Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase Is Required in Bradyrhizobium diazoefficiens for Efficient Soybean Root Colonization and Competition for Nodulation
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Relationships of Housekeeping Genes and the Cbb Operon in Hyphomicrobiales
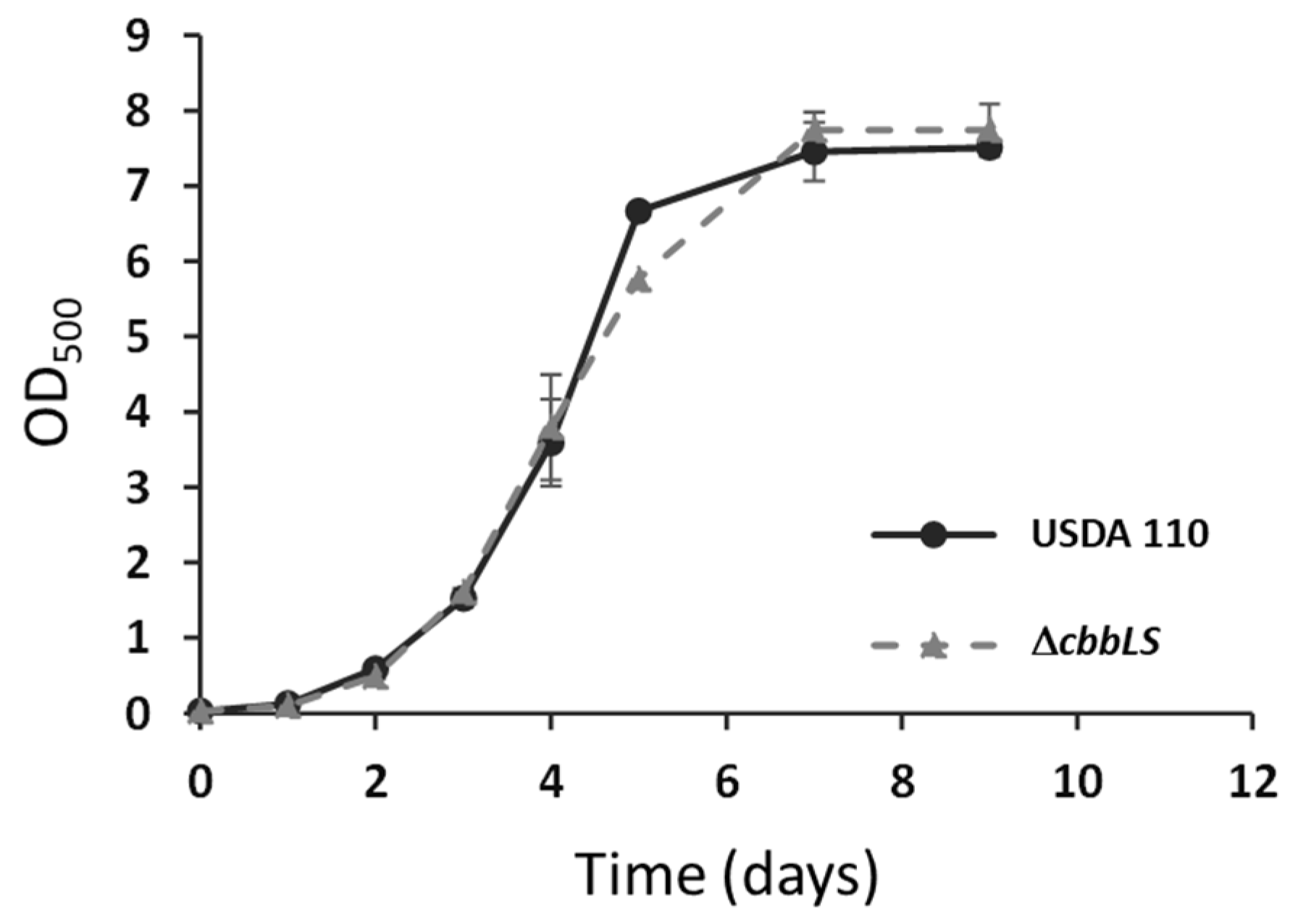
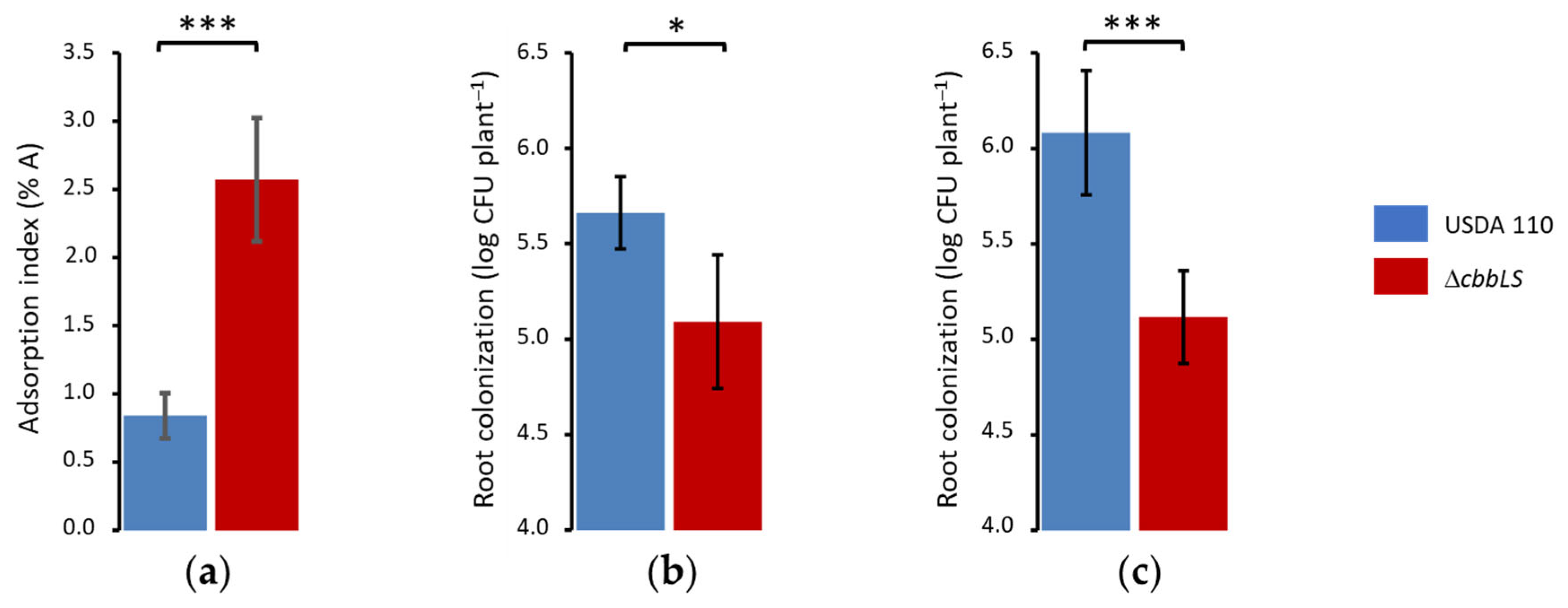
2.2. Different Effects of ΔcbbLS Mutation on Early and Late Root Adhesion and Colonization
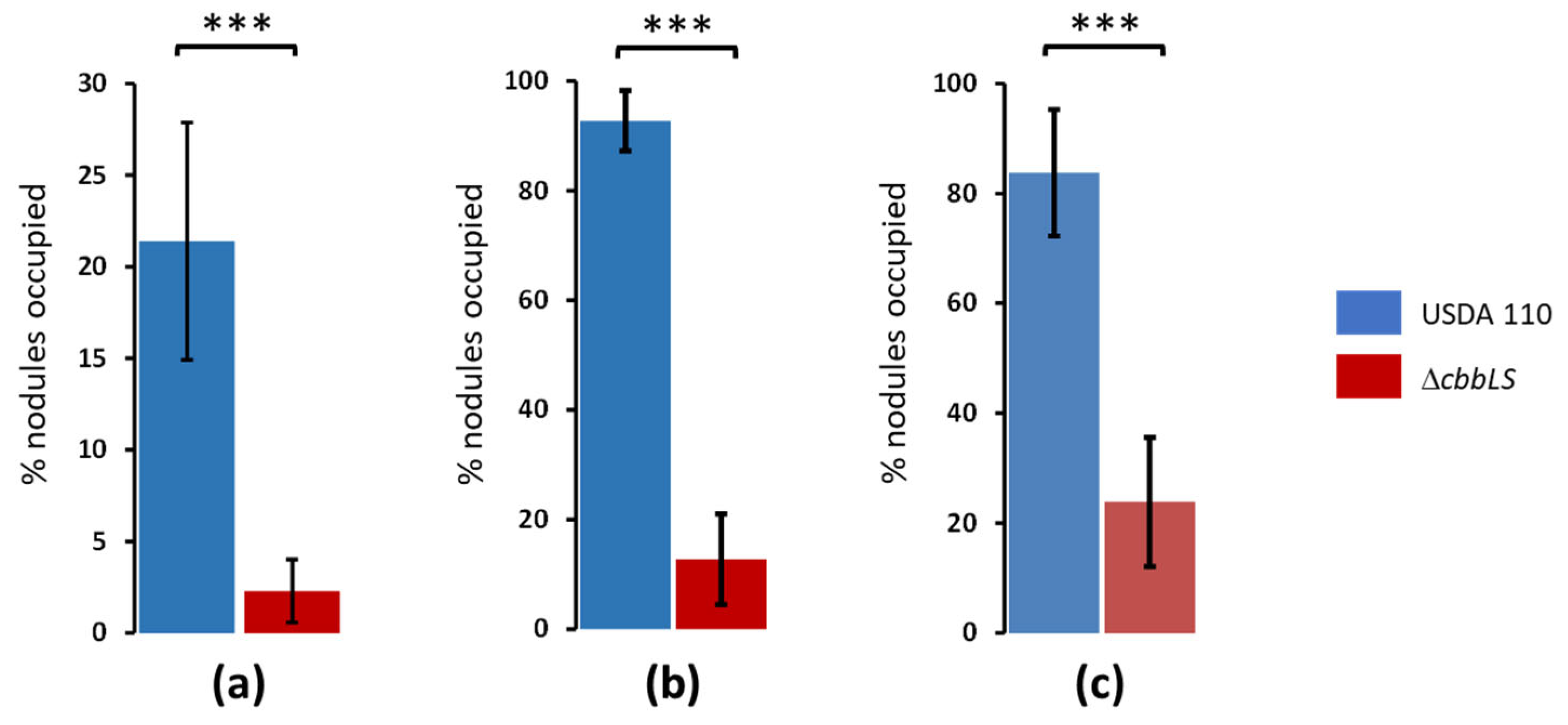
2.3. ΔcbbLS Mutation Affects Competition for Nodulation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Construction of the ΔcbbLS Mutant
4.3. Sequence Comparisons and Construction of Phylogenetic Trees
4.4. Plant Assays
4.5. Extracellular Polysaccharide Determinations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delamuta, J.R.M.; Ribeiro, R.A.; Ormeño-Orrillo, E.; Melo, I.S.; Martínez-Romero, E.; Hungria, M. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3342–3351. [Google Scholar] [CrossRef] [PubMed]
- Aroney, S.T.N.; Poole, P.S.; Sánchez-Cañizares, C. Rhizobial chemotaxis and motility systems at work in the soil. Front. Plant Sci. 2021, 12, e725338. [Google Scholar] [CrossRef]
- Keyser, H.H.; van Berkum, P.; Weber, D.F. A comparative study of the physiology of symbioses formed by Rhizobium japonicum with Glycine max, Vigna unguiculata, and Macroptilium atropurpurem. Plant Physiol. 1982, 70, 1626–1630. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.; Hennecke, H. Cloning and mapping of a novel nodulation region from Bradyrhizobium japonicum by genetic complementation of a deletion mutant. Appl. Environ. Microbiol. 1988, 54, 55–61. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Minamisawa, K.; Uchiumi, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Iriguchi, M.; Kawashima, K.; et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef]
- Badger, M.R.; Bek, E.J. Multiple Rubisco forms in proteobacteria: Their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008, 59, 1525–1541. [Google Scholar] [CrossRef]
- Novak, J.S.; Tabita, F.R. Molecular approaches to probe differential NADH activation of phosphoribulokinase isozymes from Rhodobacter sphaeroides. Arch. Biochem. Biophys. 1999, 363, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Cajar, O.; Stotz, M.; Wendler, P.; Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 2011, 479, 194–199. [Google Scholar] [CrossRef]
- Yoo, J.G.; Bowien, B. Analysis of the cbbF genes from Alcaligenes eutrophus that encode fructose-1,6-/sedoheptulose-1,7-bisphosphatase. Curr. Microbiol. 1995, 31, 55–61. [Google Scholar] [CrossRef]
- Schäferjohann, J.; Yoo, J.G.; Kusian, B.; Bowien, B. The cbb operons of the facultative chemoautotroph Alcaligenes eutrophus encode phosphoglycolate phosphatase. J. Bacteriol. 1993, 175, 7329–7340. [Google Scholar] [CrossRef]
- van den Bergh, E.R.; Baker, S.C.; Raggers, R.J.; Terpstra, P.; Woudstra, E.C.; Dijkhuizen, L.; Meijer, W.G. Primary structure and phylogeny of the Calvin cycle enzymes transketolase and fructosebisphosphate aldolase of Xanthobacter flavus. J. Bacteriol. 1996, 178, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Lyngstadaas, A.; Sprenger, G.A.; Boye, E. Impaired growth of an Escherichia coli rpe mutant lacking ribulose-5-phosphate epimerase activity. Biochim. Biophys. Acta 1998, 1381, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Kusian, B.; Bowien, B. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 1997, 21, 135–155. [Google Scholar] [CrossRef]
- Dangel, A.W.; Tabita, F.R. CbbR, the master regulator for microbial carbon dioxide fixation. J. Bacteriol. 2015, 197, 3488–3498. [Google Scholar] [CrossRef]
- Delwiche, C.F.; Palmer, J.D. Rampant horizontal transfer and duplication of Rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 1996, 13, 873–882. [Google Scholar] [CrossRef]
- Jaffe, A.L.; Castelle, C.J.; Dupont, C.L.; Banfield, J.F. Lateral gene transfer shapes the distribution of RuBisCO among Candidate Phyla Radiation bacteria and DPANN archaea. Mol. Biol. Evol. 2018, 36, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Asplund-Samuelsson, J.; Hudson, E.P. Wide range of metabolic adaptations to the acquisition of the Calvin cycle revealed by comparison of microbial genomes. PLoS Comput. Biol. 2021, 17, e1008742. [Google Scholar] [CrossRef]
- Lepo, J.E.; Hanus, F.J.; Evans, H.J. Chemoautotrophic growth of hydrogen-uptake-positive strains of Rhizobium japonicum. J. Bacteriol. 1980, 141, 664–670. [Google Scholar] [CrossRef]
- Franck, W.L.; Chang, W.S.; Qiu, J.; Sugawara, M.; Sadowsky, M.J.; Smith, S.A.; Stacey, G. Whole-genome transcriptional profiling of Bradyrhizobium japonicum during chemoautotrophic growth. J. Bacteriol. 2008, 190, 6697–6705. [Google Scholar] [CrossRef]
- Masuda, S.; Eda, S.; Sugawara, C.; Mitsui, H.; Minamisawa, K. The cbbL gene is required for thiosulfate-dependent autotrophic growth of Bradyrhizobium japonicum. Microbes Environ. 2010, 25, 220–223. [Google Scholar] [CrossRef]
- Cogo, C.; Pérez-Giménez, J.; Rajeswari, C.B.; Luna, M.F.; Lodeiro, A.R. Induction by Bradyrhizobium diazoefficiens of different pathways for growth in D-mannitol or L-arabinose leading to pronounced differences in CO2 fixation, O2 consumption, and lateral-flagellum production. Front. Microbiol. 2018, 9, e1189. [Google Scholar] [CrossRef] [PubMed]
- Gourion, B.; Delmotte, N.; Bonaldi, K.; Nouwen, N.; Vorholt, J.A.; Giraud, E. Bacterial RuBisCO is required for efficient Bradyrhizobium/Aeschynomene symbiosis. PLoS ONE 2011, 6, e21900. [Google Scholar] [CrossRef]
- Lodeiro, A.R.; Favelukes, G. Early interactions of Bradyrhizobium japonicum and soybean roots: Specificity in the process of adsorption. Soil Biol. Biochem. 1999, 31, 1405–1411. [Google Scholar] [CrossRef]
- López-García, S.L.; Vázquez, T.E.; Favelukes, G.; Lodeiro, A.R. Improved soybean root association of N-starved Bradyrhizobium japonicum. J. Bacteriol. 2001, 183, 7241–7252. [Google Scholar] [CrossRef] [PubMed]
- Dazzo, F.B.; Truchet, G.L.; Sherwood, J.E.; Hrabak, E.M.; Abe, M.; Pankratz, S.H. Specific phases of root hair attachment in the Rhizobium trifolii-clover symbiosis. Appl. Environ. Microbiol. 1984, 48, 1140–1150. [Google Scholar] [CrossRef]
- Robledo, M.; Rivera, L.; Jiménez-Zurdo, J.I.; Rivas, R.; Dazzo, F.; Velázquez, E.; Martínez-Molina, E.; Hirsch, A.M.; Mateos, P.F. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb. Cell Fact. 2012, 12, 125. [Google Scholar] [CrossRef]
- López-García, S.L.; Vázquez, T.E.; Favelukes, G.; Lodeiro, A.R. Rhizobial position as a main determinant in the problem of competition for nodulation in soybean. Environ. Microbiol. 2002, 4, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Lozano, M.J.; Redondo-Nieto, M.; Garrido-Sanz, D.; Mongiardini, E.; Quelas, J.I.; Mengucci, F.; Dardis, C.; Lodeiro, A.; Althabegoiti, M.J. Comparative analysis of three Bradyrhizobium diazoefficiens genomes show specific mutations acquired during selection for a higher motility phenotype and adaption to laboratory conditions. Microbiol. Spectr. 2021, 9, e0056921. [Google Scholar] [CrossRef]
- Vasil’eva, L.V.; Semenov, A.M. Labrys monahos, a new budding prosthecate bacterium with radial cell symmetry. Microbiology 1984, 53, 68–75. [Google Scholar]
- Rangel, W.M.; de Oliveira Longatti, S.M.; Ferreira, P.A.A.; Bonaldi, D.S.; Guimarães, A.A.; Thijs, S.; Weyens, N.; Vangronsveld, J.; Moreira, F.M.S. Leguminosae native nodulating bacteria from a gold mine As-contaminated soil: Multi-resistance to trace elements, and possible role in plant growth and mineral nutrition. Int. J. Phytoremed. 2017, 19, 925–936. [Google Scholar] [CrossRef]
- Chou, Y.-J.; Elliott, G.N.; James, E.K.; Lin, K.-Y.; Chou, J.-H.; Sheu, S.-Y.; Sheu, D.-S.; Sprent, J.I.; Chen, W.-M. Labrys neptuniae sp. nov., isolated from root nodules of the aquatic legume Neptunia oleraceaInt. J. Syst. Evol. Microbiol. 2007, 57, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Passari, A.K.; Mishra, V.K.; Leo, V.V.; Gupta, V.K.; Singh, B.P. Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol. Res. 2016, 193, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.S.; Zianni, M.; Bobst, C.E.; Tabita, F.R. Regulatory twist and synergistic role of metabolic coinducer- and response regulator-mediated CbbR-cbbI interactions in Rhodopseudomonas palustris CGA010. J. Bacteriol. 2013, 195, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Mongiardini, E.J.; Parisi, G.D.; Quelas, J.I.; Lodeiro, A.R. The tight-adhesion proteins TadGEF of Bradyrhizobium diazoefficiens USDA 110 are involved in cell adhesion and infectivity on soybean roots. Microbiol. Res. 2016, 182, 80–88. [Google Scholar] [CrossRef]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.M.; Hennecke, H. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef]
- Delmotte, N.; Ahrens, C.H.; Knief, C.; Qeli, E.; Koch, M.; Fischer, H.M.; Vorholt, J.A.; Hennecke, H.; Pessi, G. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 2010, 10, 1391–1400. [Google Scholar] [CrossRef]
- Franck, S.; Strodtman, K.N.; Qiu, J.; Emerich, D.W. Transcriptomic characterization of Bradyrhizobium diazoefficiens bacteroids reveals a post-symbiotic, hemibiotrophic-like lifestyle of the bacteria within senescing soybean nodules. Int. J. Mol. Sci. 2018, 19, 3918. [Google Scholar] [CrossRef]
- Barbour, W.M.; Hattermann, D.R.; Stacey, G. Chemotaxis of Bradyrhizobium japonicum to soybean exudates. Appl. Environ. Microbiol. 1991, 57, 2635–2639. [Google Scholar] [CrossRef]
- Timotiwu, P.B.; Sakurai, N. Identification of mono-, oligo-, and polysaccharides secreted from soybean roots. J. Plant Res. 2002, 115, 77–85. [Google Scholar] [CrossRef]
- Yaryura, P.M.; León, M.; Correa, O.S.; Kerber, N.L.; Pucheu, N.L.; García, A.F. Assessment of the role of chemotaxis and biofilm formation as requirements for colonization of roots and seeds of soybean plants by Bacillus amyloliquefaciens BNM339. Curr. Microbiol. 2008, 56, 625–632. [Google Scholar] [CrossRef]
- Pérez-Giménez, J.; Lodeiro, A.R. Two effects of combined nitrogen on the adhesion of Rhizobium etli to bean roots. Symbiosis 2013, 59, 157–163. [Google Scholar] [CrossRef]
- Rocha, R.O.; Morais, J.K.S.; Oliveira, J.T.A.; Oliveira, H.D.; Sousa, D.O.B.; Souza, C.E.A.; Moreno, F.B.; Monteiro-Moreira, A.C.O.; de Souza Júnior, J.D.A.; Grossi de Sá, M.F.; et al. Proteome of soybean seed exudates contains plant defense-related proteins active against the root-knot nematode Meloidogyne incognita. J. Agric. Food Chem. 2015, 63, 5335–5343. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, A. The soybean rhizosphere: Metabolites, microbes, and beyond—A review. J. Adv. Res. 2019, 19, 67–73. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Mongiardini, E.J.; Lodeiro, A.R. IBBM, UNLP and CCT-La Plata CONICET. La Plata, Argentina. 2023; manuscript in preparation. [Google Scholar]
- Sugiyama, A.; Yamazaki, Y.; Yamashita, K.; Takahashi, S.; Nakayama, T.; Yazaki, K. Developmental and nutritional regulation of isoflavone secretion from soybean roots. Biosci. Biotechnol. Biochem. 2016, 80, 89–94. [Google Scholar] [CrossRef]
- Aufrecht, J.; Khalid, M.; Walton, C.L.; Tate, K.; Cahill, J.F.; Retterer, S.T. Hotspots of root-exuded amino acids are created within a rhizosphere-on-a-chip. Lab Chip 2022, 22, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.N.; Schachtman, D.P. Root exudates impact plant performance under abiotic stress. Trends Plant Sci. 2022, 27, 80–91. [Google Scholar] [CrossRef]
- Walton, C.L.; Khalid, M.; Bible, A.N.; Kertesz, V.; Retterer, S.T.; Morrell-Falvey, J.; Cahill, J.F. In situ detection of amino acids from bacterial biofilms and plant root exudates by liquid microjunction surface-sampling probe mass spectrometry. J. Am. Soc. Mass Spectrom. 2022, 33, 1615–1625. [Google Scholar] [CrossRef]
- Blossfeld, S.; Schreiber, C.M.; Liebsch, G.; Kuhn, A.J.; Hinsinger, P. Quantitative imaging of rhizosphere pH and CO2 dynamics with planar optodes. Ann. Bot. 2013, 112, 267–276. [Google Scholar] [CrossRef]
- Bereswill, S.; Rudolph-Mohr, N.; Oswald, S.E. Imaging rhizosphere CO2 and O2 concentration to localize respiration hotspots linked to root type and soil moisture dynamics. Research Square, 2021. Available online: https://assets-eu.researchsquare.com/files/rs-1145795/v1/298bfe92-9310-4757-ae95-ffdb625e2107.pdf?c=1648417512 (accessed on 11 June 2024).
- Vincent, J.M. A Manual for the Practical Study of the Root Nodule Bacteria; IBP Handbook 15; Blackwell Scientific Publications: Oxford, UK, 1970. [Google Scholar] [CrossRef]
- Regensburger, B.; Hennecke, H. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 1983, 135, 103–109. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Mengucci, F.; Dardis, C.; Mongiardini, E.J.; Althabegoiti, M.J.; Partridge, J.D.; Kojima, S.; Homma, M.; Quelas, J.I.; Lodeiro, A.R. Characterization of FliL proteins in Bradyrhizobium diazoefficiens: Lateral FliL supports swimming motility, and subpolar FliL modulates the lateral flagellar system. J. Bacteriol. 2020, 202, e00708-19. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, e421. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Lodeiro, A.R.; González, P.; Hernández, A.; Balagué, L.J.; Favelukes, G. Comparison of drought tolerance in nitrogen-fixing and inorganic nitrogen-grown common beans. Plant Sci. 2000, 154, 31–41. [Google Scholar] [CrossRef]
- Althabegoiti, M.J.; Covelli, J.M.; Pérez-Giménez, J.; Quelas, J.I.; Mongiardini, E.J.; López, M.F.; López-García, S.L.; Lodeiro, A.R. Analysis of the role of the two flagella of Bradyrhizobium japonicum in competition for nodulation of soybean. FEMS Microbiol. Lett. 2011, 319, 133–139. [Google Scholar] [CrossRef]
- Brignoli, D.; Frickel-Critto, E.; Sandobal, T.J.; Balda, R.S.; Castells, C.B.; Mongiardini, E.J.; Pérez-Giménez, J.; Lodeiro, A.R. Quality control of Bradyrhizobium inoculant strains: Detection of nosZ and correlation of symbiotic efficiency with soybean leaf chlorophyll levels. Front. Agron. 2024, 6, 1336433. [Google Scholar] [CrossRef]


| Primer | Restriction Site | Sequence |
|---|---|---|
| FwUp_RBC | EcoRI | AAAAGAATTCGCTATTGGGAGCCCGACTAC |
| RvUp_RBC | BamHI | GCCGTCGACGGATCCGAGGCAAACACGTTGCCGATGATCG |
| FwDw_RBC | BamHI | TGCCTCGGATCCGTCGACGGCATGAAACTGACCCAGGGCTG |
| RvDw_RBC | HindIII | AAAAAAGCTTAAGGAGATTCGCACCGACTC |
| FwExt_RBC | GCCGCCTAGTCATTCAGAGA | |
| RvExt_RBC | ACGGTGGTGTAGCGGATTG | |
| M13 Fw | GTAAAACGACGGCCAGT | |
| M13 Rv | GCGGATAACAATTTCACACAGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balda, R.S.; Cogo, C.; Falduti, O.; Bongiorno, F.M.; Brignoli, D.; Sandobal, T.J.; Althabegoiti, M.J.; Lodeiro, A.R. Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase Is Required in Bradyrhizobium diazoefficiens for Efficient Soybean Root Colonization and Competition for Nodulation. Plants 2024, 13, 2362. https://doi.org/10.3390/plants13172362
Balda RS, Cogo C, Falduti O, Bongiorno FM, Brignoli D, Sandobal TJ, Althabegoiti MJ, Lodeiro AR. Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase Is Required in Bradyrhizobium diazoefficiens for Efficient Soybean Root Colonization and Competition for Nodulation. Plants. 2024; 13(17):2362. https://doi.org/10.3390/plants13172362
Chicago/Turabian StyleBalda, Rocío S., Carolina Cogo, Ornella Falduti, Florencia M. Bongiorno, Damián Brignoli, Tamara J. Sandobal, María Julia Althabegoiti, and Aníbal R. Lodeiro. 2024. "Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase Is Required in Bradyrhizobium diazoefficiens for Efficient Soybean Root Colonization and Competition for Nodulation" Plants 13, no. 17: 2362. https://doi.org/10.3390/plants13172362
APA StyleBalda, R. S., Cogo, C., Falduti, O., Bongiorno, F. M., Brignoli, D., Sandobal, T. J., Althabegoiti, M. J., & Lodeiro, A. R. (2024). Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase Is Required in Bradyrhizobium diazoefficiens for Efficient Soybean Root Colonization and Competition for Nodulation. Plants, 13(17), 2362. https://doi.org/10.3390/plants13172362








