Comparison of Root Transcriptomes against Clubroot Disease Pathogens in a Resistant Chinese Cabbage Cultivar (Brassica rapa cv. ‘Akimeki’)
Abstract
1. Introduction
2. Results
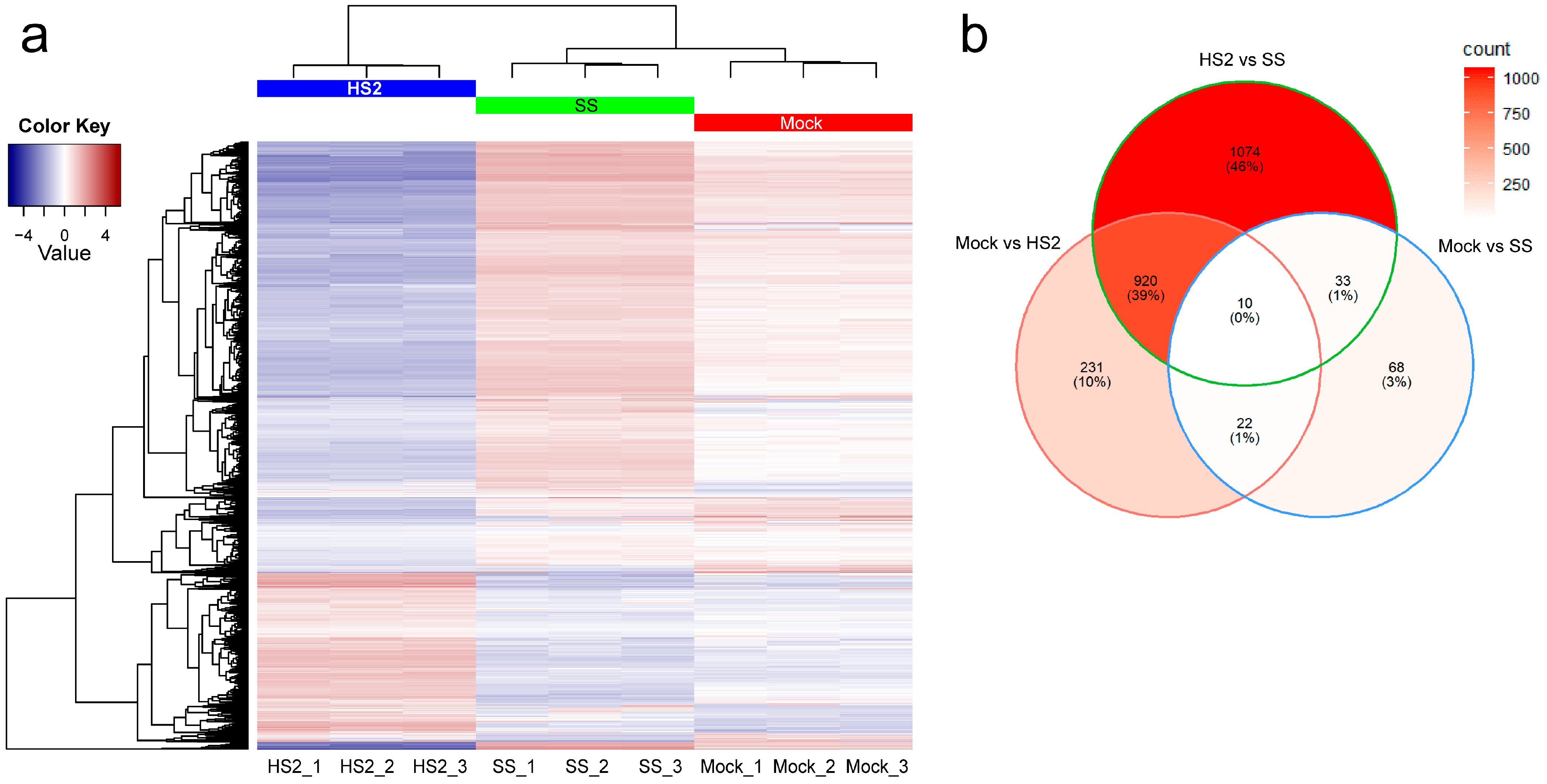
2.1. P. brassicae Infection and RNA Sequencing
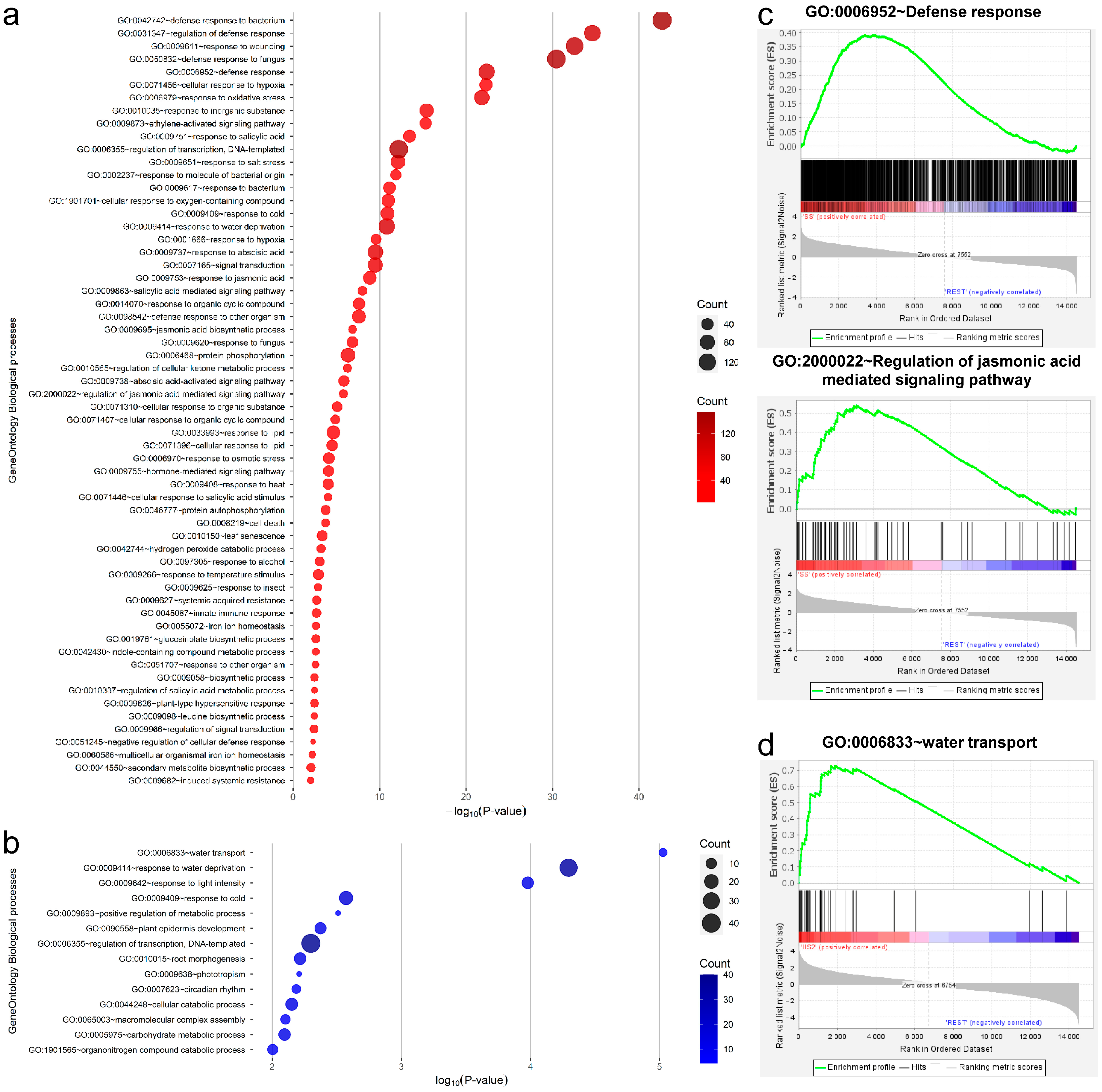
2.2. Functional Enrichment Analyses of Differentially Expressed Genes
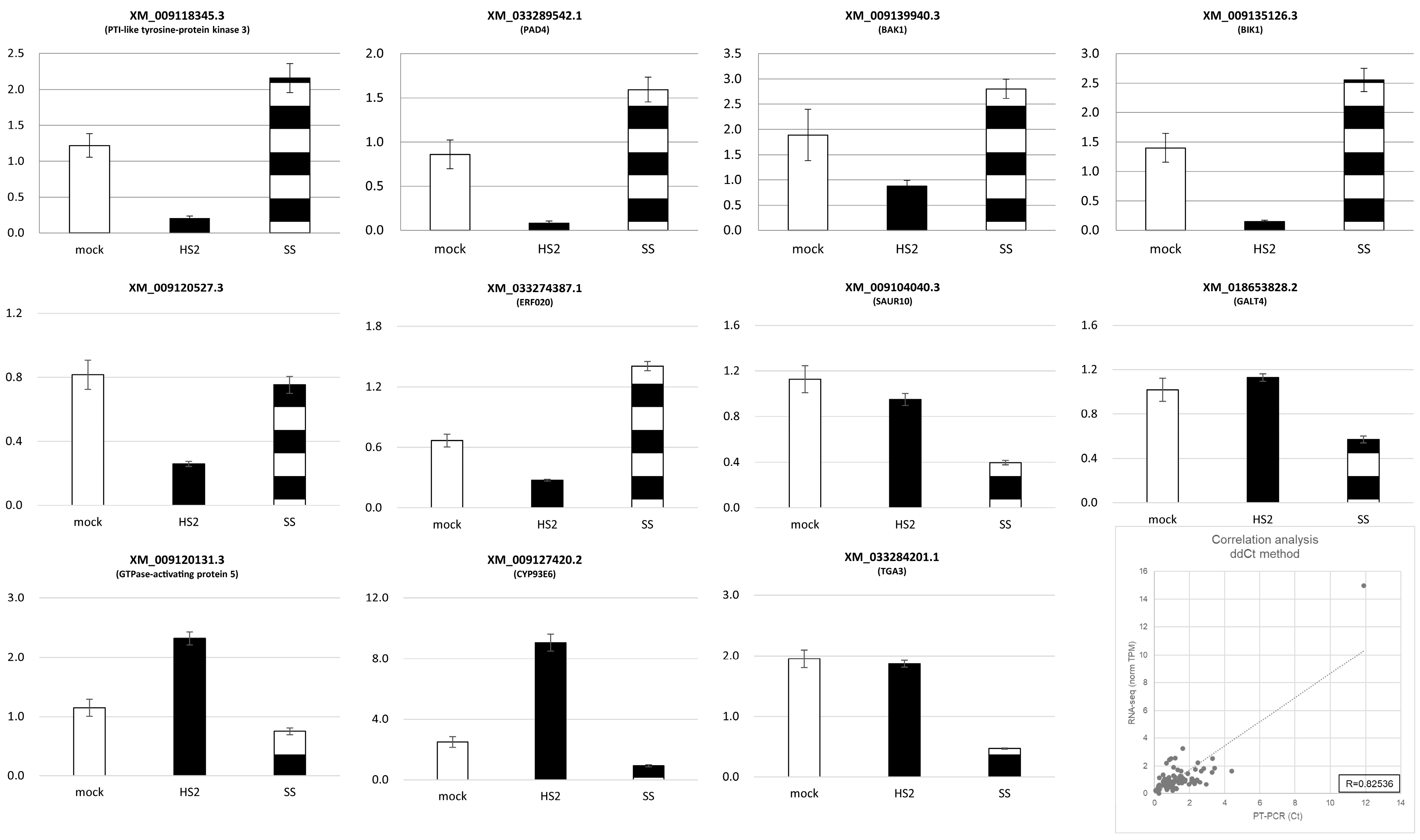
2.3. Quantitative RT-PCR Validation
3. Discussion
4. Materials and Methods
4.1. Plant Growth and P. brassicae Field Isolates
4.2. Plant Inoculation
4.3. RNA Isolation and Library Construction
4.4. RNA Sequencing and Analysis
4.5. Functional Annotation Analysis of DEGs
4.6. Correlation Analysis of Quantitative PCR and NGS Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dixon, G.R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Howard, R.J.; Strelkov, S.E.; Harding, M.W. Clubroot of cruciferous crops-new perspectives on an old disease. Can. J. Plant Pathol. 2010, 32, 43–57. [Google Scholar] [CrossRef]
- Ingram, D.S.; Tommerup, I.C. The life history of Plasmodiophora brassicae Woron. Proc. R. Soc. Lond. B 1972, 180, 103–112. [Google Scholar]
- McDonald, M.R.; Sharma, K.; Gossen, B.D.; Deora, A.; Feng, J.; Hwang, S.F. The role of primary and secondary infection in host response to Plasmodiophora brassicae. Phytopathology 2014, 104, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, K.; Asano, T. Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203–211. [Google Scholar] [CrossRef]
- Chai, A.L.; Xie, X.W.; Shi, Y.X.; Li, B.J. Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 2014, 36, 142–153. [Google Scholar] [CrossRef]
- Kim, H.; Jo, E.J.; Choi, Y.H.; Jang, K.S.; Choi, G.J. Pathotype Classification of Plasmodiophora brassicae Isolates Using Clubroot-Resistant Cultivars of Chinese Cabbage. Plant Pathol. J. 2016, 32, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Lahlali, R.; Hwang, S.F.; Pageau, D.; Hynes, R.K. Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Can. J. Plant Pathol. 2014, 36, 99–112. [Google Scholar] [CrossRef]
- Kim, M.-J.; Shim, C.-K.; Kim, Y.-K.; Hong, S.-J.; Park, J.-H.; Han, E.-J.; Lee, M.-H.; Jee, H.-J. Screening of Resistance Cultivar to Clubroot Caused by Plasmodiophora brassicae for Organic Cultivation of Chinese Cabbage. Res. Plant Dis. 2012, 18, 123–128. [Google Scholar] [CrossRef][Green Version]
- Zamani-Noor, N.; Krohne, I.; Koopmann, B. Greenhouse Evaluation of Clubroot Resistant-Brassica napus cv. Mendel and Its Efficacy Concerning Virulence and Soil Inoculum Levels of Plasmodiophora brassicae. Pathogens 2021, 10, 151. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Zhang, Y.; Dai, Z.; He, Z.; Qiu, Y.; Alharbi, S.A.; Wei, F.; Wei, L.; Ahmed, W.; et al. Pre-soil fumigation with ammonium bicarbonate and lime modulates the rhizosphere microbiome to mitigate clubroot disease in Chinese cabbage. Front. Microbiol. 2024, 15, 1376579. [Google Scholar] [CrossRef]
- Pang, W.; Fu, P.; Li, X.; Zhan, Z.; Yu, S.; Piao, Z. Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front Plant Sci. 2018, 9, 653. [Google Scholar] [CrossRef] [PubMed]
- Gravot, A.; Liégard, B.; Quadrana, L.; Veillet, F.; Aigu, Y.; Bargain, T.; Benejam, J.; Laragon, C.; Lemoine, J.; Colot, V.; et al. Two adjacent NLR genes conferring quantitative resistance to clubroot disease in Arabidopsis are regulated by a stably inherited epiallelic variation. Plant Commun. 2024, 5, 100824. [Google Scholar] [CrossRef]
- Paul, P.; Chhapekar, S.S.; Rameneni, J.J.; Oh, S.H.; Dhandapani, V.; Subburaj, S.; Shin, S.Y.; Ramchiary, N.; Shin, C.; Choi, S.R.; et al. MiR1885 Regulates Disease Tolerance Genes in Brassica rapa during Early Infection with Plasmodiophora brassicae. Int. J. Mol. Sci. 2021, 22, 9433. [Google Scholar] [CrossRef]
- Zou, Y.; Wang, S.; Zhou, Y.; Bai, J.; Huang, G.; Liu, X.; Zhang, Y.; Tang, D.; Lu, D. Transcriptional Regulation of the Immune Receptor FLS2 Controls the Ontogeny of Plant Innate Immunity. Plant Cell 2018, 30, 2779–2794. [Google Scholar] [CrossRef]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI Continuum in Phytophthora-Plant Interactions. Front Plant Sci. 2020, 11, 2030. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease Resistance Mechanisms in Plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef]
- Zhan, Z.; Nwafor, C.C.; Hou, Z.; Gong, J.; Zhu, B.; Jiang, Y.; Zhou, Y.; Wu, J.; Piao, Z.; Tong, Y.; et al. Cytological and morphological analysis of hybrids between Brassicoraphanus, and Brassica napus for introgression of clubroot resistant trait into Brassica napus L. PLoS ONE 2017, 12, e0177470. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Birch, P.R.J. The zigzag model of plant-microbe interactions: Is it time to move on? Mol. Plant Pathol. 2014, 15, 865–870. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, F.; Jiang, L.; Chen, C.; Wu, L.; Liu, Z. Different Pathogen Defense Strategies in Arabidopsis: More than Pathogen Recognition. Cells 2018, 7, 252. [Google Scholar] [CrossRef] [PubMed]
- Sagi, M.S.; Deokar, A.A.; Bunyamin, T. Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to ascochyta blight infection. Front. Plant Sci. 2017, 8, 838. [Google Scholar] [CrossRef] [PubMed]
- Bücherl, C.A.; van Esse, G.W.; Kruis, A.; Luchtenberg, J.; Westphal, A.H.; Aker, J.; van Hoek, A.; Albrecht, C.; Borst, J.W.; de Vries, S.C. Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant. Physiol. 2013, 162, 1911–1925. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Wang, Y.; Fang, Z.; Zhuang, M.; Zhang, Y.; Lv, H.; Liu, Y.; Li, Z.; Yang, L. Comparative transcriptome analysis of cabbage (Brassica oleracea var. capitata) infected by Plasmodiophora brassicae reveals drastic defense response at secondary infection stage. Plant Soil 2019, 443, 167–183. [Google Scholar]
- Fu, P.; Piao, Y.; Zhan, Z.; Zhao, Y.; Pang, W.; Li, X. Transcriptome Profile of Brassica rapa L. Reveals the Involvement of Jasmonic Acid, Ethylene, and Brassinosteroid Signaling Pathways in Clubroot Resistance. Agronomy 2019, 9, 589. [Google Scholar] [CrossRef]
- Hamel, L.-P.; Beaudoin, N. Chitooligosaccharide sensing and downstream signaling: Contrasted outcomes in pathogenic and beneficial plant-microbe interactions. Planta 2010, 232, 787–806. [Google Scholar] [CrossRef] [PubMed]
- Laila, R.; Park, J.-I.; Robin, A.H.K.; Natarajan, S.; Vijayakumar, H.; Shirasawa, K.; Isobe, S.; Kim, H.-T.; Nou, I.-S. Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage (Brassica rapa L.). BMC Plant Biol. 2019, 19, 13. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Hwang, S.; Strelkov, S.E. Studies into primary and secondary infection processes by Plasmodiophora brassicae on canola. Plant Pathol. 2013, 62, 177–183. [Google Scholar] [CrossRef]
- Yuan, Y.; Qin, L.; Su, H.; Yang, S.; Wei, X.; Wang, Z.; Zhao, Y.; Li, L.; Liu, H.; Tian, B.; et al. Transcriptome and Coexpression Network Analyses Reveal Hub Genes in Chinese Cabbage (Brassica rapa L. ssp. pekinensis) During Different Stages of Plasmodiophora brassicae Infection . Front. Plant Sci. 2021, 12, 650252. [Google Scholar] [CrossRef]
- Jia, H.; Wei, X.; Yang, Y.; Yuan, Y.; Wei, F.; Zhao, Y.; Yang, S.; Yao, Q.; Wang, Z.; Tian, B.; et al. Root RNA-seq analysis reveals a distinct transcriptome landscape between clubroot-susceptible and clubroot-resistant Chinese cabbage lines after Plasmodiophora brassicae infection. Plant Soil 2017, 421, 93–105. [Google Scholar] [CrossRef]
- Di, X.; Gomila, J.; Takken, F.L.W. Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the sus-ceptibility of tomato to Fusarium oxysporum. Mol. Plant Pathol. 2017, 18, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Denance, N.; Sanchez-Vallet, A.; Goner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J. Plant defence—What can we learn from clubroots? Austral. Plant Pathol. 2009, 38, 318–324. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Konradyova, V.; Knirsch, V.; Gaudinova, A.; Kramna, B.; Kazda, J.; Ludwig-Müller, J.; Vankova, R. Hormonal Responses to Plasmodiophora brassicae Infection in Brassica napus Cultivars Differing in Their Pathogen Resistance. Int. J. Mol. Sci. 2018, 19, 4024. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Zhang, X.; Yuan, Y. Root Transcriptome and Metabolome Profiling Reveal Key Phytohormone-Related Genes and Pathways Involved Clubroot Resistance in Brassica rapa L. Front. Plant Sci. 2021, 12, 759623. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, H.; Li, R.; Chen, W.; Liu, L.; Liu, F.; Zeng, L.; Yan, R.; Chen, K.; Fang, X. Jasmonic acid mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Front. Plant Sci. 2018, 9, 750. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The jaz family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Dong, X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004, 7, 547–552. [Google Scholar] [CrossRef]
- Weigel, R.R.; Pfitzner, U.M.; Gatz, C. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 2005, 17, 1279–1291. [Google Scholar] [CrossRef]
- Garcion, C.; Lohmann, A.; Lamodière, E.; Catinot, J.; Buchala, A.; Métraux, D.J.P. Characterization and biological function of ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef]
- Gonzalez, L.; Manolii, V.; Hwang, S.F.; Strelkov, S.E. Response of Brassica napus to Plasmodiophora brassicae involves salicylic acid-mediated immunity: An RNA-Seq-based study. Front. Plant Sci. 2020, 11, 1025. [Google Scholar] [CrossRef]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 2015, 56, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.R.; Godiard, L.; Straube, E.; Ashfield, T.; Lewald, J.; Sattler, A.; Innes, R.W.; Dangl, J.L. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 1995, 269, 843–846. [Google Scholar] [CrossRef]
- Swiderski, M.R.; Innes, R.W. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001, 26, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J.; Prinsen, E.; Rolfe, S.A.; Scholes, J.D. Metabolism and plant hormone action during clubroot disease. J. Plant Growth Regul. 2009, 28, 229–244. [Google Scholar] [CrossRef]
- Siemens, J.; Keller, I.; Sarx, J.; Kunz, S.; Schuller, A.; Nagel, W.; Schmülling, T.; Parniske, M.; Ludwig-Müller, J. Transcriptome analysis of Arabidopsis clubroots and disease resistance of cytokinin oxidase/dehydrogenase gene overexpressing plants indicate a key role for cytokinin in disease development. Mol. Plant Microbe Interact. 2006, 19, 480–494. [Google Scholar] [CrossRef]
- Devos, S.; Vissenberg, K.; Verbelen, J.P.; Prinsen, E. Infection of Chinese cabbage by Plasmodiophora brassicae leads to a stimulation of plant growth: Impacts on cell wall metabolism and hormone balance. New Phytol. 2005, 166, 241–250. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Chen, J.; Pang, W.; Chen, B.; Zhang, C.; Piao, Z. Transcriptome Analysis of Brassica rapa Near-Isogenic Lines Carrying Clubroot- Resistant and –Susceptible Alleles in Response to Plasmodiophora brassicae during Early Infection. Front. Plant Sci. 2016, 6, 1183. [Google Scholar] [CrossRef]
- Bent, A.; Kunkel, B.; Dahlbeck, D.; Brown, K.; Schmidt, R.; Giraudat, J.; Leung, J.; Staskawicz, B. RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef] [PubMed]
- Sohn, K.H.; Segonzac, C.; Rallapalli, G.; Sarris, P.F.; Woo, J.Y.; Williams, S.J.; Newman, T.E.; Paek, K.H.; Kobe, B.; Jones, J.D. The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 2014, 10, e100465. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.; Cai, C.; Liu, Z.; Liu, B.; et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, E.-S.; Park, H.; Lee, K.; Shim, D.; Oh, M.-H. Comparison of Root Transcriptomes against Clubroot Disease Pathogens in a Resistant Chinese Cabbage Cultivar (Brassica rapa cv. ‘Akimeki’). Plants 2024, 13, 2167. https://doi.org/10.3390/plants13152167
Oh E-S, Park H, Lee K, Shim D, Oh M-H. Comparison of Root Transcriptomes against Clubroot Disease Pathogens in a Resistant Chinese Cabbage Cultivar (Brassica rapa cv. ‘Akimeki’). Plants. 2024; 13(15):2167. https://doi.org/10.3390/plants13152167
Chicago/Turabian StyleOh, Eun-Seok, Hyeonseon Park, Kwanuk Lee, Donghwan Shim, and Man-Ho Oh. 2024. "Comparison of Root Transcriptomes against Clubroot Disease Pathogens in a Resistant Chinese Cabbage Cultivar (Brassica rapa cv. ‘Akimeki’)" Plants 13, no. 15: 2167. https://doi.org/10.3390/plants13152167
APA StyleOh, E.-S., Park, H., Lee, K., Shim, D., & Oh, M.-H. (2024). Comparison of Root Transcriptomes against Clubroot Disease Pathogens in a Resistant Chinese Cabbage Cultivar (Brassica rapa cv. ‘Akimeki’). Plants, 13(15), 2167. https://doi.org/10.3390/plants13152167







