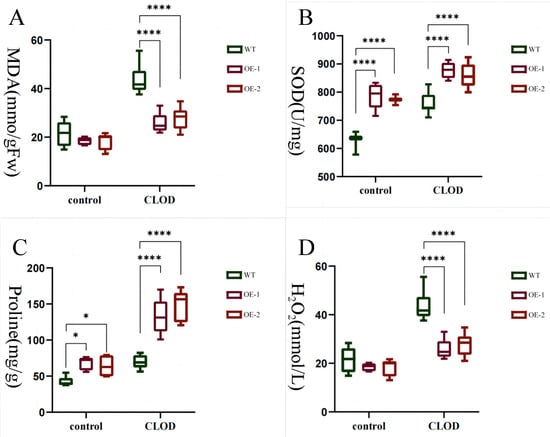
Figure 11.
Measurement of various physiological indicators of seedlings treated at −20 °C for 0 and 30 min: (A) malondialdehyde (n = 9); (B) superoxide dismutase activity (n = 9); (C) proline content (n = 9); (D) hydrogen peroxide (n = 9). * on the bar chart indicates that the difference between the two control groups reached a significant level, that is, p < 0.05; **** means the difference between the two control groups reached a very significant level, that is, p < 0.0001.
The level of H2O2 increases rapidly, which can function as a signaling molecule to activate the expression of cold resistance-related genes in plants. However, excessive accumulation of H2O2 may lead to a spike in intracellular ROS levels and result in oxidative stress. Under normal circumstances, the overexpression of LhDof6 resulted in an increase in SOD enzyme activity in Liriodendron hybrid. Subsequently, exposure to cold temperatures further amplified the SOD enzyme activity, with consistently higher levels observed in the overexpression lines compared to the wild type (Figure 11B). Under freezing stress, wild-type plants exhibited excessive accumulation of H2O2, which can lead to irreversible damage to the plants (Figure 11D).
The physiological changes observed before and after exposure to cold stress further validate the ability of LhDof6 to enhance the cold tolerance of Liriodendron hybrid seedlings, thereby increasing their survival rate in extreme environments. These analyses underscore the potential beneficial role played by the LhDof6 gene in facilitating plant adaptation to cold stress.
3. Discussion
The Dof transcription factors possess a highly conserved single-finger domain, known as a zinc finger domain, consisting of 52 amino acid residues. This specific domain facilitates precise binding to DNA []. In this study, we identified 17 Dof genes in the L. chinense genome, designated as LcDof1~17 based on their chromosomal location. The number of LcDof genes is relatively small compared to some higher angiosperms, i.e., Arabidopsis thaliana (36), Oryza sativa (30), Sorghum bicolor (28), Glycine max (78), Hordeum vulgare (21), Nicotiana tobacum (17) and Populus trichocarpa (43) []. The findings suggest a potential association between the number of Dof gene family members and their evolutionary states as well as patterns of family expansion.
The protein length, molecular weight (MW), and isoelectric point (pI) of LcDof proteins exhibit significant variations among gene families, indicating their structural diversity and potential adaptation to diverse environments. These characteristics suggest that LcDofs may possess distinct biological regulatory functions in different environments or conditions, particularly under abiotic stress. Furthermore, the variability in length, molecular weight, and isoelectric point of Dof proteins across species implies divergent biological functions of Dof genes between different organisms. This divergence may be attributed to differences in physicochemical properties and spatial structures of these proteins.
The collinearity analysis revealed six pairs of gene duplication events in the LcDof genes, with only one pair attributed to tandem duplication, while the remaining pairs were a result of segmental duplication. The fact that all duplicated genes belonged to the same category suggests that L. chinense has undergone at least one whole-genome duplication event since its divergence. The chromosomal localization analysis of the LcDof genes revealed an uneven distribution pattern, with most genes located at the terminal regions of 11 chromosomes and no more than two genes per chromosome. This spatial arrangement suggests active expression of the Dof gene family []. The involvement of several transcription factors (TFs) in plant stress responses has been identified, with some participating in intricate regulatory networks. These TFs are predominantly encoded by polygenic families that have undergone multiple rounds of gene replication throughout the evolution of land plants [,,]. The clustered distribution of LcDof genes at the chromosome termini may be associated with the typically more accessible chromatin structure in these regions, which could facilitate active gene expression. Conversely, the chromatin structure near the centromere is generally more compacted, potentially constraining gene expression [].
The regulation of plant hormones is achieved through the binding and coordinated interaction of various transcription factors with cis-acting elements present in the promoters of plant hormone response genes []. The Dof proteins function as regulators of plant hormone response genes and have been demonstrated to mediate the gibberellin response []. The Dof transcription factor also exhibits circadian rhythms and plays a pivotal role in perceiving plant photoperiods and regulating flowering time. Analysis of JcDof1 and JcDof3 in Jatropha curcas seedlings revealed their expression patterns under long-day, short-day, and continuous light conditions, as well as their interaction with F-box proteins to modulate photoperiodic flowering []. The cis-elements present in the promoters of LcDof genes predominantly consist of light-responsive elements, plant hormone regulatory elements, stress-related elements, and growth and development regulatory elements. This implies that the LcDof genes exert comparable effects on photoperiodic response, abiotic stress response, and plant hormones. Temporal transcriptome analysis of Liriodendron hybrid under low-temperature, high-temperature, and drought conditions revealed that several LcDof genes exhibited a transcriptional profile characterized by an initial upregulation followed by downregulation in response to low-temperature conditions. Notably, the expression dynamics of LhDof4 and LhDof6 were particularly remarkable. Quantitative Real-Time PCR (qRT-PCR) analysis demonstrated that the expression levels of LhDof4 and LhDof6 increased approximately tenfold within 12 to 24 h after exposure to low temperatures compared to their pre-treatment levels. This strongly suggests that these two genes may play a pivotal role in the physiological response of Liriodendron hybrid to low temperatures.
The Cycling Dof Factor (CDF) is capable of regulating various aspects of plant growth and development, including the photoperiodic control of flowering as well as root and shoot growth. While most functional characteristics of CDFs have been extensively studied in Arabidopsis, recent data indicate that their diverse roles also extend to other plant species [,,,]. The role of DOF transcription factors (TFs) has been extensively investigated in numerous plant species, including important crops such as maize, wheat, rice, potato, and bananas, in response to various environmental stress conditions [,]. Transcriptomic analysis has revealed a limited overlap in the stress response genes regulated by GI and CDF3, indicating that these two proteins have distinct functions specifically under low temperature and osmotic stress conditions []. Further comprehensive and functional analyses are imperative to elucidate the precise roles of these factors in plant responses to diverse environmental stress conditions. Moreover, it has been reported that overexpressing AtCDF3 or SlCDF3 in Solanum lycopersicum enhances tolerance to salt stress []. These reports suggest that SlCDFs may have a crucial regulatory function in the upstream pathways of salinity and drought response, similar to their counterparts in Arabidopsis. Furthermore, the overexpression of LhDof6 significantly mitigated the mortality rate of Liriodendron hybrid seedlings exposed to extreme temperatures as low as −20 °C. Physiological changes observed before and after cold stress exposure also provide evidence that LhDof6 can enhance the cold tolerance of Liriodendron hybrid seedlings, thereby increasing their survival rate in harsh environments. These analyses indicate an active role for the LhDof6 gene in facilitating plant adaptation to low temperature stress.
4. Materials and Methods
4.1. Identification of Dof Gene in L. chinense
To identify the Dof gene in L. chinense, 37 typical and atypical Dof protein sequences of Arabidopsis thaliana were downloaded from the TAIR database (https://www.arabidopsis.org/Blast/index.jsp, accessed on 21 November 2021). From the pfam website (http://pfam-legacy.xfam.org/, accessed on 22 November 2021), the Dof hidden Markov number is PF02701, and this number has been used as the query condition. Blastp and HMMER were used to query the target sequence in the L. chinense protein database, and candidate sequences were obtained. Based on the Dof conserved domains, CDD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 22 November 2021) was used to check the conserved domains of candidate sequences to further screen out redundant sequences, and finally obtain the target sequence. Gene properties, including length, molecular weight, and isoelectric point of each protein, were determined using the ExPASy website (https://web.expasy.org/protparam, accessed on 3 January 2022) tool. Subcellular localization of LcDof genes was predicted by Cell-PLoc 2.0 (www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2, accessed on 3 January 2022).
4.2. Phylogenetic Analysis and Conserved Domains and Gene Structure Analysis
Arabidopsis thaliana, Amborella trichopoda, Zea mays, and Oryza sativa Dof protein sequences used to construct the phylogenetic tree were downloaded from Phytozome (https://phytozome-next.jgi.doe.gov, accessed on 3 January 2022). Multiple sequence alignment of Dof gene family members was performed using MAFFT software (https://mafft.cbrc.jp/alignment/software, accessed on 25 January 2022) with default parameters. MAGE 7.0 was utilized to construct the phylogenetic tree, employing the neighbor-joining method with a bootstrap value of 1000 to analyze the evolution of the Dof gene in L. chinense.
The gene structure information of each LhDof6 gene was acquired from the genomic feature file (GFF3) and displayed using Tbtools software (https://github.com/CJ-Chen/TBtools/releases, accessed on 12 February 2022), while the chromosomal location and microsynteny of LhDof6 were visualized using the Tbtools software. The Multiple Collinearity Scan toolkit (MCScanX) program was used to verify putative paralogous genes (blast hits E-value cutoff < 1 × 10−6, collinearity > 70%). The cis-acting elements of LhDof6 genes were analyzed by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 15 February 2022) and displayed using TBtool software. The conserved motifs of LcDof proteins were predicted using MEME (v5.4.1) (https://meme-suite.org/meme/tools/meme, accessed on 21 February 2022) with the following settings: the discovery mode was classic, site distribution was zero or one occurrence per sequence, the background is 0-order background model, the maximum number of different motifs: 20, minimum motif width: 6, and maximum motif width: 50, and displayed using the TBtool software.
4.3. Plant Materials and Genetic Transformation of Liriodendron Hybrid
Liriodendron hybrid seedlings generated through somatic embryogenesis (SE) were used as the starting material throughout this study []. Before any experiments were performed, plantlets were taken out of the culture medium vessel and acclimatized in a greenhouse for 2 weeks (22 °C, long day photoperiod of 16 h light/8 h dark and 75% relative humidity). For various abiotic stress treatments, plants were transferred to a growth chamber (long time photoperiod of 16 h light/8 h dark and 75% relative humidity): to simulate cold or heat or drought stress, plantlets were subjected to a 4 °C or 15% PEG6000 treatment, respectively, for 1 h, 6 h, 12 h and 1 d in the growth chamber [].
The full-length coding sequence (CDSs) of LhDof6 was amplified from Liriodendron hybrid by PCR, and cloned into the pBI121 vector with overexpression of LhDof6 (hereinafter referred to as LhDof6-OE) under the control of CaMV 35S promoter and ScaI and XbalI, respectively. After the vector was constructed, the vector was transformed into Agrobacterium receptive cell EHA105, and the monoclonal colony was selected to verify its correctness before transfection []. Positive callus was obtained through genomycin screening, and mutants identified via PCR and sequencing were used for subsequent experiments.
4.4. RNA Extraction and qRT-PCR Analysis
Based on the transcriptome data of Liriodendron hybrid under different abiotic stresses, a heatmap of LhDof6 gene expression was generated using Tbtools. The transcriptome data used in this study has been archived and can also be obtained on the NCBI website, cold and heat stress accession numbers were PRJNA679089 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA679089/, accessed on 19 June 2022), and drought stress accession number was PRJNA679101 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA679101/, accessed on 19 June 2022).
RNA degradation and contamination were monitored on 1% agarose gel. RNA purity was detected using the NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA). Using the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA), The RNA Nano 6000 Assay Kit (Agilent Technologies, CA, USA) assesses RNA integrity and synthesizes cDNA using the HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China), using the extracted RNA as a template. Using primer3 website (https://www.yeastgenome.org/primer3, accessed on 30 June 2022) design quantitative expression LcDofs primers, RT-qPCR using Roche Lightcyler® 480 instrument II, use 2× AceQ® qPCR SYBR® Green Master Mix (Without ROX) (Vazyme, Nanjing, China). The PCR mixture consists of 2× AceQ® qPCR SYBR® Green Master mix (Without ROX) 10 μL, each primer 0.4 μL, cDNA template 1 μL (10 ng/μL), 8.2 μL ddH2O added, the final volume was 20 μL. The internal reference gene was L. chinense 18S gene. The reaction process is as follows: 95 °C—10 min, 95 °C—10 s, 57 °C—30 s, 40 cycles. All reactions were performed in 96-well plates. Biological replicates were performed for each reaction, as well as three technical replicates. All data generated by real-time PCR amplification were analyzed by 2−ΔΔCT [].
4.5. Subcellular Localization
To verify the subcellular localisation of LhDof4 and LhDof6, we obtained the LcDof4 and LcDof6 target fragment by PCR using the LhDof4 and LhDof6 sequence as the reference sequence and the cDNA of Liriodendron hybrid seedling as the template. The plasmid pJIT166-GFP was digested with XbaI and BamHI enzymes, and the linear vector fragment was ligated with the target gene fragment to construct the p35S:LhDof4/6-GFP fusion expression vector. Plasmids were extracted using an endotoxin-free plasmid extraction kit (TIANGEN, Beijing, China). The callus of Liriodendron hybrid cultured for 20 days was used to prepare protoplasts, and protoplasts were slowly and gently dissolved into a solution (10 mL) containing 0.5 M mannitol, 20 mM MES, pH 5.7, 20 mM KCl, 0.1% (w/v) bovine serum albumin, 10 mM CaCl2 and digested at 28 °C under dark conditions for 3 h [], the protoplasts were transformed by PEG6000, pipetted into a 6-well cell culture plate, and cultured at 23 °C under dark for 16~48 h [], then the fluorescence effect of protoplasts was observed by ZEISS LSM 800 fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
4.6. p35S: LhDof6 Positive Seedlings and Cold Stress Tolerance Assay
LhDof6-OE seedlings generated through SE. Each group selected 28 uniformly growing and healthy 6-month-old Liriodendron hybrid seedlings for a freeze tolerance test. The seedlings were exposed to −20 °C for 20 min and then transferred to a greenhouse environment (26 °C with 16 h of light/8 h of darkness) for observation. After the treatment, the plant leaves were washed with distilled water, dried with paper towels, weighed, and then flash-frozen in liquid nitrogen before being stored at −80 °C for future use. The assay kits for hydrogen peroxide (H2O2), lipid peroxidation malondialdehyde (MDA), proline (PRO), and superoxide dismutase activity (SOD) were purchased from Nanjing Jiancheng Bioengineering Institute, Jiangsu provenience, China.
4.7. Data Analysis
Data were plotted using Prism 8.0 software, and the significance of differences between samples was assessed using t-tests and one-way ANOVA. * on the bar chart indicates that the difference between the two control groups reached a significant level, that is, p < 0.05; ** means p < 0.01, *** means p < 0.001, **** means the difference between the two control groups reached a very significant level, that is, p < 0.0001. The content of malondialdehyde, superoxide dismutase activity, proline content and hydrogen peroxide in plants before and after freezing stress were significantly analyzed.
5. Conclusions
Our research findings indicate that the L. chinense genome harbors a total of 17 Dof genes. The physicochemical properties of the proteins encoded by these genes exhibit considerable variation. Specifically, the protein lengths span from 160 to 635 amino acids, while their molecular weights range between 17.01 and 71.60 kDa. The majority of LcDof proteins are categorized as unstable, rendering them susceptible to alterations caused by external environmental factors. The DOF domains within the LcDof gene family exhibit complete conservation. The six clades are characterized by distinct motif structures and gene architectures, implying potential functional similarities among LcDof genes within each clade. Phylogenetic analysis revealed that the Dof genes can be classified into eight distinct subfamilies, with the notable absence of LcDof genes in the C3 subfamily. Collinearity analysis suggests that segmental duplications are primarily responsible for the expansion of the LcDof gene family, which underwent five such events along with one tandem duplication event. Analysis of promoter regions also indicates a rich presence of cis-acting elements. The elements are classified into four categories: growth and development-related elements, light-responsive elements, abiotic stress-responsive elements, and plant hormone-responsive elements. Moreover, there is a relatively abundant presence of abiotic stress-responsive elements and plant hormone-responsive elements. The transcriptome data of Liriodendron hybrid under various abiotic stresses reveals a high expression level of Dof genes from the D1 subfamily, particularly highlighting the significant responses of LcDof4 and LcDof6. This finding was further confirmed through RT-qPCR analysis, suggesting that LcDof4 and LcDof6 may play a crucial role in positively regulating the response to cold stress. We successfully cloned the LcDof4 and LcDof6 genes, and observed their expression in the cell nuclei of protoplasts derived from Liriodendron hybrid callus. Furthermore, we conducted overexpression experiments with LhDof6 in Liriodendron hybrid, which resulted in a significant increase in the survival rate of six-month-old seedlings under −20 °C conditions. Furthermore, a series of physiological measurements have confirmed that the overexpression of LhDof6 significantly enhances the plant’s cold tolerance. In summary, we have successfully identified and analyzed the Dof gene family in L. chinense for the first time, screened for genes associated with cold tolerance, and conducted an initial functional analysis on the role of LhDof6 in enhancing cold tolerance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13142009/s1, Figure S1: Construction and verification of LhDof6-OE, 35S-LhDof4-GFP and 35S-LhDof6-GFP expression vector; Table S1: Primer Sequence.
Author Contributions
Conceptualization, J.C. and Z.H.; Data Curation, Z.H.; Formal Analysis, B.L. and P.L.; Funding Acquisition, Z.H.; Investigation, B.L. and P.L.; Methodology, L.L., Y.L., R.Z. and X.Z.; Software, B.L. and L.T.; Supervision, Z.H. and J.C.; Visualization, B.L and P.L.; Writing—Original Draft, B.L. and P.L.; Writing—Review and Editing, J.C. and Z.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Key R&D Program of China during the 14th Five-year Plan Period (2021YFD2200102), the Natural Science Foundation of China (32071784) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Data Availability Statement
The data and results are available to every reader upon reasonable request.
Acknowledgments
I would like to express my heartfelt gratitude to my laboratory members for their unwavering support and invaluable contributions. A special thanks to Ziming Lian and Yan Pan for your insightful guidance and dedication, and to Siqi Liu for your tireless efforts and innovative ideas. Your collaboration and hard work have been instrumental in the success of our projects. Thank you all for your commitment and teamwork.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zandalinas, S.I.; Mittler, R. Plant responses to multifactorial stress combination. New Phytol. 2022, 234, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Hassan, M.J.; Qi, H.; Cheng, B.; Hussain, S.; Peng, Y.; Liu, W.; Feng, G.; Zhao, J.; Li, Z. Enhanced adaptability to limited water supply regulated by diethyl aminoethyl hexanoate (DA-6) associated with lipidomic reprogramming in two white clover genotypes. Front. Plant Sci. 2022, 13, 879331–879350. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Ahuja, I.; de Vos, R.C.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Diaz, I.; Martinez, M.; Isabel-LaMoneda, I.; Rubio-Somoza, I.; Carbonero, P. The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J. 2005, 42, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, T.; Sun, X.; Wang, Y.; Du, C.; Zhu, Z.; Gichuki, D.K.; Wang, Q.; Li, S.; Xin, H. Overexpression of VaWRKY12, a transcription factor from Vitis amurensis with increased nuclear localization under low temperature, enhances cold tolerance of plants. Plant Mol. Biol. 2019, 100, 95–110. [Google Scholar] [CrossRef]
- Yanagisawa, S. A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 1995, 23, 3403–3410. [Google Scholar] [CrossRef]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef]
- Krebs, J.; Mueller-Roeber, B.; Ruzicic, S. A novel bipartite nuclear localization signal with an atypically long linker in DOF transcription factors. J. Plant Physiol. 2010, 167, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ahmad, M.; Rim, Y.; Lucas, W.J.; Kim, J.Y. Evolutionary and molecular analysis of Dof transcription factors identified a conserved motif for intercellular protein trafficking. New Phytol. 2013, 198, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, Z.; Xie, L.; Li, X.; Gao, J. Multifaceted role of PheDof12-1 in the regulation of flowering time and abiotic stress responses in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 20, 424. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Ni, Z.; Yao, Y.; Nie, X.; Sun, Q. Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol. Biol. 2007, 63, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Santopolo, S.; Boccaccini, A.; Lorrai, R.; Ruta, V.; Capauto, D.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P. DOF AFFECTING GERMINATION 2 is a positive regulator of light-mediated seed germination and is repressed by DOF AFFECTING GERMINATION 1. BMC Plant Biol. 2015, 15, 72. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Rubio-Somoza, I.; Fuentes, R.; Lara, P.; Carbonero, P.; Díaz, I. The barley cystatin gene (Icy) is regulated by DOF transcription factors in aleurone cells upon germination. J. Exp. Bot. 2005, 56, 547–556. [Google Scholar] [CrossRef]
- Papi, M.; Sabatini, S.; Altamura, M.M.; Hennig, L.; Schäfer, E.; Costantino, P.; Vittorioso, P. Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 2002, 128, 411–417. [Google Scholar] [CrossRef]
- Park, D.H.; Lim, P.O.; Kim, J.S.; Cho, D.S.; Hong, S.H.; Nam, H.G. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 2003, 34, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Ewas, M.; Khames, E.; Ziaf, K.; Shahzad, R.; Nishawy, E.; Ali, F.; Subthain, H.; Amar, M.H.; Ayaad, M.; Ghaly, O.; et al. The Tomato DOF Daily Fluctuations 1, TDDF1 acts as flowering accelerator and protector against various stresses. Sci. Rep. 2017, 7, 10299. [Google Scholar] [CrossRef] [PubMed]
- Zang, D.; Wang, L.; Zhang, Y.; Zhao, H.; Wang, Y. ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol. Biol. 2017, 94, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Chromatin structure: A repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Pancaldi, V. Network models of chromatin structure. Curr. Opin. Genet. Dev. 2023, 80, 102051–102059. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cao, Y.; Wang, S. Point mutations with positive selection were a major force during the evolution of a receptor-kinase resistance gene family of rice. Plant Physiol. 2006, 140, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Habib, F.; Kulkarni, A.; Ratnaparkhi, G.S. Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge. Genetics 2014, 196, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef]
- Melcher, K.; Zhou, X.E.; Xu, H.E. Thirsty plants and beyond: Structural mechanisms of abscisic acid perception and signaling. Curr. Opin. Struc. Biol. 2010, 20, 722–729. [Google Scholar] [CrossRef]
- Lim, J.; Lim, C.W.; Lee, S.C. Core Components of Abscisic Acid Signaling and Their Post-translational Modification. Front. Plant Sci. 2022, 13, 895698–895710. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fernie, A.R.; Shinozaki, K.; Takahashi, F. Long-distance stress and developmental signals associated with abscisic acid signaling in environmental responses. Plant J. 2021, 105, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Renau-Morata, B.; Molina, R.V.; Carrillo, L.; Cebolla-Cornejo, J.; Sánchez-Perales, M.; Pollmann, S.; Domínguez-Figueroa, J.; Corrales, A.R.; Flexas, J.; Vicente-Carbajosa, J.; et al. Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front. Plant Sci. 2017, 8, 660–687. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004, 45, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.K.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; Creelman, R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, A.; Lister, R.; Bogdanovic, O. Evolution of DNA methylome diversity in eukaryotes. J. Mol. Biol. 2020, 432, 1687–1705. [Google Scholar] [CrossRef]
- Wray, G.A.; Hahn, M.W.; Abouheif, E.; Balhoff, J.P.; Pizer, M.; Rockman, M.V.; Romano, L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. [Google Scholar] [CrossRef]
- Jia, Y.Y.; Xu, Y.; Wang, B.; Guo, L.; Guo, M.; Che, X.; Ye, K. The tissue-specific chromatin accessibility landscape of Papaver somniferum. Front. Genet. 2023, 14, 1136736–1136743. [Google Scholar] [CrossRef]
- Aerts, N.; Pereira, M.M.; Van Wees, S. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef]
- Washio, K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003, 133, 850–863. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, M.F.; Zhang, W.P.; Chen, F.; Shen, S.H. A putative flowering-time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas. Plant Sci. 2011, 181, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.M.; Sun, H.M. DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 14, 1044918–1044930. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; de Montaigu, A.; Sanchez-Villarreal, A.; Takahashi, Y.; Ver Loren van Themaat, E.; Huettel, B.; Davis, S.J.; Coupland, G. The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 2015, 81, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Goralogia, G.S.; Liu, T.K.; Zhao, L.; Panipinto, P.M.; Groover, E.D.; Bains, Y.S.; Imaizumi, T. CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J. 2017, 92, 244–262. [Google Scholar] [CrossRef]
- Ramirez, G.L.; Shi, L.; Bergonzi, S.B.; Oortwijn, M.; Franco-Zorrilla, J.M.; Solano-Tavira, R.; Visser, R.G.F.; Abelenda, J.A.; Bachem, C.W.B. Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 2021, 105, 855–869. [Google Scholar] [CrossRef] [PubMed]
- Li, M.P.; Wang, D.; Long, X.F.; Hao, Z.D.; Lu, Y.; Zhou, Y.W.; Peng, Y.; Cheng, T.L.; Shi, J.S.; Chen, J.H. Agrobacterium-mediated genetic transformation of embryogenic callus in a Liriodendron Hybrid (L. Chinense × L. Tulipifera). Front. Plant Sci. 2022, 13, 802128–802140. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhu, S.; Xu, L.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Hao, Z.; Lu, Y.; Yang, L.; et al. Genome-wide identification of the Liriodendron chinense WRKY gene family and its diverse roles in response to multiple abiotic stress. BMC Plant Biol. 2022, 22, 25. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huo, A.; Chen, Z.; Wang, P.; Yang, L.; Wang, G.; Wang, D.; Liao, S.; Cheng, T.; Chen, J.; Shi, J. Establishment of transient gene expression systems in protoplasts from Liriodendron hybrid mesophyll cells. PLoS ONE 2017, 12, e172475–e172483. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).