Epigenetics Regulation in Responses to Abiotic Factors in Plant Species: A Systematic Review
Abstract
:1. Introduction
2. Results
2.1. Number of Articles and Species Studied
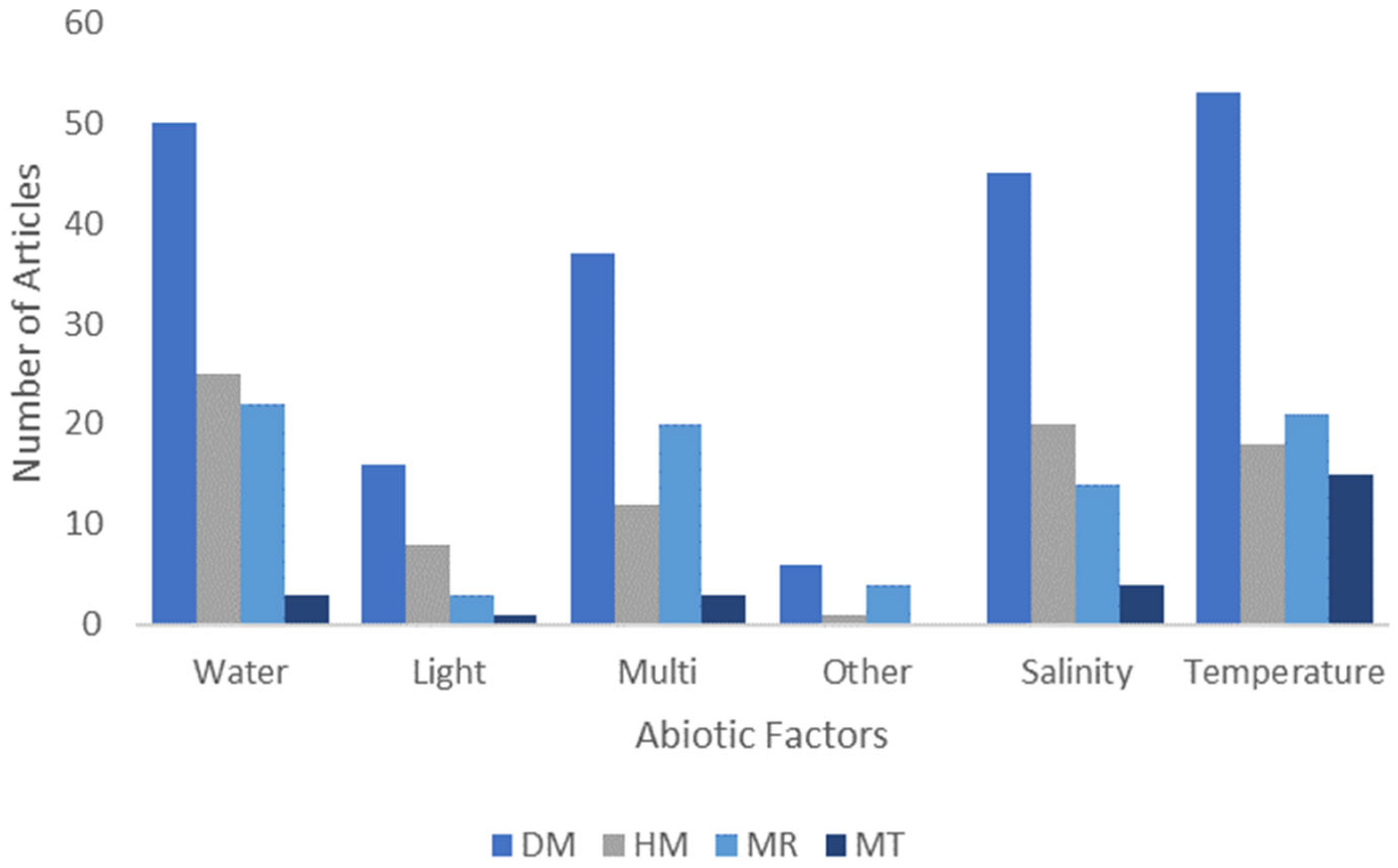
2.2. Epigenetic Technique and Abiotic Factors Best Studied
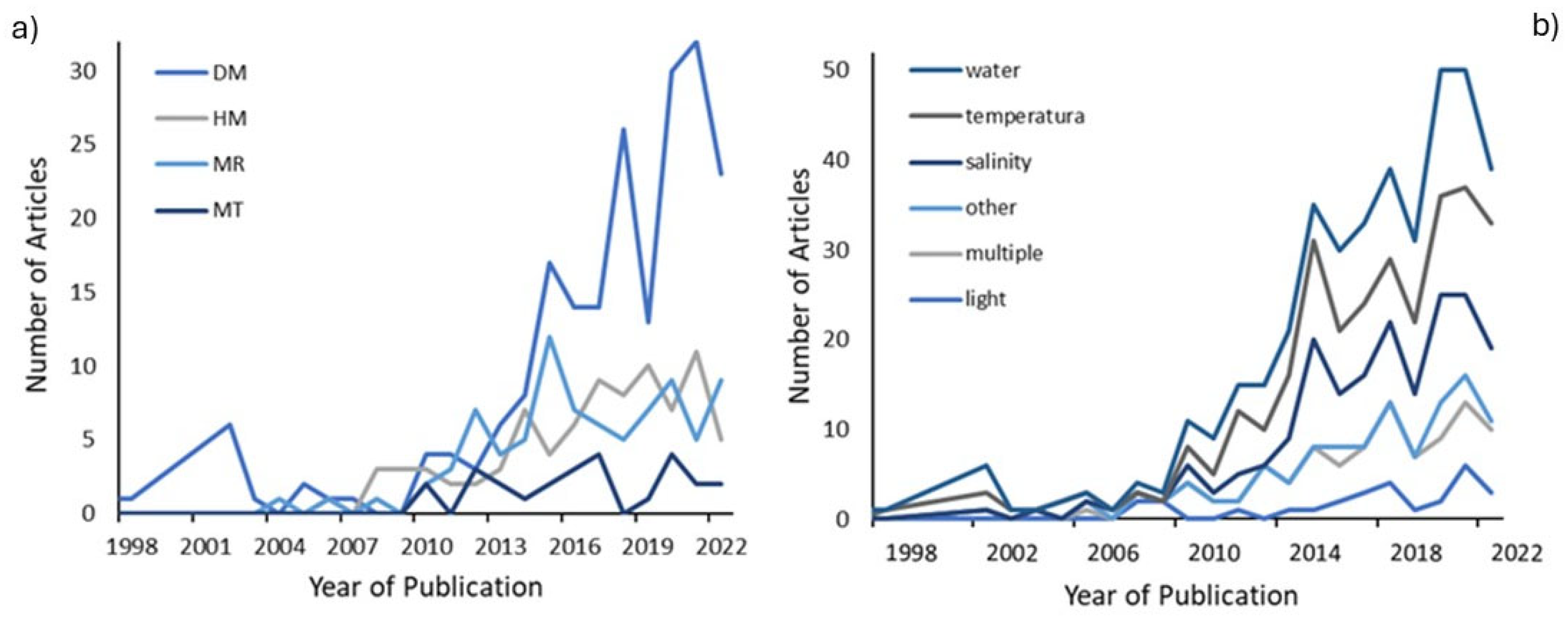
2.3. Time Trend of the Publications
2.4. Geographical Distribution of the Studies
2.5. Overlapping Techniques, Factors and Species
3. Discussion
3.1. The Influence of Epigenetic Responses to Abiotic Stress on Plants
3.2. Most of the Studied Species Are of Economic Interest
3.3. An Increase in Research in the Last Decade
3.4. Leading Country of Publication
3.5. DNA Methylation Is the Most Studied Epigenetic
3.6. Temperature and Water Proportionately Studied
3.7. Research Deficits in the Tropics and in Native Species
4. Materials and Methods
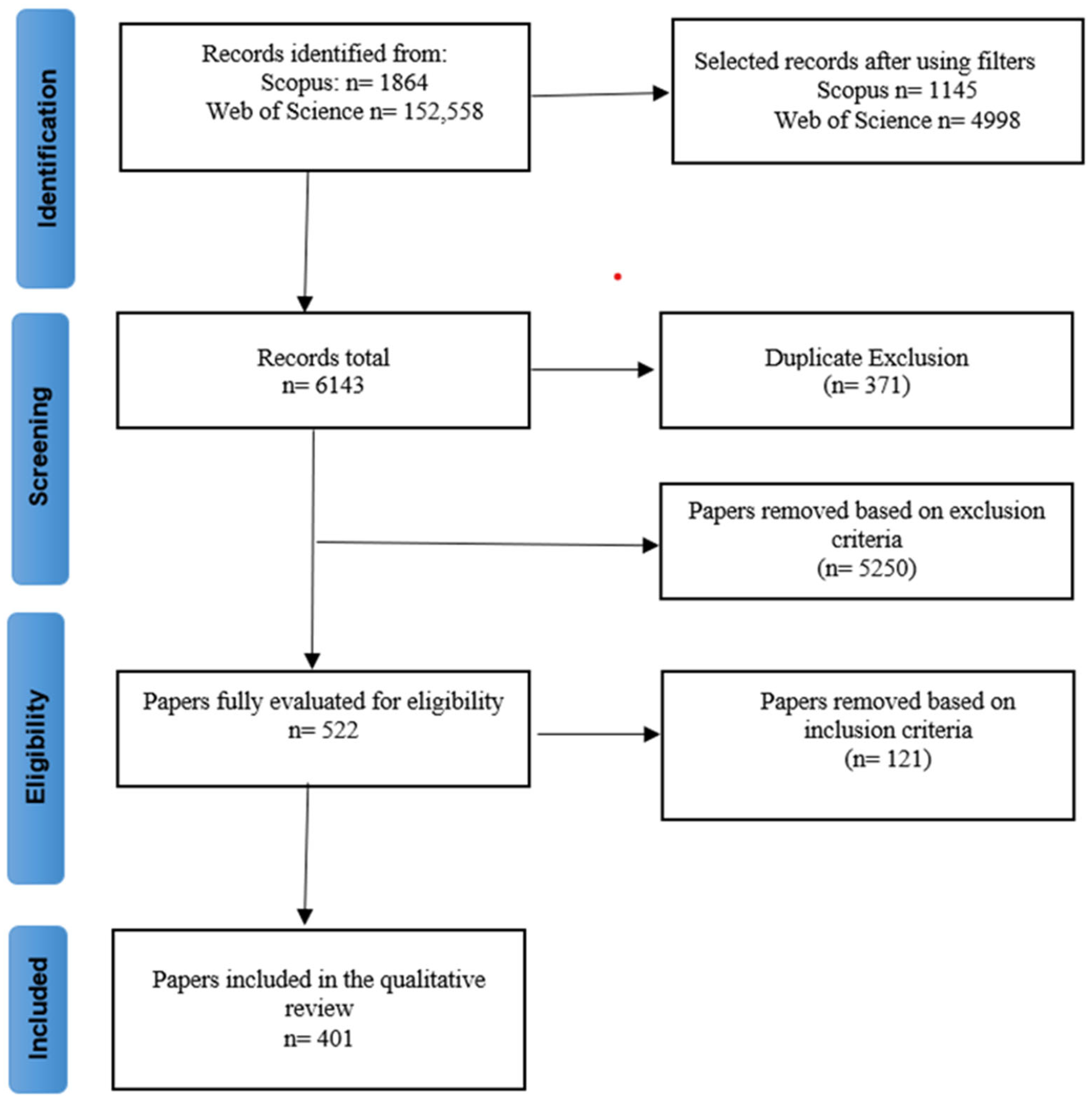
4.1. Search and Database
| (epigenetic* OR “DNA methylation” OR “epigenetic inheritance” OR “transgenerational epigenetic” OR “epigenetic regulation*” OR “ecological epigenetic*” OR “heritable epigenetic” OR “epigenetic change*” OR “epigenetic variation” OR “histone modification” OR “MicroRNA”) AND (plant* OR tree*) AND (phenotypic* OR “phenotypic plasticity” OR “environmental change*” OR “morphological variation*” OR “environmental stress*”)) |
4.2. Categorization and Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waddington, C.H. Towards a Theoretical Biology. Nature 1968, 218, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, A.P.; Guschin, D. Review: Chromatin Structural Features and Targets That Regulate Transcription. J. Struct. Biol. 2000, 129, 102–122. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Fei, Z.; Chen, Y.R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-Base Resolution Methylomes of Tomato Fruit Development Reveal Epigenome Modifications Associated with Ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Robert, A. Martienssen* and Vincent Colo DNA Methylation and Epigenetic Inheritancein Plants and Filamentous Fungi. Science 2001, 293, 1070–1074. [Google Scholar]
- Rossella, F.; Polledri, E.; Bollati, V.; Baccarelli, A.; Fustinoni, S. Development and Validation of a Gas Chromatography/Mass Spectrometry Method for the Assessment of Genomic DNA Methylation. Rapid Commun. Mass. Spectrom. 2009, 23, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Lennartsson, A. Dynamics and Function of DNA Methylation During Development. In Chromatin Regulation and Dynamics; Elsevier: Amsterdam, The Netherlands, 2017; pp. 65–94. [Google Scholar]
- Millar, D.S.; Holliday, R.; Grigg, G.W. Five Not Four: History and Significance of the Fifth Base. In The Epigenome; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005; pp. 1–20. [Google Scholar]
- Ronemus, M.J.; Galbiati, M.; Ticknor, C.; Chen, J.; Dellaporta, S.L. Demethylation-Induced Developmental Pleiotropy in Arabidopsis. Science 1996, 273, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jacobsen, S.E. Role of the Arabidopsis DRM Methyltransferases in De Novo DNA Methylation and Gene Silencing. Curr. Biol. 2002, 12, 1138–1144. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Hsieh, P.-H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Agarwal, G.; Kudapa, H.; Ramalingam, A.; Choudhary, D.; Sinha, P.; Garg, V.; Singh, V.K.; Patil, G.B.; Pandey, M.K.; Nguyen, H.T.; et al. Epigenetics and Epigenomics: Underlying Mechanisms, Relevance, and Implications in Crop Improvement. Funct. Integr. Genom. 2020, 20, 739–761. [Google Scholar] [CrossRef]
- Yan, C.; Boyd, D.D. Histone H3 Acetylation and H3 K4 Methylation Define Distinct Chromatin Regions Permissive for Transgene Expression. Mol. Cell. Biol. 2006, 26, 6357–6371. [Google Scholar] [CrossRef]
- Liu, C.; Xin, Y.; Xu, L.; Cai, Z.; Xue, Y.; Liu, Y.; Xie, D.; Liu, Y.; Qi, Y. Arabidopsis ARGONAUTE 1 Binds Chromatin to Promote Gene Transcription in Response to Hormones and Stresses. Dev. Cell 2018, 44, 348–361.e7. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.F. Non-Coding RNAs, Epigenetics and Complexity. Gene 2008, 410, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-Coding RNAs as Regulators in Epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Boquete, M.T.; Muyle, A.; Alonso, C. Plant Epigenetics: Phenotypic and Functional Diversity beyond the DNA Sequence. Am. J. Bot. 2021, 108, 553–558. [Google Scholar] [CrossRef]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In Response to Abiotic Stress, Dna Methylation Confers Epigenetic Changes in Plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef] [PubMed]
- Miryeganeh, M. Plants’ Epigenetic Mechanisms and Abiotic Stress. Genes 2021, 12, 1106. [Google Scholar] [CrossRef]
- García-García, I.; Méndez-Cea, B.; Martín-Gálvez, D.; Seco, J.I.; Gallego, F.J.; Linares, J.C. Challenges and Perspectives in the Epigenetics of Climate Change-Induced Forests Decline. Front. Plant Sci. 2022, 12, 797958. [Google Scholar] [CrossRef]
- Saeed, F.; Chaudhry, U.K.; Bakhsh, A.; Raza, A.; Saeed, Y.; Bohra, A.; Varshney, R.K. Moving Beyond DNA Sequence to Improve Plant Stress Responses. Front. Genet. 2022, 13, 874648. [Google Scholar] [CrossRef]
- Gavrilets, S. The Maynard Smith Model of Sympatric Speciation. J. Theor. Biol. 2006, 239, 172–182. [Google Scholar] [CrossRef]
- Meyerowitz, E.M. Arabidopsis thaliana. Annu. Rev. Genet. 1987, 21, 93–111. [Google Scholar] [CrossRef] [PubMed]
- FAO-ESS. Estatísticas de Culturas—Conceitos, Definições e Classificações; Divisão de Estatísticas Da FAO (ESS): 2021. Available online: https://www.fao.org/about/who-we-are/departments/statistics-division/en (accessed on 15 April 2022).
- World Rice Statistics 2021. International Rice Research Institute. Available online: https://www.irri.org/rice-information-gateway (accessed on 15 April 2022).
- Erenstein, O.; Jaleta, M.; Sonder, K.; Mottaleb, K.; Prasanna, B.M. Global Maize Production, Consumption and Trade: Trends and R&D Implications. Food Secur. 2022, 14, 1295–1319. [Google Scholar]
- Wang, Y.; Li, J. The Plant Architecture of Rice (Oryza sativa). Plant Mol. Biol. 2005, 59, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Boškovi’c, A.B.; Rando, O.J. Transgenerational Epigenetic Inheritance. Annu. Rev. Genet. 2018, 52, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Morgan, K.R.; Petryk, N.; Groth, A. Chromatin Replication and Epigenetic Cell Memory. Nat. Cell Biol. 2020, 22, 361–371. [Google Scholar] [CrossRef]
- Ashe, A.; Colot, V.; Oldroyd, B.P. How Does Epigenetics Influence the Course of Evolution? Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200111. [Google Scholar] [CrossRef]
- Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. A Role for Epigenetic Regulation in the Adaptation and Stress Responses of Non-Model Plants. Front. Plant Sci. 2019, 10, 413504. [Google Scholar] [CrossRef]
- Geange, S.R.; Arnold, P.A.; Catling, A.A.; Coast, O.; Cook, A.M.; Gowland, K.M.; Leigh, A.; Notarnicola, R.F.; Posch, B.C.; Venn, S.E.; et al. The Thermal Tolerance of Photosynthetic Tissues: A Global Systematic Review and Agenda for Future Research. New Phytol. 2021, 229, 2497–2513. [Google Scholar] [CrossRef]
- Lazarev, G.I.; Krasova, E.V. Research and Development in China: Scope and Specifics of Innvovation Process. Amazon Investig. 2018, 7, 73–83. [Google Scholar]
- Jenkins, M.F.; White, E.P.; Hurlbert, A.H. The Proportion of Core Species in a Community Varies with Spatial Scale and Environmental Heterogeneity. PeerJ 2018, 6, e6019. [Google Scholar] [CrossRef]
- Varotto, S.; Tani, E.; Abraham, E.; Krugman, T.; Kapazoglou, A.; Melzer, R.; Radanović, A.; Miladinović, D. Epigenetics: Possible Applications in Climate-Smart Crop Breeding. J. Exp. Bot. 2020, 71, 5223–5236. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Birdsey, R.A.; Phillips, O.L.; Jackson, R.B. The Structure, Distribution, and Biomass of the World’s Forests. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 593–622. [Google Scholar] [CrossRef]
- Rezende, C.L.; Scarano, F.R.; Assad, E.D.; Joly, C.A.; Metzger, J.P.; Strassburg, B.B.N.; Tabarelli, M.; Fonseca, G.A.; Mittermeier, R.A. From Hotspot to Hopespot: An Opportunity for the Brazilian Atlantic Forest. Perspect. Ecol. Conserv. 2018, 16, 208–214. [Google Scholar] [CrossRef]
- Gaston, K.J. Global Patterns in Biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How Much Is Left, and How Is the Remaining Forest Distributed? Implications for Conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mohapatra, T. Dynamics of DNA Methylation and Its Functions in Plant Growth and Development. Front. Plant Sci. 2021, 12, 596236. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Ecker, J.R. Diversity and Dynamics of DNA Methylation: Epigenomic Resources and Tools for Crop Breeding. Breed. Sci. 2019, 69, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Ali, K.; Yan, K.; Fiaz, S.; Dormatey, R.; Bi, Z.; Bai, J. Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review. Plants 2021, 10, 1226. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Ashapkin, V.V. DNA Methylation in Higher Plants: Past, Present and Future. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 360–368. [Google Scholar] [CrossRef]
- Feng, S.; Rubbi, L.; Jacobsen, S.E.; Pellegrini, M. Determining DNA Methylation Profiles Using Sequencing. Methods Mol. Biol. 2011, 733, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Baccarelli, A. Environmental Epigenetics. Heredity 2010, 105, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, A.F.; Rocha-Santos, L.; Benchimol, M.; Mielke, M.S. Habitat Loss and Canopy Openness Mediate Leaf Trait Plasticity of an Endangered Palm in the Brazilian Atlantic Forest. Oecologia 2021, 196, 619–631. [Google Scholar] [CrossRef]
- O’sullivan, O.S.; Heskel, M.A.; Reich, P.B.; Tjoelker, M.G.; Weerasinghe, L.K.; Penillard, A.; Zhu, L.; Egerton, J.J.G.; Bloomfield, K.J.; Creek, D.; et al. Thermal Limits of Leaf Metabolism across Biomes. Glob. Chang. Biol. 2017, 23, 209–223. [Google Scholar] [CrossRef] [PubMed]
- McCaw, B.A.; Stevenson, T.J.; Lancaster, L.T. Epigenetic Responses to Temperature and Climate. Integr. Comp. Biol. 2020, 60, 1469–1480. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, S.; Derkacheva, M.; Ramström, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 Domain Protein MSI1 Functions in a Histone Deacetylase Complex to Fine-Tune Abscisic Acid Signaling. Plant Cell 2016, 28, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xie, W.; Xu, D.; Miki, D.; Tang, K.; Huang, C.F.; Zhu, J.K. DNA Demethylase ROS1 Negatively Regulates the Imprinting of DOGL4 and Seed Dormancy in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E9962–E9970. [Google Scholar] [CrossRef]
- Hutchison, C.; Gravel, D.; Guichard, F.; Potvin, C. Effect of Diversity on Growth, Mortality, and Loss of Resilience to Extreme Climate Events in a Tropical Planted Forest Experiment. Sci. Rep. 2018, 8, 15443. [Google Scholar] [CrossRef]
- Álvarez-Yépiz, J.C. Restoration Ecology in the Anthropocene: Learning from Responses of Tropical Forests to Extreme Disturbance Events. Restor. Ecol. 2020, 28, 271–276. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Plant Development. In Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Piacentini, L.; Fanti, L.; Specchia, V.; Bozzetti, M.P.; Berloco, M.; Palumbo, G.; Pimpinelli, S. Transposons, Environmental Changes, and Heritable Induced Phenotypic Variability. Chromosoma 2014, 123, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.E. Plant Breeding: Past, Present and Future. Euphytica 2017, 213, 60. [Google Scholar] [CrossRef]
- Sobral, M.; Sampedro, L. Phenotypic, Epigenetic, and Fitness Diversity within Plant Genotypes. Trends Plant Sci. 2022, 27, 843–846. [Google Scholar] [CrossRef] [PubMed]

| Species | Family | Number of Studies |
|---|---|---|
| Arabidopsis thaliana | Brassicaceae | 90 |
| Oryza sativa | Poaceae | 38 |
| Zea mays | Poaceae | 26 |
| Hordeum vulgare | Poaceae | 11 |
| Triticum aestivum | Poaceae | 11 |
| Glycine max | Leguminosae | 9 |
| Gossypium hirsutum | Malvaceae | 8 |
| Brassica napus | Brassicaceae | 7 |
| Nicotiana tabacum | Solanaceae | 6 |
| Solanum lycopersicum | Solanaceae | 6 |
| Country | Number of Articles |
|---|---|
| China | 158 |
| USA | 30 |
| India | 21 |
| Germany | 17 |
| Japan | 16 |
| Italy | 15 |
| Spain | 14 |
| Poland | 10 |
| Canada | 9 |
| France | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa, G.S.; Cerqueira, A.F.; de Brito, C.R.; Mielke, M.S.; Gaiotto, F.A. Epigenetics Regulation in Responses to Abiotic Factors in Plant Species: A Systematic Review. Plants 2024, 13, 2082. https://doi.org/10.3390/plants13152082
da Costa GS, Cerqueira AF, de Brito CR, Mielke MS, Gaiotto FA. Epigenetics Regulation in Responses to Abiotic Factors in Plant Species: A Systematic Review. Plants. 2024; 13(15):2082. https://doi.org/10.3390/plants13152082
Chicago/Turabian Styleda Costa, Geane Santos, Amanda Freitas Cerqueira, Carolina Reis de Brito, Marcelo Schramm Mielke, and Fernanda Amato Gaiotto. 2024. "Epigenetics Regulation in Responses to Abiotic Factors in Plant Species: A Systematic Review" Plants 13, no. 15: 2082. https://doi.org/10.3390/plants13152082
APA Styleda Costa, G. S., Cerqueira, A. F., de Brito, C. R., Mielke, M. S., & Gaiotto, F. A. (2024). Epigenetics Regulation in Responses to Abiotic Factors in Plant Species: A Systematic Review. Plants, 13(15), 2082. https://doi.org/10.3390/plants13152082







