Revolutionizing Tomato Cultivation: CRISPR/Cas9 Mediated Biotic Stress Resistance
Abstract
1. Introduction
2. Mechanism and Mode of Action of CRISPR/Cas9 Technology

| Type/Effector | Nuclease Domains | Target | Cut Structure | tracrRNA Requirement | PAM/PFS | References |
|---|---|---|---|---|---|---|
| II/Cas9 | RuvC, HNH | dsDNA | blunt | Yes | 3′ GC-rich PAM | [49] |
| V/CasX | RuvC | dsDNA | Staggered, 5′-overhangs | Yes | 5′TTCN | [50,51] |
| V/Cas12 | RuvC, NUC | dsDNA, ssDNA | Staggered, 5′-overhangs | No | 5′-T-rich | [47,51] |
| V-A/Cas12a | RuvC, NUC | dsDNA | Staggered, 5′-overhangs | No | 5′ AT-rich PAM | [47,49] |
| VI-A/Cas13a(C2c2) | 2x HEPN | ssRNA | Guide-dependent RNA cuts + collateral RNA cleavage | No | 3′ PFS: non-G | [49,52,53,54] |
| VI-B/Cas13b | 2x HEPN | ssRNA | Guide-dependent RNA cuts + collateral RNA cleavage | No | 5′ PFS: non-C; 30 PFS: NAN/NNA | |
| V/Cas14 | RuvC | ssDNA | Collateral cleavage of ssDNA sequences | Yes | none | [47,55,56] |
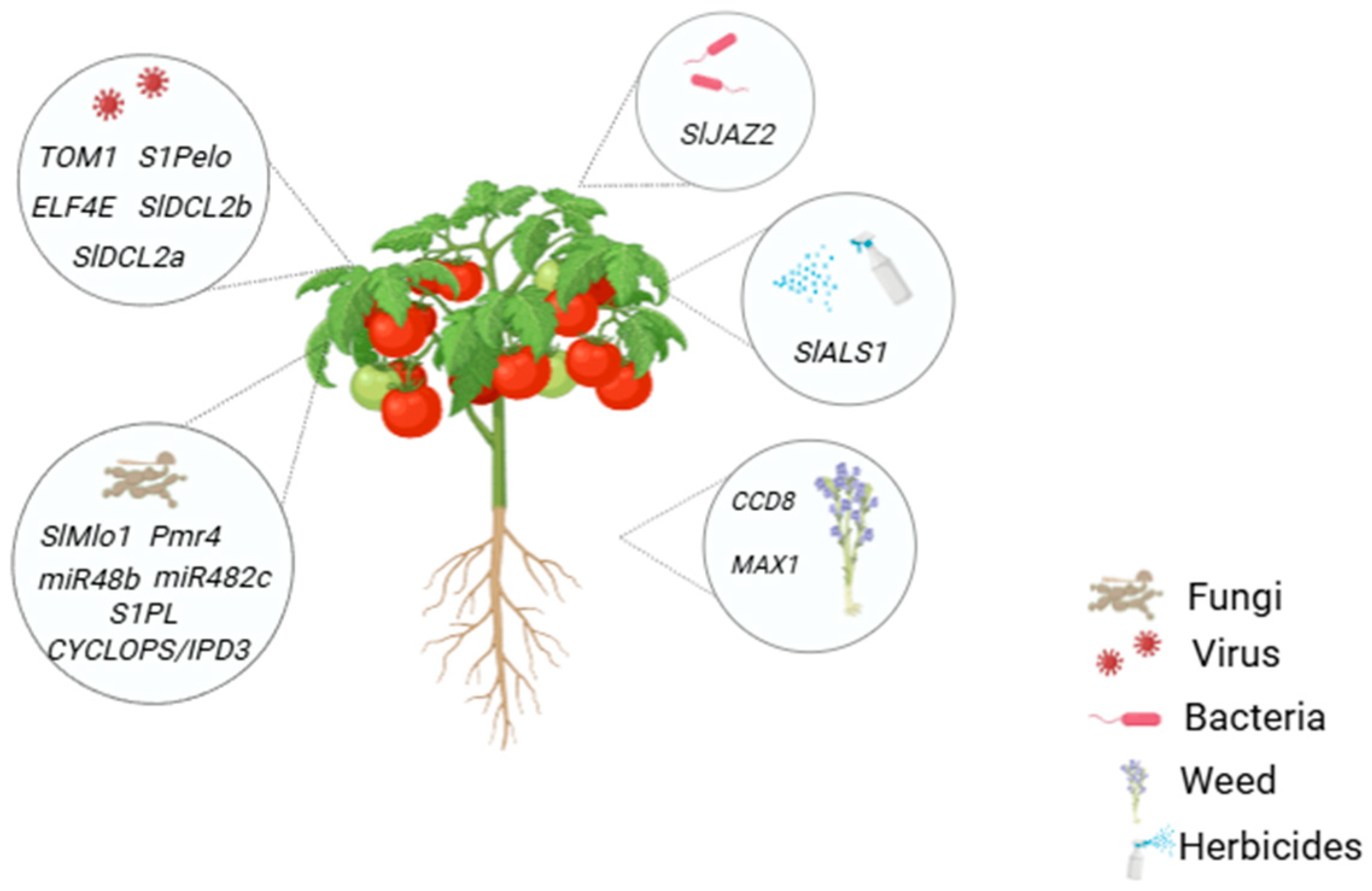
3. Applications of CRISPR/Cas9 in Editing Genes Related to Tomato Biotic Stress Resistance
3.1. CRIPSR/Cas9 Mediated Development and Understanding of Bacterial Resistance in Tomato
3.2. CRIPSR/Cas9 Mediated Development of Fungal Resistant Tomato
3.2.1. CRIPSR/Cas9 Mediated Development of Powdery Mildew Resistant Tomato
3.2.2. Targeted CRISPR/Cas9 Editing of Susceptibility Genes to Enhance Resistance Late Blight Disease
3.2.3. Targeting Genes for Fusarium Wilt Resistant
3.2.4. Enhancing Tomato Resistance to Grey Mold Disease
3.3. CRIPSR/Cas9 Mediated Development of Viral Resistant Tomato
3.3.1. Targeting TOM1 for Tomato Brown Rugose Fruit Virus Resistance
3.3.2. Targeting ELF4E Gene for Potyvirus Resistance in Tomato
3.3.3. Targeting SIPelo Gene for Tomato Yellow Leaf Curl Virus Resistance
3.3.4. Studying the Importance of Dicer-like Protein (DCL) Genes in Resistance to Mosaic Viruses Using CRISPR/Cas9 System
3.4. CRIPSR/Cas9 Mediated Development of Weed Resistant Tomato
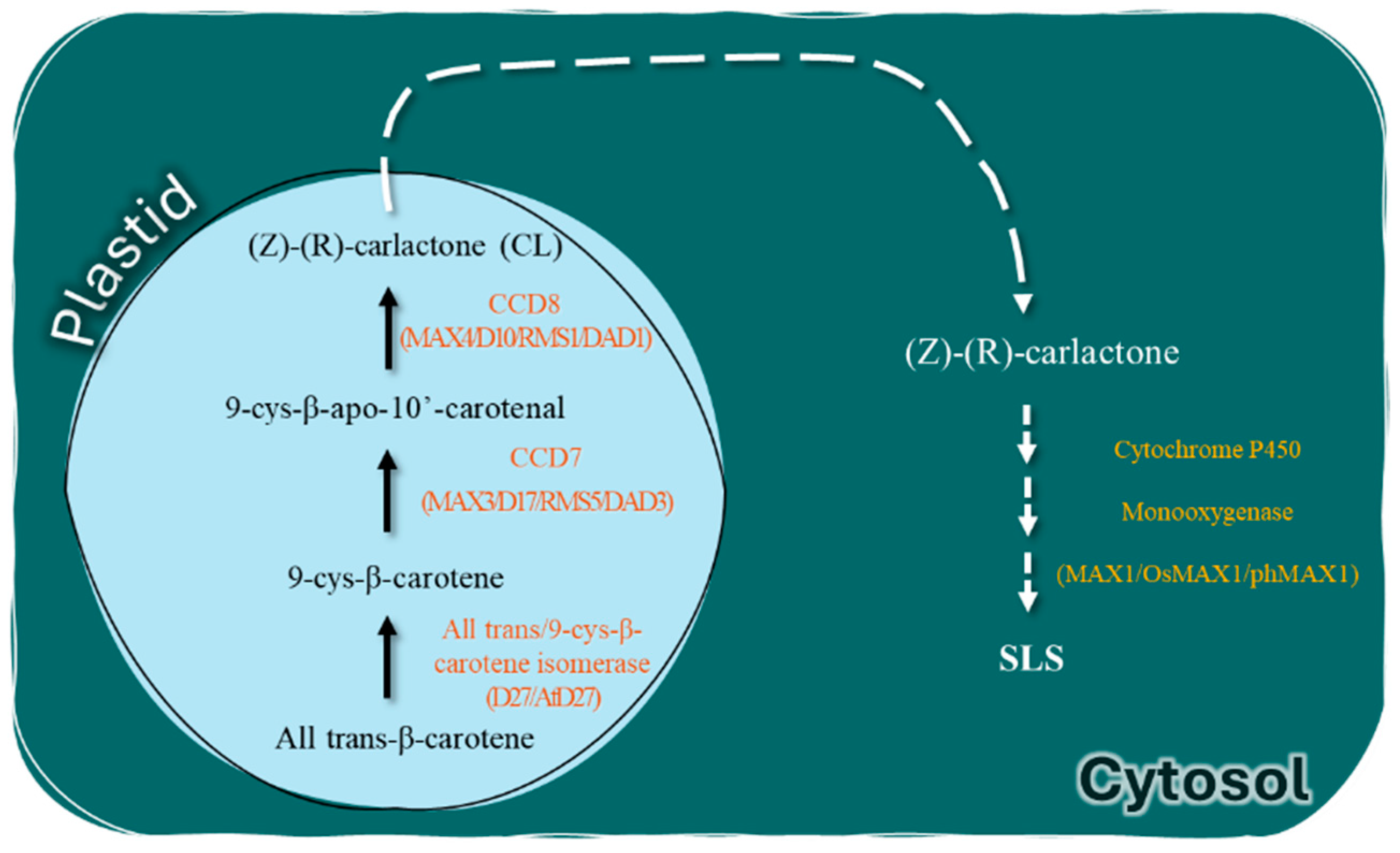
3.4.1. Targeting CCD8 Gene in Tomato for Phelipanche aegyptiaca Weed Resistance
3.4.2. Targeting MAX1 Gene in Tomato for Phelipanche aegyptiaca Weed Resistance
3.5. Enhancing Herbicide Resistance in Tomato via CRISPR/Cas9
4. De Novo Domestication and Multiplex Genome Editing in Tomato for Biotic Stress Resistance
5. Conclusions and Future Perspective
| Infection | Disease | Editing Tool | Target | Editing Method | Effect | References |
|---|---|---|---|---|---|---|
| Bacterial | Bacterial speck | CRISPR/Cas9 | SlJAZ2 | Knockout | Bacterial speck resistance in tomato | [61] |
| Bacterial wilt disease | RNA- interference (RNAi) | DPS | ----------- | Soil-borne pathogen resistance in root | [28] | |
| Herbicidal | Chlorsulfuron | Base editing (CBE) | SlALS1 | ----------- | Chlorsulfuron resistant tomato | [123] |
| Fungal | Powdery mildew | CRISPR/Cas9 | SlMlo1 | Knockout | powdery mildew-resistant tomato | [70,71] |
| Pmr4 | [140] | |||||
| Fusarium wilt | CYCLOPS/IPD3 | Fusarium wilt-resistant tomato | [141] | |||
| XSP10 and SAMT | Multiplexing both genes showed significant Fusarium wilt resistance | [42] | ||||
| Late blight | miR48b | Late blight-resistant tomato | [77] | |||
| miR482c | ||||||
| Gray mold | S1PL | Reduced susceptibility of tomato by >50%. | [95] | |||
| Viral | Tomato brown rugose fruit virus resistance (ToBRFV) | CRISPR/Cas9 | TOM1 | Knockout | ToBRFV resistant tomato | [107] |
| Tomato Yellow Leaf Curl Virus (TYLCV) | SlPelo | Decreases accumulation of TYLCV | [11] | |||
| Potyvirus | ELF4E | Conferred resistance to one type of potyvirus, Pepper Mottle Virus (PepMoV), but was susceptible to Tobacco etch virus (TEV) | [108] | |||
| Tomato mosaic virus | ----------- | SlDCL2b | ------------ | -------------- | [111] | |
| Potato virus X | ----------- | SlDCL2a | [112] | |||
| SlDCL2b | ||||||
| Weed | Broomrapes | CRISPR/Cas9 | CCD8 | Knockout | Remarkable reduction in parasite infection | [117] |
| Root parasitic weed | MAX1 | Root parasitic weed-resistant tomato | [115] |
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.P.; Lutts, S. Tomato Fruit Development and Metabolism. Front. Plant Sci. 2019, 10, 1554. [Google Scholar] [CrossRef]
- Behiry, S.; Soliman, S.A.; Massoud, M.A.; Abdelbary, M.; Kordy, A.M.; Abdelkhalek, A.; Heflish, A. Trichoderma pubescens Elicit Induced Systemic Resistance in Tomato Challenged by Rhizoctonia solani. J. Fungi 2023, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- El-Sappah, A.H.; Qi, S.; Soaud, S.A.; Huang, Q.; Saleh, A.M.; Abourehab, M.A.S.; Wan, L.; Cheng, G.-T.; Liu, J.; Ihtisham, M.; et al. Natural resistance of tomato plants to Tomato yellow leaf curl virus. Front. Plant Sci. 2022, 13, 1081549. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.C.; Kumar, A.U.; Wong, Y.P.; Ling, A.P.K. Bioinformatics approaches and applications in plant biotechnology. J. Genet. Eng. Biotechnol. 2022, 20, 106. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Jayalakshmi, K.; Tilgam, J.; Gupta, A.; Nagaraju, Y.; Kumar, A.; Hamid, S.; Singh, H.V.; Minkina, T.; Rajput, V.D.; et al. ROS generated from biotic stress: Effects on plants and alleviation by endophytic microbes. Front. Plant Sci. 2022, 13, 1042936. [Google Scholar] [CrossRef]
- Singla, J.; Krattinger, S.G. Biotic Stress Resistance Genes in Wheat. In Encyclopedia of Food Grains, 2nd ed.; Wrigley, C., Corke, H., Seetharaman, K., Faubion, J., Eds.; Academic Press: Oxford, UK, 2016; pp. 388–392. [Google Scholar] [CrossRef]
- Mertens, D.; Boege, K.; Kessler, A.; Koricheva, J.; Thaler, J.S.; Whiteman, N.K.; Poelman, E.H. Predictability of Biotic Stress Structures Plant Defence Evolution. Trends Ecol. Evol. 2021, 36, 444–456. [Google Scholar] [CrossRef]
- Kovalchuk, I. Chapter 35-Transgenerational genome instability in plants. In Genome Stability, 2nd ed.; Kovalchuk, I., Kovalchuk, O., Eds.; Academic Press: Boston, MA, USA, 2021; Volume 26, pp. 659–678. [Google Scholar]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Maioli, A.; Yan, Z.; Bai, Y.; Valentino, D.; Milani, A.M.; Pompili, V.; Comino, C.; Lanteri, S.; Moglia, A.; et al. CRISPR/Cas9-Based Knock-Out of the PMR4 Gene Reduces Susceptibility to Late Blight in Two Tomato Cultivars. Int. J. Mol. Sci. 2022, 23, 14542. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.Y. CRISPR/Cas9-Mediated Generation of Pathogen-Resistant Tomato against Tomato Yellow Leaf Curl Virus and Powdery Mildew. Int. J. Mol. Sci. 2021, 22, 1878. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Abdelrahman, M.; Gao, Y.; Ji, Z.; Mishra, R.; Sun, H.; Sui, Y.; Wu, C.; Wang, C.; Zhao, K. Engineering broad-spectrum resistance to bacterial blight by CRISPR-Cas9-mediated precise homology directed repair in rice. Mol. Plant 2021, 14, 1215–1218. [Google Scholar] [CrossRef]
- Zhang, M.; Coaker, G. Harnessing Effector-Triggered Immunity for Durable Disease Resistance. Phytopathology 2017, 107, 912–919. [Google Scholar] [CrossRef]
- Yu, R.-M.; Zhu, J.-H.; Li, C.-L. Gene Modification of Medicinal Plant Germplasm Resources; Springer: Singapore, 2019; pp. 145–190. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd Allah, E.F.; Bae, H. Genome Editing Tools in Plants. Genes 2017, 8, 399. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.H.; Yu, M.; Lai, E.M. Agrobacterium-mediated plant transformation: Biology and applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef] [PubMed]
- Tzfira, T.; Citovsky, V. Agrobacterium-mediated genetic transformation of plants: Biology and biotechnology. Curr. Opin. Biotechnol. 2006, 17, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Matres, J.M.; Hilscher, J.; Datta, A.; Armario-Najera, V.; Baysal, C.; He, W.; Huang, X.; Zhu, C.; Valizadeh-Kamran, R.; Trijatmiko, K.R.; et al. Genome editing in cereal crops: An overview. Transgenic Res. 2021, 30, 461–498. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Wei, Z.; Rohila, J.S.; Zhao, K. Multiplex Genome-Editing Technologies for Revolutionizing Plant Biology and Crop Improvement. Front. Plant Sci. 2021, 12, 721203. [Google Scholar] [CrossRef]
- Rodriguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.e8. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Kwon, C.-T. Trait Improvement of Solanaceae Fruit Crops for Vertical Farming by Genome Editing. J. Plant Biol. 2023, 66, 1–14. [Google Scholar] [CrossRef]
- Xia, X.; Cheng, X.; Li, R.; Yao, J.; Li, Z.; Cheng, Y. Advances in application of genome editing in tomato and recent development of genome editing technology. Theor. Appl. Genet. 2021, 134, 2727–2747. [Google Scholar] [CrossRef]
- Amoroso, C.G.; Panthee, D.R.; Andolfo, G.; Ramìrez, F.P.; Ercolano, M.R. Genomic Tools for Improving Tomato to Biotic Stress Resistance. In Genomic Designing for Biotic Stress Resistant Vegetable Crops; Kole, C., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–35. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Singh, A.K.; Behera, T.K. CRISPR/Cas genome editing in tomato improvement: Advances and applications. Front. Plant Sci. 2023, 14, 1121209. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.A.; Kottam, S.K.; Narasu, M.L.; Kumari, P.H. Recent Trends in Targeting Genome Editing of Tomato for Abiotic and Biotic Stress Tolerance. In Genome Editing: Current Technology Advances and Applications for Crop Improvement; Wani, S.H., Hensel, G., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 273–285. [Google Scholar] [CrossRef]
- Salava, H.; Thula, S.; Mohan, V.; Kumar, R.; Maghuly, F. Application of Genome Editing in Tomato Breeding: Mechanisms, Advances, and Prospects. Int. J. Mol. Sci. 2021, 22, 682. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Boopathi, T.; Paramasivan, M. A status-quo review on CRISPR-Cas9 gene editing applications in tomato. Int. J. Biol. Macromol. 2021, 190, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.E.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadan, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Lipinski, D.; Zeyland, J.; Slomski, R. CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Arch. Immunol. Ther. Exp. 2017, 65, 233–240. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Asmamaw, M.; Zawdie, B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics 2021, 15, 353–361. [Google Scholar] [CrossRef]
- Almiron, M.; Link, A.J.; Furlong, D.; Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes. Dev. 1992, 6, 2646–2654. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Zhao, K. Genome Editing and Rice Grain Quality. In The Future of Rice Demand: Quality Beyond Productivity; Costa de Oliveira, A., Pegoraro, C., Ebeling Viana, V., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 395–422. [Google Scholar] [CrossRef]
- Stemmer, M.; Thumberger, T.; Del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef]
- Weber, E.; Gruetzner, R.; Werner, S.; Engler, C.; Marillonnet, S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE 2011, 6, e19722. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, A.; Li, K.; Wang, B.; Jin, S.; Reiser, M.; Lockey, R.F. CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. Biotechniques 2015, 58, 161–170. [Google Scholar] [CrossRef]
- Debbarma, J.; Saikia, B.; Singha, D.L.; Das, D.; Keot, A.K.; Maharana, J.; Velmurugan, N.; Arunkumar, K.P.; Reddy, P.S.; Chikkaputtaiah, C. CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum lycopersicum L.). Genes 2023, 14, 488. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, D.; Jogam, P.; Venkatapuram, A.K.; Savitikadi, P.; Peddaboina, V.; Allini, V.R.; Abbagani, S. Highly efficient Agrobacterium-mediated transformation and plant regeneration system for genome engineering in tomato. Saudi J. Biol. Sci. 2022, 29, 103292. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillo, L.; Valencia-Lozano, E.; Priego-Ranero, N.A.; Cabrera-Ponce, J.L.; Duarte-Ake, F.P.; Vizuet-de-Rueda, J.C.; Rivera-Toro, D.M.; Herrera-Ubaldo, H.; de Folter, S.; Alvarez-Venegas, R. CRISPRa-mediated transcriptional activation of the SlPR-1 gene in edited tomato plants. Plant Sci. 2023, 329, 111617. [Google Scholar] [CrossRef]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovic, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef]
- Tian, X.; Gu, T.; Patel, S.; Bode, A.M.; Lee, M.-H.; Dong, Z. CRISPR/Cas9—An evolving biological tool kit for cancer biology and oncology. npj Precis. Oncol. 2019, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Patel, D.J. CasX: A new and small CRISPR gene-editing protein. Cell Res. 2019, 29, 345–346. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Munir, A.; Aslam, H.M.U.; Khan, S.H.; Ahmad, A. Genome Editing Advances in Soybean Improvement against Biotic and Abiotic Stresses; Springer International Publishing: Cham, Switzerland, 2022; pp. 241–274. [Google Scholar] [CrossRef]
- Bot, J.F.; van der Oost, J.; Geijsen, N. The double life of CRISPR–Cas13. Curr. Opin. Biotechnol. 2022, 78, 102789. [Google Scholar] [CrossRef]
- Palaz, F.; Kalkan, A.K.; Can, Ö.; Demir, A.N.; Tozluyurt, A.; Özcan, A.; Ozsoz, M. CRISPR-Cas13 System as a Promising and Versatile Tool for Cancer Diagnosis, Therapy, and Research. ACS Synth. Biol. 2021, 10, 1245–1267. [Google Scholar] [CrossRef]
- Savage, D.F. Cas14: Big Advances from Small CRISPR Proteins. Biochemistry 2019, 58, 1024–1025. [Google Scholar] [CrossRef]
- Aquino-Jarquin, G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomed. Nanotechnol. Biol. Med. 2019, 18, 428–431. [Google Scholar] [CrossRef]
- Hajİ Nour, S.A.M.; Horuz, S. Determination of the efficacies of different phosphites in the management of tomato bacterial speck disease caused by Pseudomonas syringae pv. tomato. Mustafa Kemal Üniversitesi Tarım Bilim. Derg. 2023, 28, 25–37. [Google Scholar] [CrossRef]
- Enow, A. Influence of Salicylate and Abscisic Acid on the Resisitance of Tomato to Biotrophic and Necrotrophic Pathogens. 2007. Available online: https://www.researchgate.net/publication/292609853_Influence_of_salicylate_and_abscisic_acid_on_the_resisitance_of_tomato_to_biotrophic_and_necrotrophic_pathogens (accessed on 4 April 2024).
- Meddya, S.; Meshram, S.; Sarkar, D.; S, R.; Datta, R.; Singh, S.; Avinash, G.; Kumar Kondeti, A.; Savani, A.K.; Thulasinathan, T. Plant Stomata: An Unrealized Possibility in Plant Defense against Invading Pathogens and Stress Tolerance. Plants 2023, 12, 3380. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bhardwaj, M.; Tran, L.P. Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence. Int. J. Mol. Sci. 2020, 21, 7482. [Google Scholar] [CrossRef] [PubMed]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 2019, 17, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Shao, Q.; Lu, Q.; Li, X.; Gao, Y. Transcriptome analysis reveals differential transcription in tomato (Solanum lycopersicum) following inoculation with Ralstonia solanacearum. Sci. Rep. 2022, 12, 22137. [Google Scholar] [CrossRef] [PubMed]
- Yeom, S.I.; Seo, E.; Oh, S.K.; Kim, K.W.; Choi, D. A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J. 2012, 69, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Saikia, B.; S, R.; Debbarma, J.; Maharana, J.; Sastry, G.N.; Chikkaputtaiah, C. CRISPR/Cas9-based genome editing and functional analysis of SlHyPRP1 and SlDEA1 genes of Solanum lycopersicum L. in imparting genetic tolerance to multiple stress factors. Front. Plant Sci. 2024, 15, 1304381. [Google Scholar] [CrossRef] [PubMed]
- Bishnoi, R.; Kaur, S.; Sandhu, J.S.; Singla, D. Genome engineering of disease susceptibility genes for enhancing resistance in plants. Funct. Integr. Genom. 2023, 23, 207. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Jacobsen, E.; Visser, R.G.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2010, 25, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, M.; Andel, A.; Huibers, R.P.; Panstruga, R.; Weisbeek, P.J.; Van den Ackerveken, G. Identification of arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 2005, 18, 583–592. [Google Scholar] [CrossRef]
- Thomazella, D.P.T.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2026152118. [Google Scholar] [CrossRef]
- Suzuki, T.; Murakami, T.; Takizumi, Y.; Ishimaru, H.; Kudo, D.; Takikawa, Y.; Matsuda, Y.; Kakutani, K.; Bai, Y.; Nonomura, T. Trichomes: Interaction sites of tomato leaves with biotrophic powdery mildew pathogens. Eur. J. Plant Pathol. 2018, 150, 115–125. [Google Scholar] [CrossRef]
- Paul, N.C.; Park, S.W.; Liu, H.; Choi, S.; Ma, J.; MacCready, J.S.; Chilvers, M.I.; Sang, H. Plant and Fungal Genome Editing to Enhance Plant Disease Resistance Using the CRISPR/Cas9 System. Front. Plant Sci. 2021, 12, 700925. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef]
- Vogel, J.; Somerville, S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 2000, 97, 1897–1902. [Google Scholar] [CrossRef]
- Huibers, R.P.; Loonen, A.E.; Gao, D.; Van den Ackerveken, G.; Visser, R.G.; Bai, Y. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef]
- Santillán Martínez, M.I.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.F.; Wolters, A.A.; Bai, Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020, 20, 284. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.A.; Ukladov, E.O.; Golubeva, T.S. Phytophthora infestans: An Overview of Methods and Attempts to Combat Late Blight. J. Fungi 2021, 7, 1071. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Hong, Y.; Meng, J.; He, X.; Zhang, Y.; Liu, Y.; Zhang, C.; Qi, H.; Luan, Y. Editing miR482b and miR482c Simultaneously by CRISPR/Cas9 Enhanced Tomato Resistance to Phytophthora infestans. Phytopathology 2021, 111, 1008–1016. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; Tan, Y.; Zhao, T.; Xu, X.; Yang, H.; Li, J. CRISPR/Cas9-Mediated SlMYBS2 Mutagenesis Reduces Tomato Resistance to Phytophthora infestans. Int. J. Mol. Sci. 2021, 22, 11423. [Google Scholar] [CrossRef]
- Yang, W.; Liu, C.; Fu, Q.; Jia, X.; Deng, L.; Feng, C.; Wang, Y.; Yang, Z.; Yang, H.; Xu, X. Knockout of SlbZIP68 reduces late blight resistance in tomato. Plant Sci. 2023, 336, 111861. [Google Scholar] [CrossRef] [PubMed]
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandić, A.; Noris, E.; Matić, S. A Review of the Most Common and Economically Important Diseases That Undermine the Cultivation of Tomato Crop in the Mediterranean Basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Gu, Y.Q.; Yang, C.; Thara, V.K.; Zhou, J.; Martin, G.B. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 2000, 12, 771–786. [Google Scholar] [CrossRef]
- Banno, H.; Ikeda, Y.; Niu, Q.W.; Chua, N.H. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 2001, 13, 2609–2618. [Google Scholar] [CrossRef] [PubMed]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Cas9/sgRNA-based genome editing and other reverse genetic approaches for functional genomic studies in rice. Brief. Funct. Genom. 2018, 17, 339–351. [Google Scholar] [CrossRef]
- Ijaz, S.; Haq, I.U.; Razzaq, H.A. Mutation introduced in DDTFR10/A gene of ethylene response element-binding protein (EREBP) family through CRISPR/Cas9 genome editing confers increased Fusarium wilt tolerance in tomato. Physiol. Mol. Biol. Plants 2023, 29, 1–10. [Google Scholar] [CrossRef]
- Kavi Kishor, P.B.; Hima Kumari, P.; Sunita, M.S.; Sreenivasulu, N. Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front. Plant Sci. 2015, 6, 544. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ouyang, B.; Wang, T.; Luo, Z.; Yang, C.; Li, H.; Sima, W.; Zhang, J.; Ye, Z. HyPRP1 Gene Suppressed by Multiple Stresses Plays a Negative Role in Abiotic Stress Tolerance in Tomato. Front. Plant Sci. 2016, 7, 967. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.T.; Doan, D.T.H.; Kim, J.; Song, Y.J.; Sung, Y.W.; Das, S.; Kim, E.J.; Son, G.H.; Kim, S.H.; Van Vu, T.; et al. CRISPR/Cas9-based precise excision of SlHyPRP1 domain(s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 2021, 40, 999–1011. [Google Scholar] [CrossRef]
- Tran, M.T.; Son, G.H.; Song, Y.J.; Nguyen, N.T.; Park, S.; Thach, T.V.; Kim, J.; Sung, Y.W.; Das, S.; Pramanik, D.; et al. CRISPR-Cas9-based precise engineering of SlHyPRP1 protein towards multi-stress tolerance in tomato. Front. Plant Sci. 2023, 14, 1186932. [Google Scholar] [CrossRef]
- Hanika, K.; Schipper, D.; Chinnappa, S.; Oortwijn, M.; Schouten, H.J.; Thomma, B.; Bai, Y. Impairment of Tomato WAT1 Enhances Resistance to Vascular Wilt Fungi Despite Severe Growth Defects. Front. Plant Sci. 2021, 12, 721674. [Google Scholar] [CrossRef] [PubMed]
- Matrose, N.; Obikeze, K.; Belay, Z.; Caleb, O. Plant extracts and other natural compounds as alternatives for post-harvest management of fruit fungal pathogens: A review. Food Biosci. 2020, 41, 100840. [Google Scholar] [CrossRef]
- Brito, C.; Hansen, H.; Espinoza, L.; Faúndez, M.; Olea, A.F.; Pino, S.; Díaz, K. Assessing the Control of Postharvest Gray Mold Disease on Tomato Fruit Using Mixtures of Essential Oils and Their Respective Hydrolates. Plants 2021, 10, 1719. [Google Scholar] [CrossRef] [PubMed]
- Toral, L.; Rodríguez, M.; Béjar, V.; Sampedro, I. Crop Protection against Botrytis cinerea by Rhizhosphere Biological Control Agent Bacillus velezensis XT1. Microorganisms 2020, 8, 992. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.; Vicente, A.R.; Greve, L.C.; Dewey, F.M.; Bennett, A.B.; Labavitch, J.M.; Powell, A.L. The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. USA 2008, 105, 859–864. [Google Scholar] [CrossRef]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef]
- Silva, C.J.; van den Abeele, C.; Ortega-Salazar, I.; Papin, V.; Adaskaveg, J.A.; Wang, D.; Casteel, C.L.; Seymour, G.B.; Blanco-Ulate, B. Host susceptibility factors render ripe tomato fruit vulnerable to fungal disease despite active immune responses. J. Exp. Bot. 2021, 72, 2696–2709. [Google Scholar] [CrossRef]
- Vossen, J.H.; Abd-El-Haliem, A.; Fradin, E.F.; van den Berg, G.C.; Ekengren, S.K.; Meijer, H.J.; Seifi, A.; Bai, Y.; ten Have, A.; Munnik, T.; et al. Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J. 2010, 62, 224–239. [Google Scholar] [CrossRef]
- Gonorazky, G.; Guzzo, M.C.; Abd-El-Haliem, A.M.; Joosten, M.H.; Laxalt, A.M. Silencing of the tomato phosphatidylinositol-phospholipase C2 (SlPLC2) reduces plant susceptibility to Botrytis cinerea. Mol. Plant Pathol. 2016, 17, 1354–1363. [Google Scholar] [CrossRef]
- Perk, E.A.; Arruebarrena Di Palma, A.; Colman, S.; Mariani, O.; Cerrudo, I.; D’Ambrosio, J.M.; Robuschi, L.; Pombo, M.A.; Rosli, H.G.; Villareal, F.; et al. CRISPR/Cas9-mediated phospholipase C2 knock-out tomato plants are more resistant to Botrytis cinerea. Planta 2023, 257, 117. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P. Emerging viral diseases of tomato crops. Mol. Plant Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef]
- Xu, C.; Sun, X.; Taylor, A.; Jiao, C.; Xu, Y.; Cai, X.; Wang, X.; Ge, C.; Pan, G.; Wang, Q.; et al. Diversity, Distribution, and Evolution of Tomato Viruses in China Uncovered by Small RNA Sequencing. J. Virol. 2017, 91, e00173-17. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, G.Q.; de Oliveira Silva, J.; Copati, M.G.F.; de Oliveira Dias, F.; dos Santos, M.C. Tomato breeding for disease resistance. Multi-Sci. J. 2020, 3, 8–16. [Google Scholar] [CrossRef]
- Shahriari, Z.; Su, X.; Zheng, K.; Zhang, Z. Advances and Prospects of Virus-Resistant Breeding in Tomatoes. Int. J. Mol. Sci. 2023, 24, 15448. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H. Susceptibility genes to plant viruses. Viruses 2018, 10, 484. [Google Scholar] [CrossRef]
- Kan, J.; Cai, Y.; Cheng, C.; Jiang, C.; Jin, Y.; Yang, P. Simultaneous editing of host factor gene TaPDIL5-1 homoeoalleles confers wheat yellow mosaic virus resistance in hexaploid wheat. New Phytol. 2022, 234, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Ohta, T.; Takahashi, M.; Meshi, T.; Schmidt, R.; Dean, C.; Naito, S.; Ishikawa, M. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 2000, 97, 10107–10112. [Google Scholar] [CrossRef]
- Ishikawa, M.; Yoshida, T.; Matsuyama, M.; Kouzai, Y.; Kano, A.; Ishibashi, K. Tomato brown rugose fruit virus resistance generated by quadruple knockout of homologs of TOBAMOVIRUS MULTIPLICATION1 in tomato. Plant Physiol. 2022, 189, 679–686. [Google Scholar] [CrossRef]
- Yoon, Y.J.; Venkatesh, J.; Lee, J.H.; Kim, J.; Lee, H.E.; Kim, D.S.; Kang, B.C. Genome Editing of eIF4E1 in Tomato Confers Resistance to Pepper Mottle Virus. Front. Plant Sci. 2020, 11, 1098. [Google Scholar] [CrossRef]
- Khan, Z.A.; Kumar, R.; Dasgupta, I. CRISPR/Cas-Mediated Resistance against Viruses in Plants. Int. J. Mol. Sci. 2022, 23, 2303. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Kasai, A.; Maoka, T.; Masuta, C.; Sano, T.; Nakahara, K.S. RNA silencing-related genes contribute to tolerance of infection with potato virus X and Y in a susceptible tomato plant. Virol. J. 2020, 17, 149. [Google Scholar] [CrossRef] [PubMed]
- Ullah, N.; Akhtar, K.P.; Saleem, M.Y.; Habib, M. Characterization of tomato mosaic virus and search for its resistance in Solanum species. Eur. J. Plant Pathol. 2019, 155, 1195–1209. [Google Scholar] [CrossRef]
- Wang, Z.; Hardcastle, T.J.; Canto Pastor, A.; Yip, W.H.; Tang, S.; Baulcombe, D.C. A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes. Dev. 2018, 32, 1155–1160. [Google Scholar] [CrossRef]
- Mohamed, I.A.; Abdalla, R.M. Weed Control, Growth, and Yield of Tomato After Application of Metribuzin and Different Pendimethalin Products in Upper Egypt. J. Soil Sci. Plant Nutr. 2023, 23, 924–937. [Google Scholar] [CrossRef]
- Zhang, L.; Cao, X.; Yao, Z.; Dong, X.; Chen, M.; Xiao, L.; Zhao, S. Identification of risk areas for Orobanche cumana and Phelipanche aegyptiaca in China, based on the major host plant and CMIP6 climate scenarios. Ecol. Evol. 2022, 12, e8824. [Google Scholar] [CrossRef] [PubMed]
- Bari, V.K.; Nassar, J.A.; Aly, R. CRISPR/Cas9 mediated mutagenesis of MORE AXILLARY GROWTH 1 in tomato confers resistance to root parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2021, 11, 3905. [Google Scholar] [CrossRef] [PubMed]
- Bari, V.K.; Nassar, J.A.; Kheredin, S.M.; Gal-On, A.; Ron, M.; Britt, A.; Steele, D.; Yoder, J.; Aly, R. CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2019, 9, 11438. [Google Scholar] [CrossRef]
- Rojas-Vásquez, R.; Gatica-Arias, A. Use of genome editing technologies for genetic improvement of crops of tropical origin. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 140, 215–244. [Google Scholar] [CrossRef]
- Armelina, A.d. Weed competition in direct-sown tomatoes in the lower Río Negro valley. Malezas (Buenos Aires) 1983, 11, 142–146. [Google Scholar]
- Samant, T.; Prusty, M. Effect of weed management on yield, economics and nutrient uptake in tomato (Lycopersicon esculentum Mill.). Adv. Res. J. Crop Improv. 2014, 5, 144–148. [Google Scholar] [CrossRef]
- Yang, S.H.; Kim, E.; Park, H.; Koo, Y. Selection of the high efficient sgRNA for CRISPR-Cas9 to edit herbicide related genes, PDS, ALS, and EPSPS in tomato. Appl. Biol. Chem. 2022, 65, 13. [Google Scholar] [CrossRef]
- Panozzo, S.; Mascanzoni, E.; Scarabel, L.; Milani, A.; Dalazen, G.; Merotto, A.J.; Tranel, P.J.; Sattin, M. Target-Site Mutations and Expression of ALS Gene Copies Vary According to Echinochloa Species. Genes 2021, 12, 1841. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.J.; Palma-Bautista, C.; Vazquez-Garcia, J.G.; Rojano-Delgado, A.M.; Osuna, M.D.; Torra, J.; De Prado, R. Multiple mutations in the EPSPS and ALS genes of Amaranthus hybridus underlie resistance to glyphosate and ALS inhibitors. Sci. Rep. 2020, 10, 17681. [Google Scholar] [CrossRef]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogué, F.; Mazier, M. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef]
- Zsogon, A.; Cermak, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.K. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 2013, 6, 2008–2011. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Yan, L.; Wei, S.; Wu, Y.; Hu, R.; Li, H.; Yang, W.; Xie, Q. High-Efficiency Genome Editing in Arabidopsis Using YAO Promoter-Driven CRISPR/Cas9 System. Mol. Plant 2015, 8, 1820–1823. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Mao, Y.; Ha, S.; Liu, W.; Botella, J.R.; Zhu, J.K. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2016, 35, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014, 56, 343–349. [Google Scholar] [CrossRef]
- Tang, X.; Zheng, X.; Qi, Y.; Zhang, D.; Cheng, Y.; Tang, A.; Voytas, D.F.; Zhang, Y. A Single Transcript CRISPR-Cas9 System for Efficient Genome Editing in Plants. Mol. Plant 2016, 9, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Agapito-Tenfen, S.Z.; Okoli, A.S.; Bernstein, M.J.; Wikmark, O.G.; Myhr, A.I. Revisiting Risk Governance of GM Plants: The Need to Consider New and Emerging Gene-Editing Techniques. Front. Plant Sci. 2018, 9, 1874. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.; Agapito-Tenfen, S.Z. Unintended Genomic Outcomes in Current and Next Generation GM Techniques: A Systematic Review. Plants 2022, 11, 2997. [Google Scholar] [CrossRef]
- Eckerstorfer, M.F.; Dolezel, M.; Engelhard, M.; Giovannelli, V.; Grabowski, M.; Heissenberger, A.; Lener, M.; Reichenbecher, W.; Simon, S.; Staiano, G.; et al. Recommendations for the Assessment of Potential Environmental Effects of Genome-Editing Applications in Plants in the EU. Plants 2023, 12, 1764. [Google Scholar] [CrossRef]
- Spok, A.; Sprink, T.; Allan, A.C.; Yamaguchi, T.; Daye, C. Towards social acceptability of genome-edited plants in industrialised countries? Emerging evidence from Europe, United States, Canada, Australia, New Zealand, and Japan. Front. Genome Ed. 2022, 4, 899331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, L.; Zhao, R.; Yu, W.; Li, R.; Li, Y.; Sheng, J.; Shen, L. Knockout of SlMAPK3 Reduced Disease Resistance to Botrytis cinerea in Tomato Plants. J. Agric. Food Chem. 2018, 66, 8949–8956. [Google Scholar] [CrossRef] [PubMed]
- Prihatna, C.; Larkan, N.J.; Barbetti, M.J.; Barker, S.J. Tomato CYCLOPS/IPD3 is required for mycorrhizal symbiosis but not tolerance to Fusarium wilt in mycorrhiza-deficient tomato mutant rmc. Mycorrhiza 2018, 28, 495–507. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shawky, A.; Hatawsh, A.; Al-Saadi, N.; Farzan, R.; Eltawy, N.; Francis, M.; Abousamra, S.; Ismail, Y.Y.; Attia, K.; Fakhouri, A.S.; et al. Revolutionizing Tomato Cultivation: CRISPR/Cas9 Mediated Biotic Stress Resistance. Plants 2024, 13, 2269. https://doi.org/10.3390/plants13162269
Shawky A, Hatawsh A, Al-Saadi N, Farzan R, Eltawy N, Francis M, Abousamra S, Ismail YY, Attia K, Fakhouri AS, et al. Revolutionizing Tomato Cultivation: CRISPR/Cas9 Mediated Biotic Stress Resistance. Plants. 2024; 13(16):2269. https://doi.org/10.3390/plants13162269
Chicago/Turabian StyleShawky, Abdelrahman, Abdulrahman Hatawsh, Nabil Al-Saadi, Raed Farzan, Nour Eltawy, Mariz Francis, Sara Abousamra, Yomna Y. Ismail, Kotb Attia, Abdulaziz S. Fakhouri, and et al. 2024. "Revolutionizing Tomato Cultivation: CRISPR/Cas9 Mediated Biotic Stress Resistance" Plants 13, no. 16: 2269. https://doi.org/10.3390/plants13162269
APA StyleShawky, A., Hatawsh, A., Al-Saadi, N., Farzan, R., Eltawy, N., Francis, M., Abousamra, S., Ismail, Y. Y., Attia, K., Fakhouri, A. S., & Abdelrahman, M. (2024). Revolutionizing Tomato Cultivation: CRISPR/Cas9 Mediated Biotic Stress Resistance. Plants, 13(16), 2269. https://doi.org/10.3390/plants13162269






