Biological Decline of Alfalfa Is Accompanied by Negative Succession of Rhizosphere Soil Microbial Communities
Abstract
:1. Introduction
2. Results
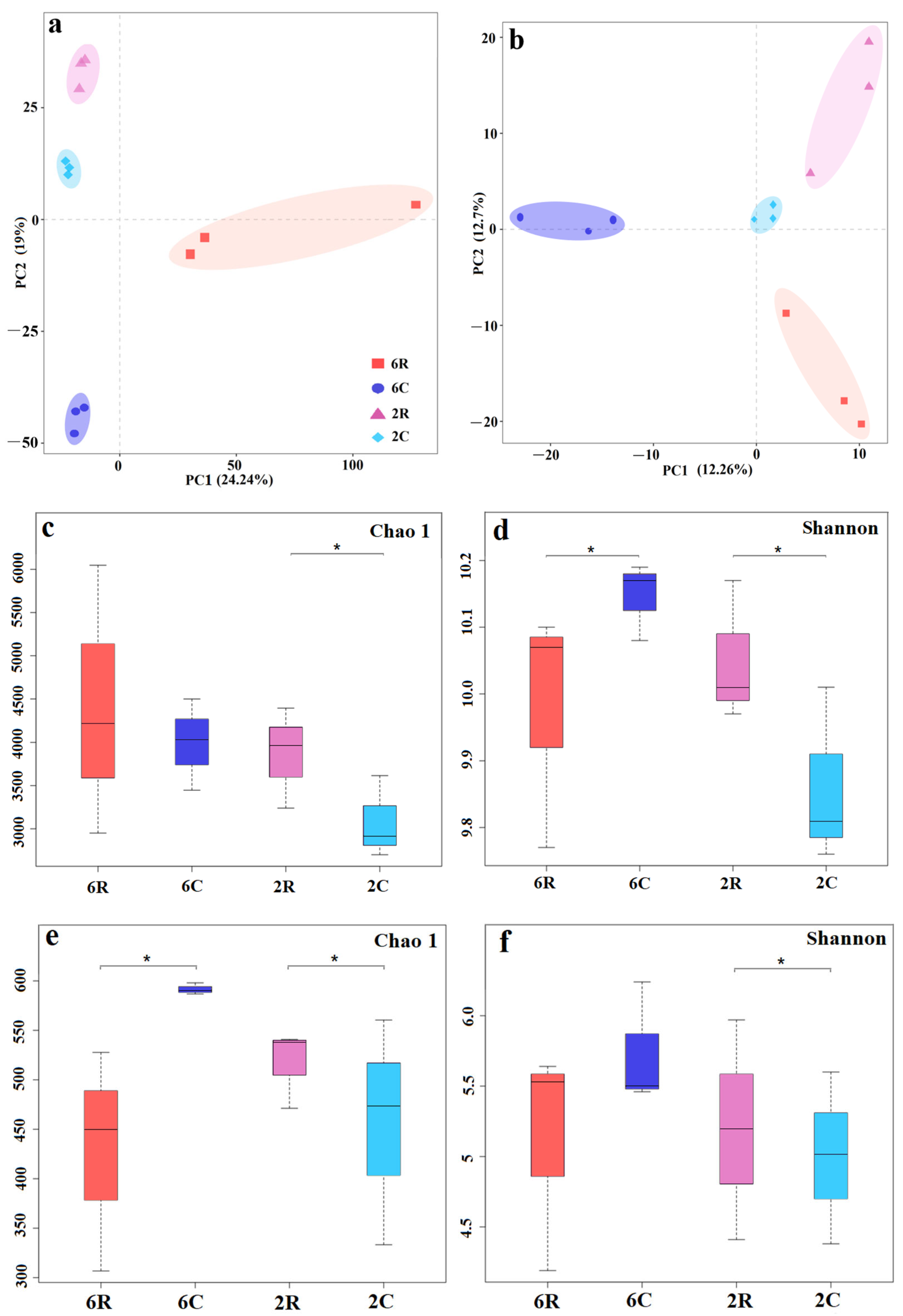
2.1. Rhizosphere Microbial Community Diversity
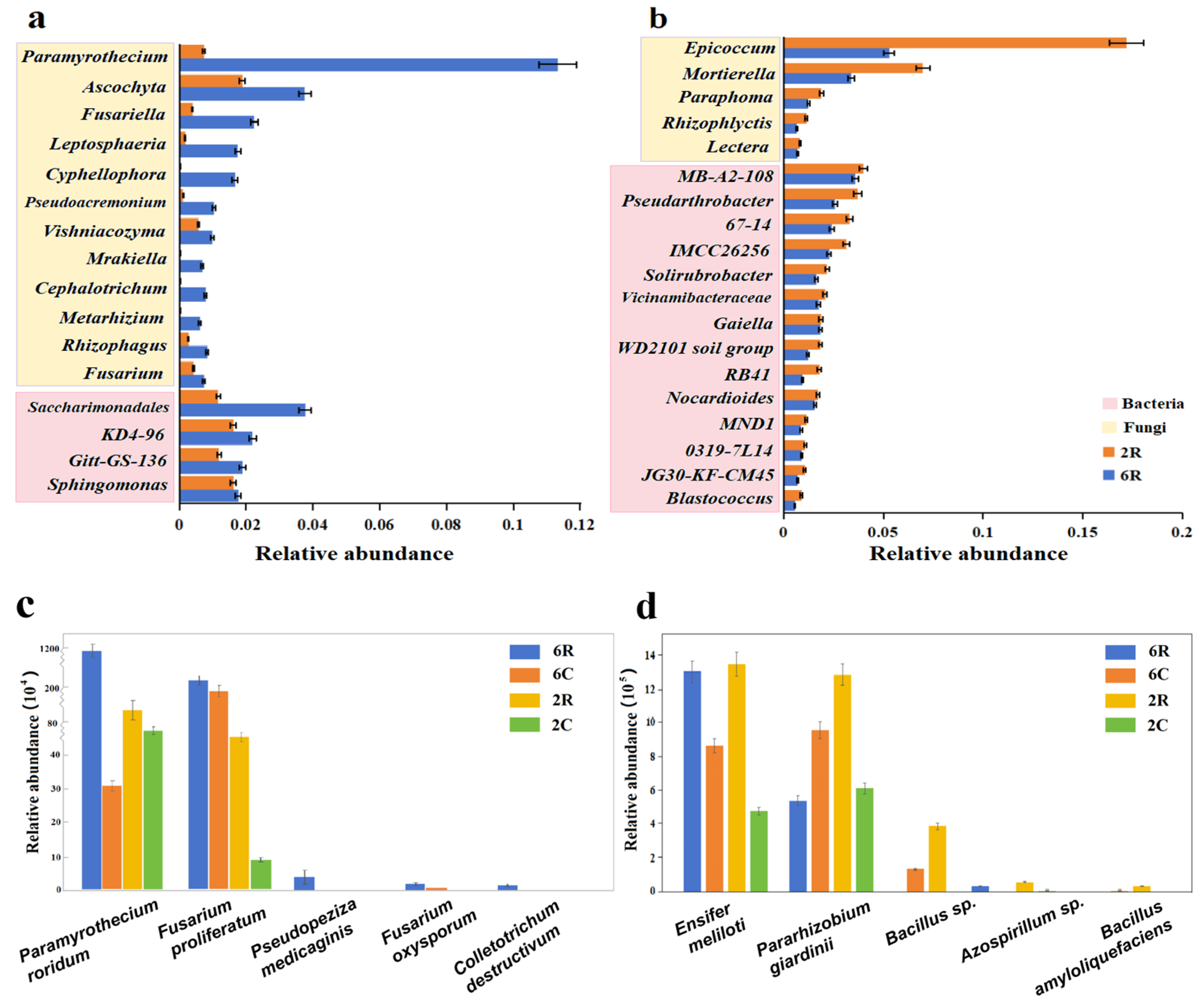
2.2. Identification of Rhizosphere-Specific Microbial Taxa
2.3. Regulation of Rhizosphere Microbial Communities by Root Activity
2.4. Effects of Stand Age on Rhizosphere Microbial Communities
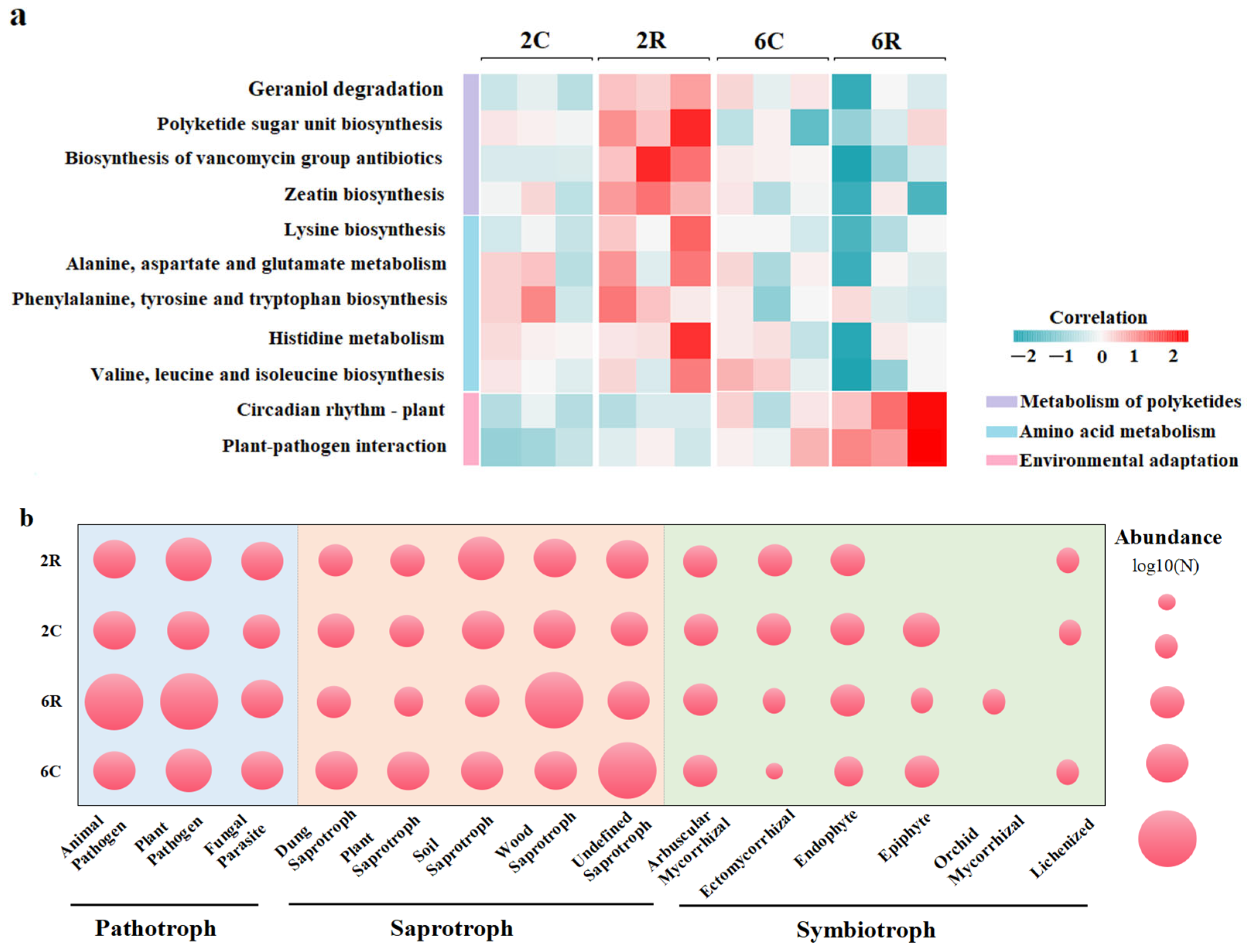
2.5. Predicted Functions of Rhizosphere Microbial Communities
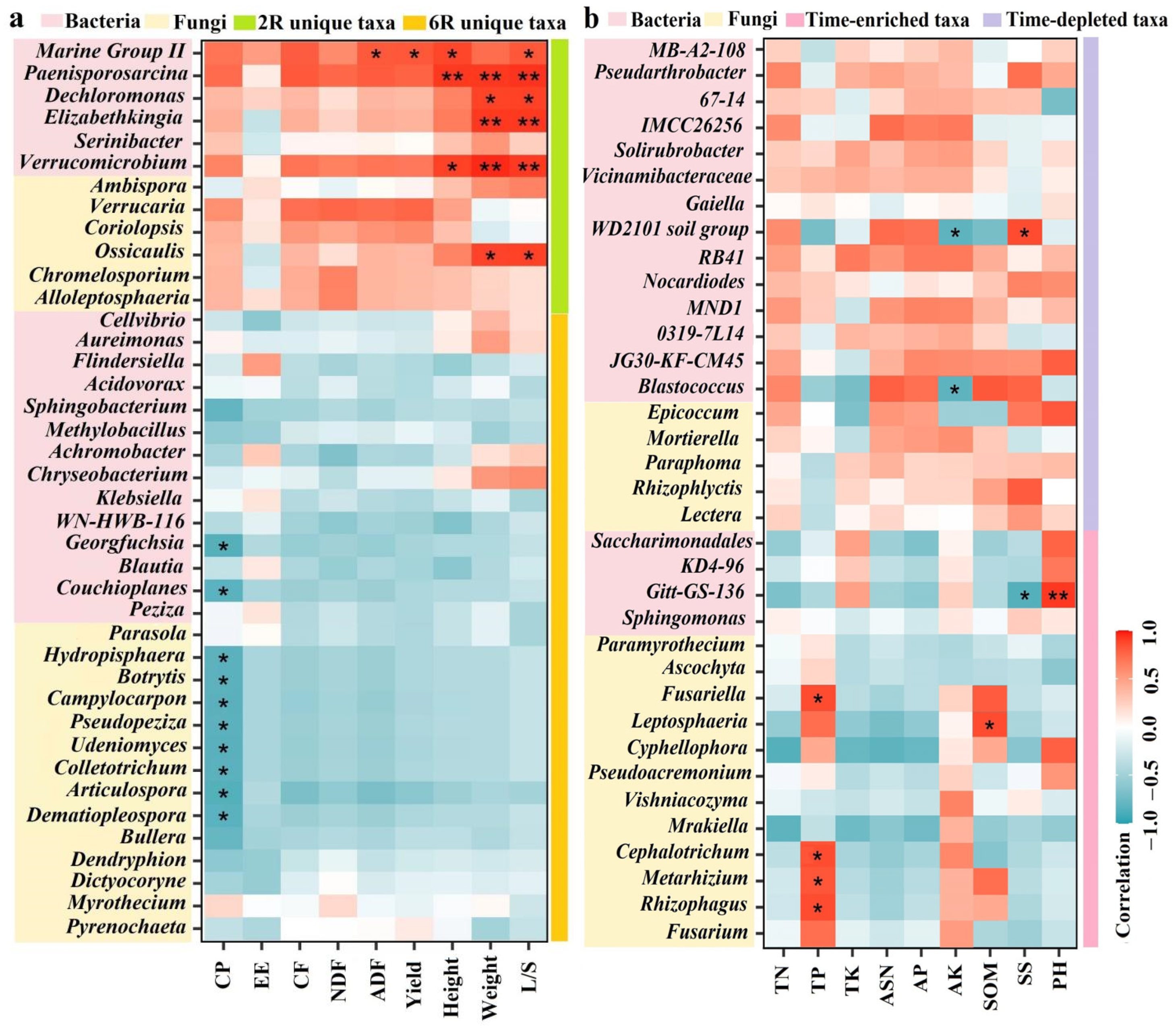
2.6. Linking Rhizosphere Microbial Taxa to Plant Traits and Soil Properties
3. Discussion
3.1. Variations in Rhizosphere-Specific Microbial Communities with Stand Age
3.2. Selective Promotion and Inhibition of Rhizosphere Microbial Taxa by Root Activity
3.3. Rhizosphere Microbial Community Succession in Relation to Alfalfa Decline
3.4. Rhizosphere Microbial Community Function Associated with Alfalfa Decline
4. Materials and Methods
4.1. Study Area
4.2. Soil Sampling and Chemical Analysis
4.3. Soil Microbial Community Analysis
4.4. Plant Sampling and Analysis
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, S.; Lehmann, M.; Zhang, Z.; Subedi, U.; Burton, H.; Lim, N.; Ortega, P.; Chen, G.; Acharya, S.; Hannoufa, A. Elucidation of physiological, transcriptomic and metabolomic salinity response mechanisms in Medicago sativa. Plants 2023, 12, 2059. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Shi, S.; Sun, C. Determination of coumarins and major phenolic acids in plant and rhizosphere soil of alfalfa (Medicago sativa L.). Soils 2016, 48, 931–938. [Google Scholar]
- Yin, G. Soil Microecological Characteristics and Reduction of Autotoxicity in Alfalfa-Wheat-Maize Rotation System. Ph.D. Thesis, Gansu Agricultural University, Lanzhou, China, 2019. [Google Scholar]
- Wang, R.; Liu, J.; Jiang, W.; Ji, P.; Li, Y. Metabolomics and microbiomics reveal impacts of rhizosphere metabolites on alfalfa continuous cropping. Front. Microbiol. 2022, 13, 833968. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ma, S.; Chen, G.; Lu, X.; Chai, Q.; Li, S. Mechanisms and mitigation strategies for the occurrence of continuous cropping obstacles of legumes in China. Agronomy 2024, 14, 104. [Google Scholar] [CrossRef]
- Li, X.; Lewis, E.; Liu, Q. Effects of long-term continuous cropping on soil nematode community and soil condition associated with replant problem in strawberry habitat. Sci. Rep. 2016, 6, 30466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, S.; Luo, D.; Mao, P.; Rosazlina, R.; Martin, F.; Xu, L. Decline in morel production upon continuous cropping is related to changes in soil mycobiome. J. Fungi 2023, 9, 492. [Google Scholar] [CrossRef]
- Yu, Y.; Qi, Z.; Zhang, Z.; Zhou, S.; Jin, M. Plants select antibiotic resistome in rhizosphere in early stage. Sci. Total Environ. 2023, 858, 159847. [Google Scholar] [CrossRef]
- Zeeshan, U.; Yu, J.; Yao, G.; Yang, H. Systematic Review on the continuous cropping obstacles and control strategies in medicinal plants. Int. J. Mol. Sci. 2023, 24, 12470. [Google Scholar] [CrossRef]
- Li, G.; Gong, P.; Zhou, J.; Wang, L.; Song, X.; Ding, P.; Jin, Y.; Zhang, Y.; Zhou, X.; Yang, J.; et al. The succession of rhizosphere microbial community in the continuous cropping soil of tobacco. Front. Environ. Sci. 2024, 11, 1251938. [Google Scholar] [CrossRef]
- Bi, Y.; Zhang, X.; Jiao, X.; Li, J.; Peng, N.; Tian, G.; Wang, Y.; Gao, W. The relationship between shifts in the rhizosphere microbial community and root rot disease in a continuous cropping American ginseng system. Front. Microbiol. 2023, 14, 1097742. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Zhang, F. Community assembly correlates with alfalfa production by mediating rhizosphere soil microbial community composition in different planting years and regimes. Plant Soil. 2022, 479, 355–370. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Z.; Chen, L.; Xun, W.; Shu, X.; Chen, Y.; Sun, X.; Wang, Z.; Ren, Y.; Shen, Q.; et al. Root colonization by beneficial rhizobacteria. FEMS Microbiol. Rev. 2024, 48, 66. [Google Scholar] [CrossRef] [PubMed]
- Roeland, L.; Corné, M.; Peter, A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Zhou, Y.; Coventry, D.; Gupta, V. The preceding root system drives the composition and function of the rhizosphere microbiome. Genome Biol. 2020, 21, 89. [Google Scholar] [CrossRef]
- Gabriele, B.; Kornelia, S. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, Y.; Yue, L. Impacts of continuous cropping on the rhizospheric and endospheric microbial communities and root exudates of Astragalus mongholicus. BMC Plant Biol. 2024, 24, 340. [Google Scholar] [CrossRef]
- Zhao, W.; Bi, X.; Peng, Y.; Bai, M. Research advances of the phosphorus-accumulating organisms of Candidatus Accumulibacter, Dechloromonas and Tetrasphaera: Metabolic mechanisms, applications and influencing factors. Chemosphere 2022, 307, 135675. [Google Scholar] [CrossRef]
- Aguirre, W.; Rocha, E.; Shapiro, L. Enrichment of Verrucomicrobia, Actinobacteria and Burkholderiales drives selection of bacterial community from soil by maize roots in a traditional milpa agroecosystem. PLoS ONE 2018, 13, e0208852. [Google Scholar] [CrossRef]
- Abeywickrama, P.; Zhang, W.; Li, X.; Jayawardena, R.; Hyde, K.; Yan, J. Campylocarpon fasciculare (Nectriaceae, Sordariomycetes); Novel emergence of black-foot causing pathogen on young grapevines in China. Pathogens 2021, 10, 1555. [Google Scholar] [CrossRef]
- Ogawa, T.; Chen, J.; Mise, K.; Takano, Y. Multiple Colletotrichum species commonly exhibit focal effector accumulation in a biotrophic interface at the primary invasion sites in their host plants. Plant Signal Behav. 2021, 16, 1935604. [Google Scholar] [CrossRef]
- Qin, F.; Liu, D.; Sun, B.; Ruan, L.; Ma, Z.; Wang, H. Identification of alfalfa leaf diseases using image recognition technology. PLoS ONE 2016, 11, e0168274. [Google Scholar] [CrossRef]
- Veloso, J.; van Kan, J. Many Shades of Grey in Botrytis-Host Plant Interactions. Trends Plant Sci. 2018, 23, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, H.; Wang, Y. Transcriptome characterization and differential expression analysis of disease-responsive genes in alfalfa leaves infected by Pseudopeziza medicaginis. Euphytica 2018, 214, 126. [Google Scholar] [CrossRef]
- Cui, J.; Zhang, M.; Chen, L.; Zhang, S.; Luo, Y.; Cao, W.; Zhao, J.; Wang, L.; Jia, Z.; Bao, Z. Methanotrophs contribute to nitrogen fixation in emergent macrophytes. Front. Microbiol. 2022, 13, 851424. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, B.; Jin, T.; Chen, H.; Zhao, G.; Qin, X.; Yang, Y.; Xu, J. Rhizospheric microbiomics integrated with plant transcriptomics provides insight into the Cd response mechanisms of the newly identified Cd accumulator Dahlia pinnata. Front. Plant Sci. 2022, 13, 1091056. [Google Scholar] [CrossRef]
- Elshafie, H.; Camele, I. Rhizospheric actinomycetes revealed antifungal and plant-growth-promoting activities under controlled environment. Plants 2022, 11, 1872. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jiang, H.; Chang, G.; Liang, S.; Ma, K.; Cai, Y.; Tian, B.; Shi, X. Effects of rhizosphere microbial communities on cucumber fusarium wilt disease suppression. Microorganisms 2023, 11, 1576. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Liu, L. Screening of phosphate-solubilizing bacteria and their abilities of phosphorus solubilization and wheat growth promotion. BMC Microbiol. 2022, 22, 296. [Google Scholar] [CrossRef]
- Lei, H.; Jin, C.; Xiao, Z. Relationship between pepper (Capsicum annuum L.) root morphology, inter-root soil bacterial community structure and diversity under water–air intercropping conditions. Planta 2023, 257, 98. [Google Scholar] [CrossRef]
- Kitahara, R.; Ikeda, Y.; Shimura, H. A unique mitovirus from Glomeromycota, the phylum of Arbuscular mycorrhizal fungi. Arch. Virol. 2014, 159, 2157–2160. [Google Scholar] [CrossRef]
- Li, Y.; Nan, Z.; Matthew, C.; Wang, Y.; Duan, T. Arbuscular mycorrhizal fungus changes alfalfa (Medicago sativa) metabolites in response to leaf spot (Phoma medicaginis) infection, with subsequent effects on pea aphid (Acyrthosiphon pisum) behavior. New Phytol. 2023, 239, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Tello, A.; Jiménez, M.; Sánchez, G. Management of sewage sludge by composting using fermented water hyacinth. Environ. Sci Pollut. Res. Int. 2015, 22, 14781–14792. [Google Scholar] [CrossRef] [PubMed]
- Foresto, E.; Carezzano, M.; Giordano, W.; Bogino, P. Ascochyta blight in chickpea: An update. J. Fungi 2023, 49, 203. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Li, Y.; Li, C.; Li, B.; Ren, Y.; Zheng, H. Identification and comparative analysis of microRNAs in barnyardgrass in response to rice allelopathy. Plant Cell Environ. 2015, 38, 1368–1381. [Google Scholar] [CrossRef]
- Fu, Y.; Ma, K.; Li, Q.; Li, G.; Cui, H. Effects of potato intercropped with maize on soil bacterial diversity. Chin. J. Eco-Agric. 2020, 28, 1715–1725. [Google Scholar] [CrossRef]
- Li, P.; Zhu, Z.; Zhang, Y. The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome 2022, 10, 56. [Google Scholar] [CrossRef]
- Ham, S.; Yoon, A.R.; Oh, H.; Park, Y. Plant growth-promoting microorganism Pseudarthrobacter sp. NIBRBAC000502770 enhances the growth and flavonoid content of Geum aleppicum. Microorganisms 2022, 10, 1241. [Google Scholar] [CrossRef]
- Mary, L. The phylogeny of plant and animal pathogens in the Ascomycota. Physiol. Mol. Plant P 2001, 59, 165–187. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.; Chen, Y.; Ariyawansa, H. Integrative approaches for species delimitation in Ascomycota. Fungal Divers. 2021, 109, 155–179. [Google Scholar] [CrossRef]
- Almario, J.; Fabiańska, I.; Saridis, G.; Bucher, M. Unearthing the plant-microbe quid pro quo in root associations with beneficial fungi. New Phytol. 2022, 234, 1967–1976. [Google Scholar] [CrossRef]
- Shelobolina, E.; Roden, E.; Benzine, J.; Xiong, M.Y. Using phyllosilicate-Fe (II)-oxidizing soil bacteria to improve Fe and K plant nutrition. U. S. Pat. Appl. 2014, 14, 509. [Google Scholar]
- Bai, Y.; Feng, P.; Chen, W.; Xu, S.; Liang, J.; Jia, J. Effect of three microbial fertilizer carriers on water infiltration and evaporation, microbial community and alfalfa growth in saline-alkaline soil. Commun. Soil Sci. Plan. 2021, 52, 2462–2470. [Google Scholar] [CrossRef]
- Ning, Q.; Chen, L.; Li, F. Effects of mortierella on soil nutrient availability and straw degradation. Acta Pedol. Sin. 2022, 59, 206–217. [Google Scholar]
- Withee, P.; Haituk, S.; Senwanna, C.; Karunarathna, A.; Tamakaew, N.; Pakdeeniti, P.; Suwannarach, N.; Kumla, J.; Suttiprapan, P.; Taylor, P. Identification and pathogenicity of Paramyrothecium species associated with leaf spot disease in northern Thailand. Plants 2022, 11, 1445. [Google Scholar] [CrossRef]
- Bingol, E.; Qi, A.; Karandeni-Dewage, C.; Ritchie, F.; Fitt, B.; Huang, Y. Co-inoculation timing affects the interspecific interactions between phoma stem canker pathogens Leptosphaeria maculans and Leptosphaeria biglobosa. Pest Manag. Sci. 2024, 80, 2443–2452. [Google Scholar] [CrossRef]
- Yang, J.; Han, J.; Jing, Y.; Li, S.; Lan, B.; Zhang, Q.; Yin, K. Virulent fusarium isolates with diverse morphologies show similar invasion and colonization strategies in alfalfa. Front. Plant Sci. 2024, 15, 1390069. [Google Scholar] [CrossRef]
- Punja, Z.; Chandanie, W.; Chen, X.; Rodríguez, G. Phoma leaf spot of wasabi (Wasabia japonica) caused by Leptosphaeria biglobosa. Plant Pathol. 2016, 66, 480–489. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, B.; Zhang, S. Research progress of alfalfa root disease and its control. J. Henan Agric. Univ. 2022, 56, 914–923. [Google Scholar] [CrossRef]
- Marek, K.; Wielbo, J.; Pawlik, A.; Skorupska, A. Nodulation competitiveness of Ensifer meliloti alfalfa nodule isolates and their potential for application as inoculants. Pol. J. Microbiol. 2014, 63, 375–386. [Google Scholar] [CrossRef]
- Debnath, S.; Das, A.; Maheshwari, D.; Pandey, P. Treatment with Atypical rhizobia, Pararhizobium giardinii and Ochrobactrum sp. modulate the rhizospheric bacterial community, and enhances Lens culinaris growth in fallow-soil. Microbiol. Res. 2023, 267, 127255. [Google Scholar] [CrossRef]
- Luo, L.; Zhao, C.; Wang, E.; Raza, A.; Yin, C. Bacillus amyloliquefaciens as an excellent agent for biofertilizer and biocontrol in agriculture: An overview for its mechanisms. Microbiol. Res. 2022, 259, 127016. [Google Scholar] [CrossRef] [PubMed]
- Galili, G.; Amir, R.; Fernie, A. The Regulation of essential amino acid synthesis and accumulation in plants. Annu. Rev. Plant Biol. 2016, 67, 153–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, D.; Liu, Q. Connections between amino acid metabolisms in plants: Lysine as an example. Front. Plant Sci. 2020, 11, 928. [Google Scholar] [CrossRef]
- Chang, J.; Tabima, J. A covert operation by a plant pathogen. Cell Host Microbe 2016, 20, 413–415. [Google Scholar] [CrossRef]
- Das, D.; Paries, M.; Hobecker, K. Phosphate starvation response transcription factors enable arbuscular mycorrhiza symbiosis. Nat. Commun. 2022, 13, 477. [Google Scholar] [CrossRef]
- Paula, E.J. Zeatin: The 60th anniversary of its identification. Plant Physiol. 2023, 192, 34–55. [Google Scholar] [CrossRef]
- Liu, S.; García, P.; Tedersoo, L. Phylotype diversity within soil fungal functional groups drives ecosystem stability. Nat. Ecol. Evol. 2022, 6, 900–909. [Google Scholar] [CrossRef]
- Ferraro, V.; Venturella, G.; Cirlincione, F.; Mirabile, G.; Gargano, M.L.; Colasuonno, P. The checklist of sicilian macrofungi: Second edition. J. Fungi 2022, 8, 566. [Google Scholar] [CrossRef]
- Gil, M.M.; López, G.; Domínguez, M.T.; Navarro, F.; Carmen, M.; Kjøller, R.; Tibbett, M.; Marañón, T. Ectomycorrhizal fungal communities and their functional traits mediate plant–soil interactions in trace element contaminated soils. Front. Plant Sci. 2018, 9, 1682. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, J. The potassium dichromate volumetric method for the determination of soil organic matter-heating method. China High Technol. Enterp. 2016, 26, 11–12. [Google Scholar]
- Zhang, W.; Fu, Y.; Li, J. Comparative study on Kjeldahl method and Dumas combustion method for total Nitrogen measurement in soil. Chin. Agric. Sci. Bull. 2015, 31, 172–175. [Google Scholar]
- Wang, Q.; Xu, Q.; Yao, Z. Determination of available phosphorus in soil. Instrum. Anal. Monit. 2009, 4, 36–38. [Google Scholar]
- Dai, Z.; Li, M. Study on determination methods for available potassium in upland field. Acta Pedol. Sin. 1997, 34, 336–343. [Google Scholar]
- Gu, X.; Zhang, F.; Xie, X. Effects of N and P addition on nutrient and stoichiometry of rhizosphere and non-rhizosphere soils of alfalfa in alkaline soil. Sci. Rep. 2023, 13, 12119. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Jiang, X.; Wu, Q.; Zhou, N. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbiol. 2013, 79, 5962–5969. [Google Scholar] [CrossRef]
- Rousk, J.; Baath, E.; Brookes, P.C. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 14th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 1995; Available online: http://lib3.dss.go.th/fulltext/scan_ebook/aoac_1995_v78_n3.pdf (accessed on 18 August 2024).
- Yang, Z.; Chen, Q.; Ai, D. Determination of crude fiber content in alfalfa by filter bag method. Hubei Agric. Sci. 2017, 56, 1739–1741. [Google Scholar] [CrossRef]
- Soest, P.; Robertson, J.; Lewis, B. Methods of dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Li, T.; Peng, L.; Wang, H.; Zhang, Y.; Wang, Y.; Cheng, Y.; Hou, F. Multi-cutting improves forage yield and nutritional value and maintains the soil nutrient balance in a rainfed agroecosystem. Front. Plant Sci. 2022, 13, 825117. [Google Scholar] [CrossRef]
- Reyon, D.; Tsai, S.Q.; Khayter, C. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012, 30, 460–465. [Google Scholar] [CrossRef]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J. QIIME allows analysis of high-throughput community sequencing data. Nat. Met-Hods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Douglas, G.; Maffei, V.; Zaneveld, J. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Nguyen, N.; Song, Z.; Bates, S.; Branco, S.; Tedersoo, L.; Menke, J. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Kruskal, W.; Wallis, W. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Shen, Y.; Zhou, X.; Ma, H.; Lan, J.; Fu, B.; Xue, Q. Biological Decline of Alfalfa Is Accompanied by Negative Succession of Rhizosphere Soil Microbial Communities. Plants 2024, 13, 2589. https://doi.org/10.3390/plants13182589
Ma Y, Shen Y, Zhou X, Ma H, Lan J, Fu B, Xue Q. Biological Decline of Alfalfa Is Accompanied by Negative Succession of Rhizosphere Soil Microbial Communities. Plants. 2024; 13(18):2589. https://doi.org/10.3390/plants13182589
Chicago/Turabian StyleMa, Yuanyuan, Yan Shen, Xiaoping Zhou, Hongbin Ma, Jian Lan, Bingzhe Fu, and Quanhong Xue. 2024. "Biological Decline of Alfalfa Is Accompanied by Negative Succession of Rhizosphere Soil Microbial Communities" Plants 13, no. 18: 2589. https://doi.org/10.3390/plants13182589
APA StyleMa, Y., Shen, Y., Zhou, X., Ma, H., Lan, J., Fu, B., & Xue, Q. (2024). Biological Decline of Alfalfa Is Accompanied by Negative Succession of Rhizosphere Soil Microbial Communities. Plants, 13(18), 2589. https://doi.org/10.3390/plants13182589




