Transcriptome Analysis Revealed ZmPTOX1 Is Required for Seedling Development and Stress Tolerance in Maize
Abstract
:1. Introduction
2. Results
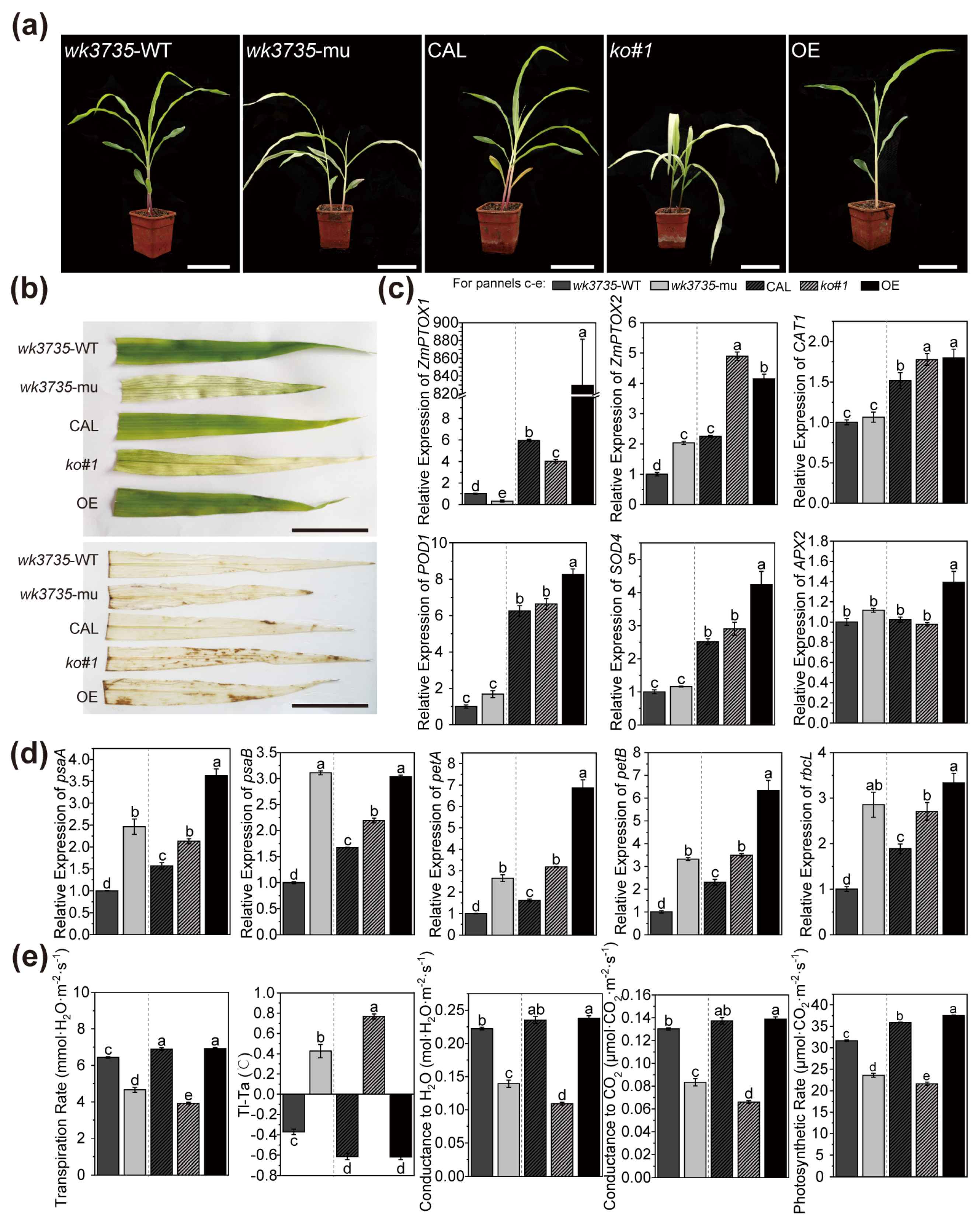
2.1. ZmPTOX1 Mutation Produces Yellow-Green Crossbanding and Affects the Photosynthetic Efficiency in Seedlings
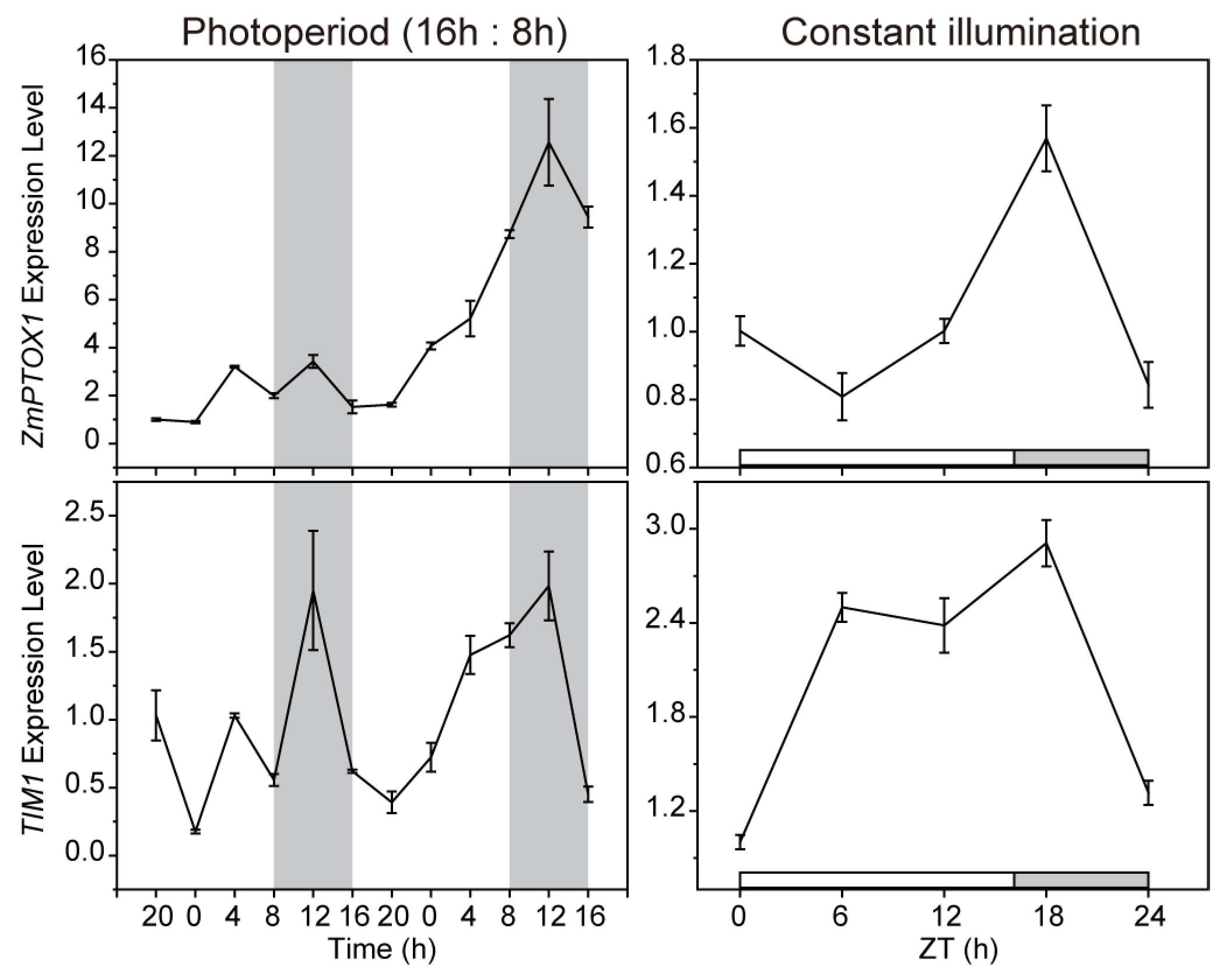
2.2. The Expression of ZmPTOX1 Is Regulated by the Circadian Clock
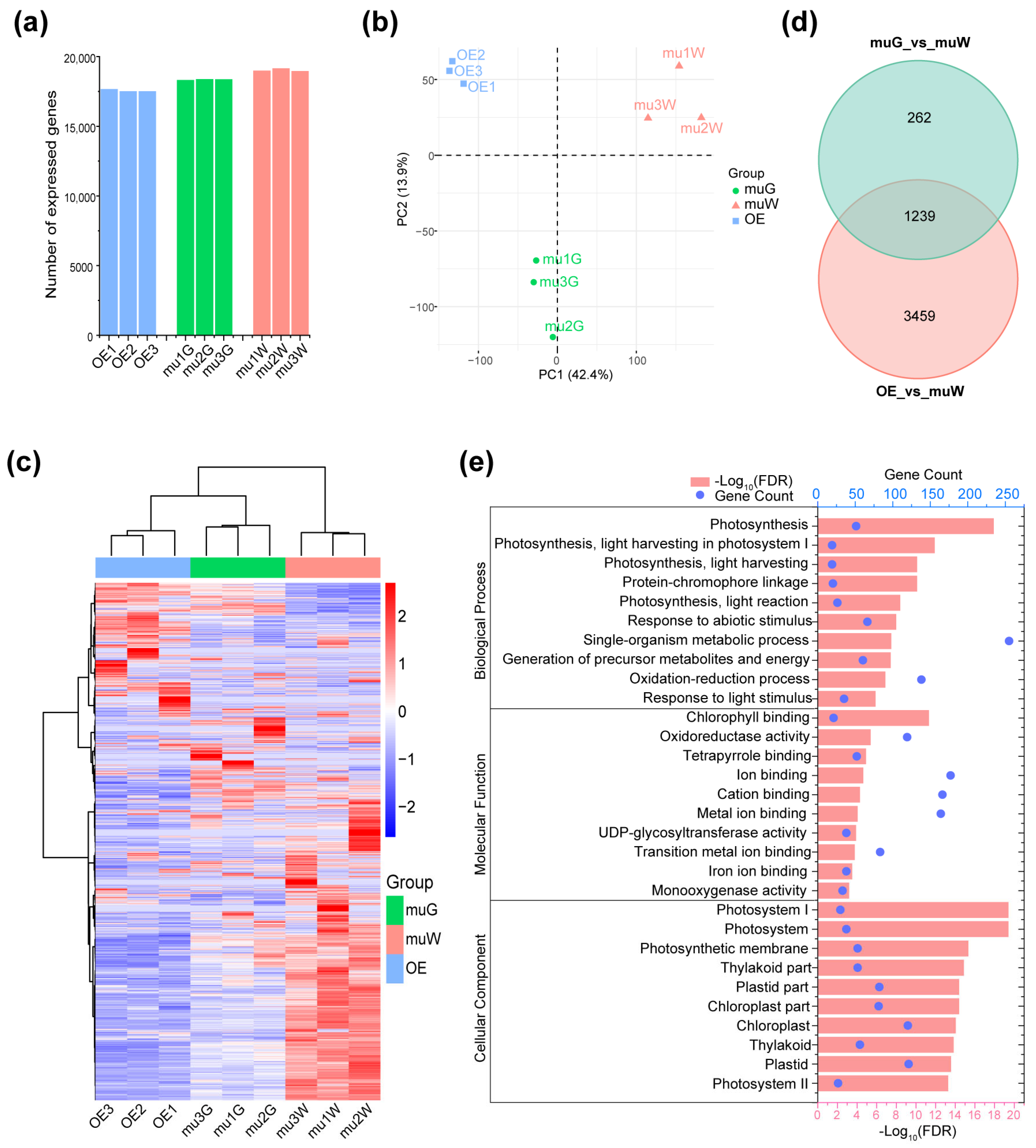
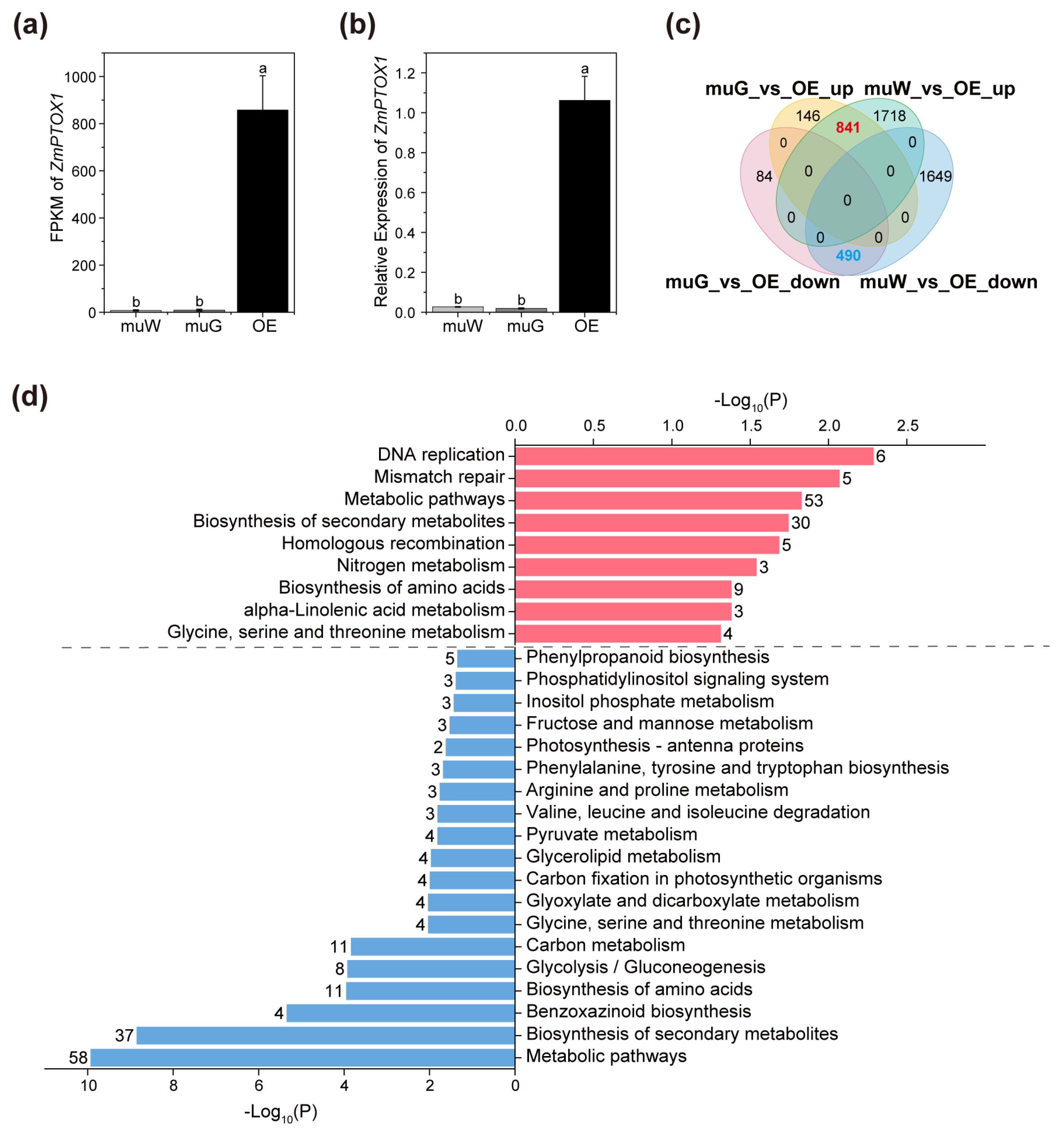
2.3. Transcriptome Analysis for OE and Segregated Etiolated and Green Sections of ko#1 Seedlings
2.4. Zebra Leaf Etiolated Sections Exhibit a Marked Defect in the Expression of Genes Involved in the Circadian Rhythm and Rhythmic Stress Response
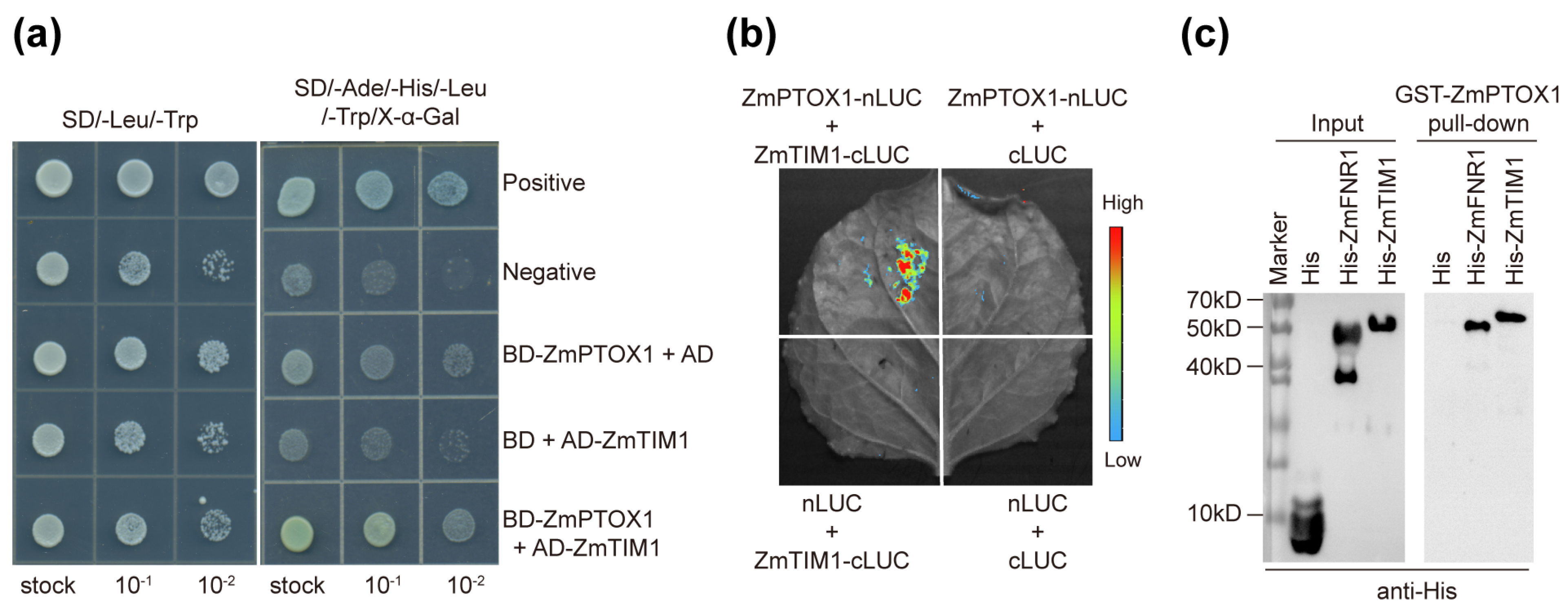
2.5. ZmPTOX1’s Expression Level Regulates Seedling Stress Resistance
3. Discussion
3.1. ZmPTOX1 Is a Novel Zebra Leaf Phenotypic Control Gene with Pleiotropic Effects
3.2. The Zebra Leaf Phenotype of wk3735 Is Co-Regulated by Internal Signaling Pathways and External Stimuli
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction, qRT-PCR, and RNA Sequencing
4.3. Sequence Data Analysis
4.4. Photosynthesis and Gas Exchange Measurements
4.5. Measurement of Starch and Superoxide
4.6. Assays of Protein–Protein Interactions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hines, P.J. Speeding up stomatal responses. Science 2019, 363, 1411–1412. [Google Scholar] [CrossRef]
- Iwashina, T. Contribution to Flower Colors of Flavonoids Including Anthocyanins: A Review. Nat. Prod. Commun. 2015, 10, 529–544. [Google Scholar] [CrossRef]
- Wang, L.; Yue, C.; Cao, H.; Zhou, Y.; Zeng, J.; Yang, Y.; Wang, X. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 2014, 14, 352. [Google Scholar] [CrossRef] [PubMed]
- Pospisil, P. Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta-Bioenerg. 2009, 1787, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K.K. Sensing and Responding to Excess Light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef]
- Wilson-Sanchez, D.; Rubio-Diaz, S.; Munoz-Viana, R.; Manuel Perez-Perez, J.; Jover-Gil, S.; Ponce, M.R.; Micol, J.L. Leaf phenomics: A systematic reverse genetic screen for Arabidopsis leaf mutants. Plant J. 2014, 79, 878–891. [Google Scholar] [CrossRef]
- Fu, M.; Cheng, S.; Xu, F.; Chen, Z.; Liu, Z.; Zhang, W.; Zheng, J.; Wang, L. Advance in mechanism of plant leaf colour mutation. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12071. [Google Scholar] [CrossRef]
- Chai, C.; Fang, J.; Liu, Y.; Tong, H.; Gong, Y.; Wang, Y.; Liu, M.; Wang, Y.; Qian, Q.; Cheng, Z.; et al. ZEBRA2, encoding a carotenoid isomerase, is involved in photoprotection in rice. Plant Mol. Biol. 2011, 75, 211–221. [Google Scholar] [CrossRef]
- Huang, M.; Slewinski, T.L.; Baker, R.F.; Janick-Buckner, D.; Buckner, B.; Johal, G.S.; Braun, D.M. Camouflage Patterning in Maize Leaves Results from a Defect in Porphobilinogen Deaminase. Mol. Plant 2009, 2, 773–789. [Google Scholar] [CrossRef]
- Liu, W.; Cheng, C.; Chen, F.; Ni, S.; Lin, Y.; Lai, Z. High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans). BMC Plant Biol. 2018, 18, 308. [Google Scholar] [CrossRef]
- Nitschke, S.; Cortleven, A.; Iven, T.; Feussner, I.; Havaux, M.; Riefler, M.; Schmuelling, T. Circadian Stress Regimes Affect the Circadian Clock and Cause Jasmonic Acid-Dependent Cell Death in Cytokinin-Deficient Arabidopsis Plants. Plant Cell 2016, 28, 1616–1639. [Google Scholar] [CrossRef] [PubMed]
- Perales, M.; Más, P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 2007, 19, 2111–2123. [Google Scholar] [CrossRef]
- He, Z.; Su, Y.; Wang, T. Full-Length Transcriptome Analysis of Four Different Tissues of Cephalotaxus oliveri. Int. J. Mol. Sci. 2021, 22, 787. [Google Scholar] [CrossRef]
- Gotter, A.L. A Timeless debate: Resolving TIM’s noncircadian roles with possible clock function. Neuroreport 2006, 17, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Yang, D.; Jin, Y.; Liu, X.; Zhang, Z.; Gu, L.; Zhang, H. Genome-Wide Identification of NAP1 and Function Analysis in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2022, 23, 6491. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liang, Z.; Cai, M.; Wang, J.; Li, D.; Chen, Q.; Du, X.; Gu, R.; Wang, G.; Schnable, P.S.; et al. ZmPTOX1, a plastid terminal oxidase, contributes to redox homeostasis during seed development and germination. Plant J. 2024, 119, 460–477. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.D.; Chiu, J.C. Timeless in animal circadian clocks and beyond. FEBS J. 2022, 289, 6559–6575. [Google Scholar] [CrossRef]
- García-Bermúdez, J.; Sánchez-Aragó, M.; Soldevilla, B.; del Arco, A.; Nuevo-Tapioles, C.; Cuezva, J.M. PKA Phosphorylates the ATPase Inhibitory Factor 1 and Inactivates Its Capacity to Bind and Inhibit the Mitochondrial Mitochondrial H+-ATP Synthase. Cell Rep. 2015, 12, 2143–2155. [Google Scholar] [CrossRef]
- Johnson, W.T.; Thomas, A.C. Copper deprivation potentiates oxidative stress in HL-60 cell mitochondria. Proc. Soc. Exp. Biol. Med. 1999, 221, 147–152. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Park, S.-I.; Kim, J.-J.; Shin, S.-Y.; Kwak, S.-S.; Lee, C.-H.; Park, H.-M.; Kim, Y.-H.; Kim, I.-S.; Yoon, H.-S. Over-Expression of Dehydroascorbate Reductase Improves Salt Tolerance, Environmental Adaptability and Productivity in Oryza sativa. Antioxidants 2022, 11, 1077. [Google Scholar] [CrossRef]
- Decena, M.A.; Galvez-Rojas, S.; Agostini, F.; Sancho, R.; Contreras-Moreira, B.; Des Marais, D.L.; Hernandez, P.; Catalan, P. Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses. Plants 2021, 10, 2664. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Li, J.; Yang, X.; Li, W.; Zhou, Z.; Xiao, S.; Xue, C. Iron redistribution induces oxidative burst and resistance in maize against Curvularia lunata. Planta 2022, 256, 46. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.-Y.; Lei, L.; Fan, X.-W.; Li, Y.-Z. Dark response genes: A group of endogenous pendulum/timing players in maize? Planta 2020, 252, 1. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, R.; Chotewutmontri, P.; Belcher, S.; Barkan, A. Correlated retrograde and developmental regulons implicate multiple retrograde signals as coordinators of chloroplast development in maize. Plant Cell 2022, 34, 4897–4919. [Google Scholar] [CrossRef]
- Zhang, T.; Feng, P.; Li, Y.; Yu, P.; Yu, G.; Sang, X.; Ling, Y.; Zeng, X.; Li, Y.; Huang, J.; et al. VIRESCENT-ALBINO LEAF 1 regulates leaf colour development and cell division in rice. J. Exp. Bot. 2018, 69, 4791–4804. [Google Scholar] [CrossRef]
- Liu, X.; Yu, H.; Han, F.; Li, Z.; Fang, Z.; Yang, L.; Zhuang, M.; Lv, H.; Liu, Y.; Li, Z.; et al. Differentially Expressed Genes Associated with the Cabbage Yellow-Green-Leaf Mutant in the ygl-1 Mapping Interval with Recombination Suppression. Int. J. Mol. Sci. 2018, 19, 2936. [Google Scholar] [CrossRef]
- Zhang, S.; Tang, S.; Tang, C.; Luo, M.; Jia, G.; Zhi, H.; Diao, X. SiSTL2 Is Required for Cell Cycle, Leaf Organ Development, Chloroplast Biogenesis, and Has Effects on C4 Photosynthesis in Setaria italica (L.) P. Beauv. Front. Plant Sci. 2018, 9, 1103. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, W.; Zhang, Y.; Sun, S.; Wang, L.; Zhong, S.; Zhao, X.; Liu, B. PPR647 Protein Is Required for Chloroplast RNA Editing, Splicing and Chloroplast Development in Maize. Int. J. Mol. Sci. 2021, 22, 11162. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; She, G.; Zhang, X.; Jordan, B.; Chen, Q.; Zhao, J.; Wan, X. Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants (Camellia sinensis) in response to shading. BMC Plant Biol. 2018, 18, 233. [Google Scholar] [CrossRef]
- Shi, D.; Zheng, X.; Li, L.; Lin, W.; Xie, W.; Yang, J.; Chen, S.; Jin, W. Chlorophyll Deficiency in the Maize elongated mesocotyl2 Mutant Is Caused by a Defective Heme Oxygenase and Delaying Grana Stacking. PLoS ONE 2013, 8, e80107. [Google Scholar] [CrossRef]
- Asakura, Y.; Hirohashi, T.; Kikuchi, S.; Belcher, S.; Osborne, E.; Yano, S.; Terashima, I.; Barkan, A.; Nakai, M. Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 2004, 16, 201–214. [Google Scholar] [CrossRef]
- Guan, Z.; Wang, W.; Yu, X.; Lin, W.; Miao, Y. Comparative Proteomic Analysis of Coregulation of CIPK14 and WHIRLY1/3 Mediated Pale Yellowing of Leaves in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2231. [Google Scholar] [CrossRef]
- Sawers, R.J.H.; Linley, P.J.; Farmer, P.R.; Hanley, N.P.; Costich, D.E.; Terry, M.J.; Brutnell, T.P. elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol. 2002, 130, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Dhami, N.; Pogson, B.J.; Tissue, D.T.; Cazzonelli, C.I. A foliar pigment-based bioassay for interrogating chloroplast signalling revealed that carotenoid isomerisation regulates chlorophyll abundance. Plant Methods 2022, 18, 18. [Google Scholar] [CrossRef] [PubMed]
- Stepien, P.; Johnson, G.N. Plastid terminal oxidase requires translocation to the grana stacks to act as a sink for electron transport. Proc. Natl. Acad. Sci. USA 2018, 115, 9634–9639. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, X.; Xie, J.; Ding, W.; Li, Y.; Yue, Y.; Wang, L. Biochemical and Comparative Transcriptome Analyses Reveal Key Genes Involved in Major Metabolic Regulation Related to Colored Leaf Formation in Osmanthus fragrans ‘Yinbi Shuanghui’ during Development. Biomolecules 2020, 10, 549. [Google Scholar] [CrossRef]
- Feng, P.; Shi, J.; Zhang, T.; Zhong, Y.; Zhang, L.; Yu, G.; Zhang, T.; Zhu, X.; Xing, Y.; Yin, W.; et al. Zebra leaf 15, a receptor-like protein kinase involved in moderate low temperature signaling pathway in rice. Rice 2019, 12, 83. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Z.; Gu, H.; You, J.; Hu, M.; Zhang, Y.; Zhu, Z.; Wang, Y.; Liu, S.; Chen, L.; et al. Identification and Phenotypic Characterization of ZEBRA LEAF16 Encoding a β-Hydroxyacyl-ACP Dehydratase in Rice. Front. Plant Sci. 2018, 9, 782. [Google Scholar] [CrossRef]
- Qiu, Y.; Li, H.; Peng, K.; Liu, Z.; Chen, S.; Liu, R.; Liang, M.; Chen, L. Observation of chloroplast ultrastructure of zebra leaf in rice mutant B411. Acta Agron. Sin. 2010, 36, 184–190. [Google Scholar] [CrossRef]
- Lu, X.; Hu, X.; Zhao, Y.; Song, W.; Zhang, M.; Chen, Z.; Chen, W.; Dong, Y.; Wang, Z.; Lai, J. Map-Based Cloning of zb7 Encoding an IPP and DMAPP Synthase in the MEP Pathway of Maize. Mol. Plant 2012, 5, 1100–1112. [Google Scholar] [CrossRef]
- Yuan, G.; Li, Y.; Chen, B.; He, H.; Wang, Z.; Shi, J.; Yang, Y.; Zou, C.; Pan, G. Identification and fine mapping of a recessive gene controlling zebra leaf phenotype in maize. Mol. Breed. 2021, 41, 9. [Google Scholar] [CrossRef]
- Wang, D.; Liu, J.; Li, C.; Kang, H.; Wang, Y.; Tan, X.; Liu, M.; Deng, Y.; Wang, Z.; Liu, Y.; et al. Genome-wide Association Mapping of Cold Tolerance Genes at the Seedling Stage in Rice. Rice 2016, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, W.; Liao, X.; Xiong, C.; Yu, G.; Yang, Q.; Yang, C.; Ye, Z. The tomato WV gene encoding a thioredoxin protein is essential for chloroplast development at low temperature and high light intensity. BMC Plant Biol. 2019, 19, 265. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.Y.; Wright, D.A.; Wetzel, C.; Voytas, D.F.; Rodermel, S. The immutans variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 1999, 11, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Murakami, K.; Ikawa, H. Trade-off between grain yield and protein concentration is modulated by canopy photosynthesis in Japanese wheat cultivars. bioRxiv 2022. [Google Scholar] [CrossRef]
- Nagata, T. Strontium-induced mineral imbalance, cell death, and reactive oxygen species generation in Arabidopsis thaliana. Plant Root 2023, 17, 36–44. [Google Scholar] [CrossRef]
- Kong, L.; Huo, H.; Mao, P. Antioxidant response and related gene expression in aged oat seed. Front. Plant Sci. 2015, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Liao, X.; Yang, X.; Luo, Y.; Lin, P.; Zeng, Q.; Bai, H.; Jiang, B.; Pan, Y.; Zhang, F.; et al. Lysine crotonylation of DgTIL1 at K72 modulates cold tolerance by enhancing DgnsLTP stability in chrysanthemum. Plant Biotechnol. J. 2021, 19, 1125–1140. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ou, S.; Mao, B.; Tang, J.; Wang, W.; Wang, H.; Cao, S.; Schläppi, M.R.; Zhao, B.; Xiao, G.; et al. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat. Commun. 2018, 9, 3302. [Google Scholar] [CrossRef] [PubMed]


| Sample Name | Group | Rep. | Total Reads | Rate of Total Mapped Reads (%) | Rate of Uniquely Mapped Reads (%) | Num. of Expressed Genes | Rate of Expressed Genes (%) |
|---|---|---|---|---|---|---|---|
| OE1 | OE | 1 | 26,921,898 | 98.22 | 65.09 | 17,678 | 38.21 |
| OE2 | OE | 2 | 22,588,229 | 97.67 | 82.77 | 17,518 | 37.86 |
| OE3 | OE | 3 | 11,001,271 | 97.27 | 82.14 | 17,517 | 37.86 |
| mu1G | muG | 1 | 24,837,626 | 97.54 | 84.64 | 18,324 | 39.6 |
| mu2G | muG | 2 | 22,968,295 | 97.56 | 78.88 | 18,388 | 39.74 |
| mu3G | muG | 3 | 22,426,755 | 97.69 | 84.3 | 18,375 | 39.71 |
| mu1W | muW | 1 | 27,333,115 | 97.47 | 83.26 | 18,992 | 41.05 |
| mu2W | muW | 2 | 25,006,640 | 97.2 | 85.83 | 19,161 | 41.41 |
| mu3W | muW | 3 | 24,363,229 | 97.47 | 83.87 | 18,963 | 40.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Liang, Z.; Qing, X.; Wen, M.; Yuan, Z.; Chen, Q.; Du, X.; Gu, R.; Wang, J.; Li, L. Transcriptome Analysis Revealed ZmPTOX1 Is Required for Seedling Development and Stress Tolerance in Maize. Plants 2024, 13, 2346. https://doi.org/10.3390/plants13172346
Peng Y, Liang Z, Qing X, Wen M, Yuan Z, Chen Q, Du X, Gu R, Wang J, Li L. Transcriptome Analysis Revealed ZmPTOX1 Is Required for Seedling Development and Stress Tolerance in Maize. Plants. 2024; 13(17):2346. https://doi.org/10.3390/plants13172346
Chicago/Turabian StylePeng, Yixuan, Zhi Liang, Xindong Qing, Motong Wen, Zhipeng Yuan, Quanquan Chen, Xuemei Du, Riliang Gu, Jianhua Wang, and Li Li. 2024. "Transcriptome Analysis Revealed ZmPTOX1 Is Required for Seedling Development and Stress Tolerance in Maize" Plants 13, no. 17: 2346. https://doi.org/10.3390/plants13172346
APA StylePeng, Y., Liang, Z., Qing, X., Wen, M., Yuan, Z., Chen, Q., Du, X., Gu, R., Wang, J., & Li, L. (2024). Transcriptome Analysis Revealed ZmPTOX1 Is Required for Seedling Development and Stress Tolerance in Maize. Plants, 13(17), 2346. https://doi.org/10.3390/plants13172346








