Enzymatic Characterization of SpPAL Genes in S. polyrhiza and Overexpression of the SpPAL3
Abstract
:1. Introduction
2. Results
2.1. Cloning of SpPALs
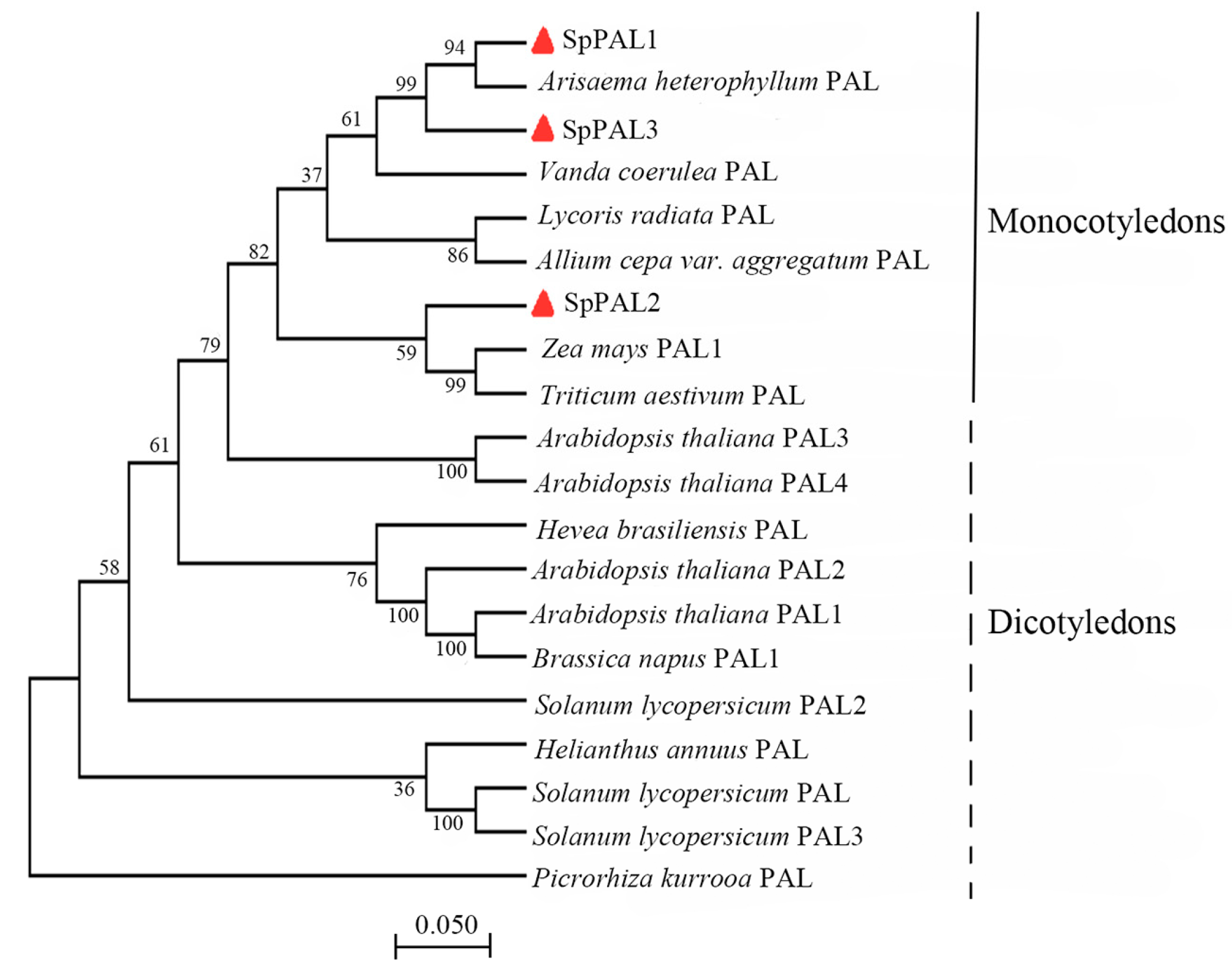
2.2. Bioinformatics and Phylogenetic Analyses of SpPALs
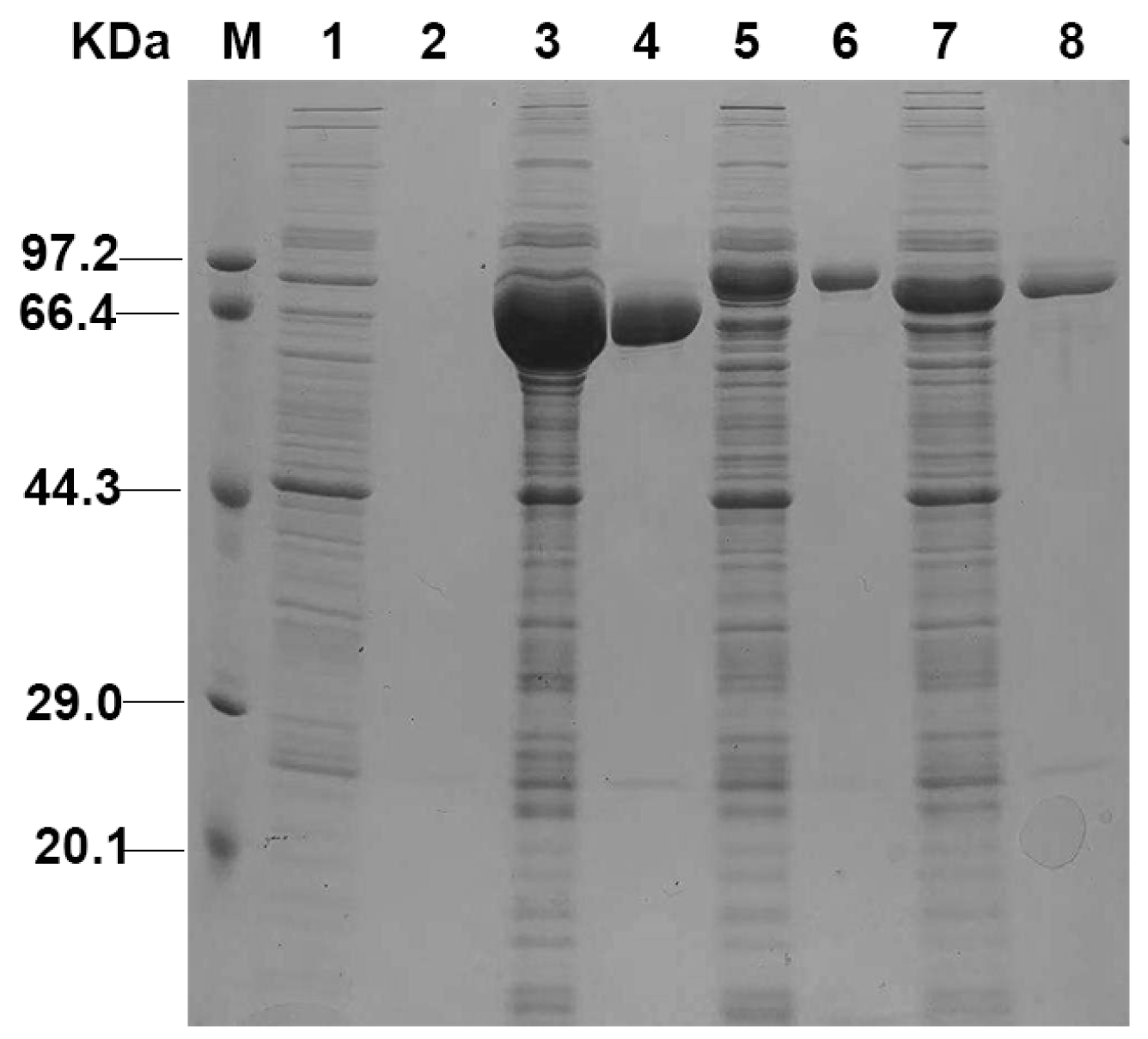
2.3. Expression and Purification of Recombinant SpPALs
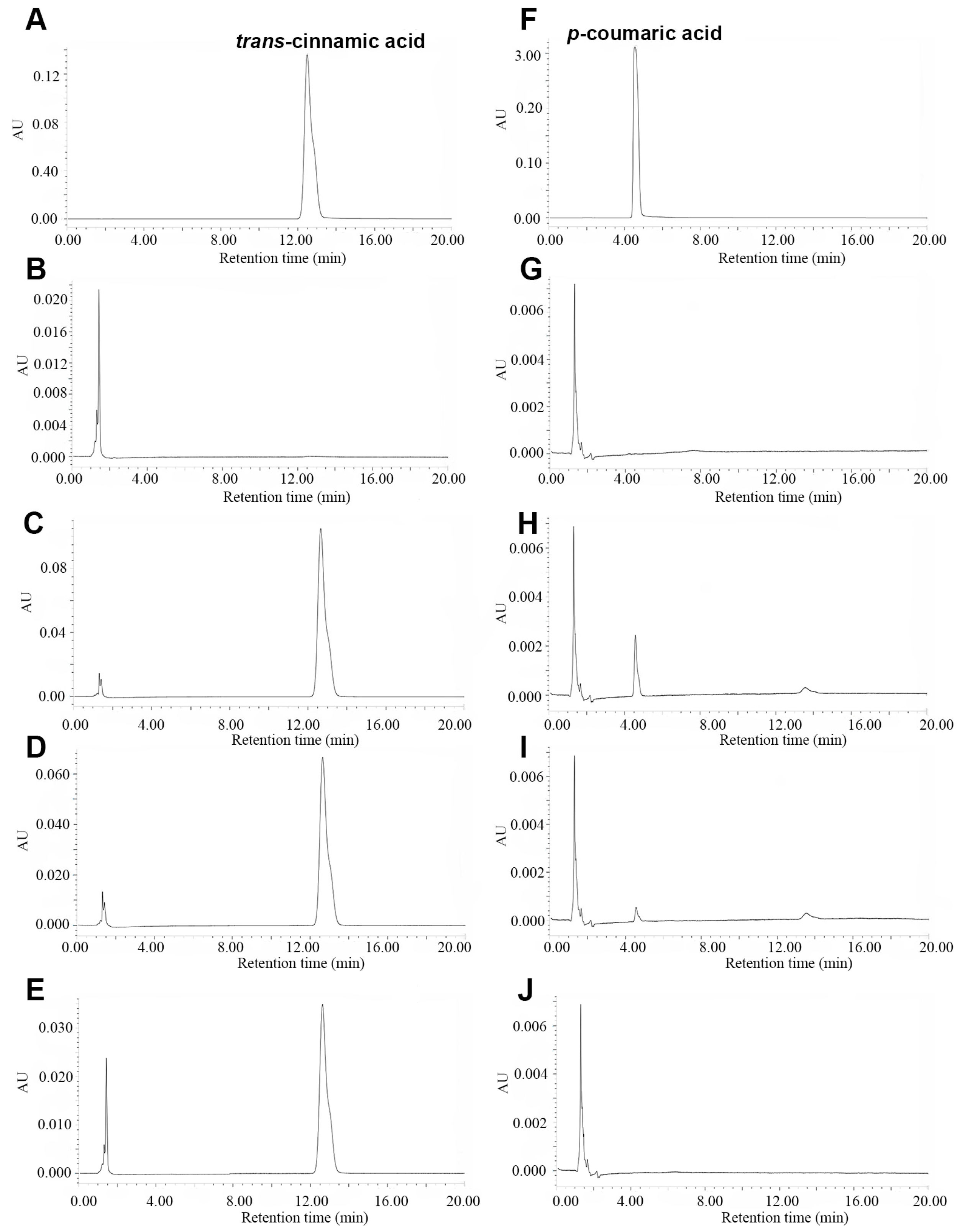
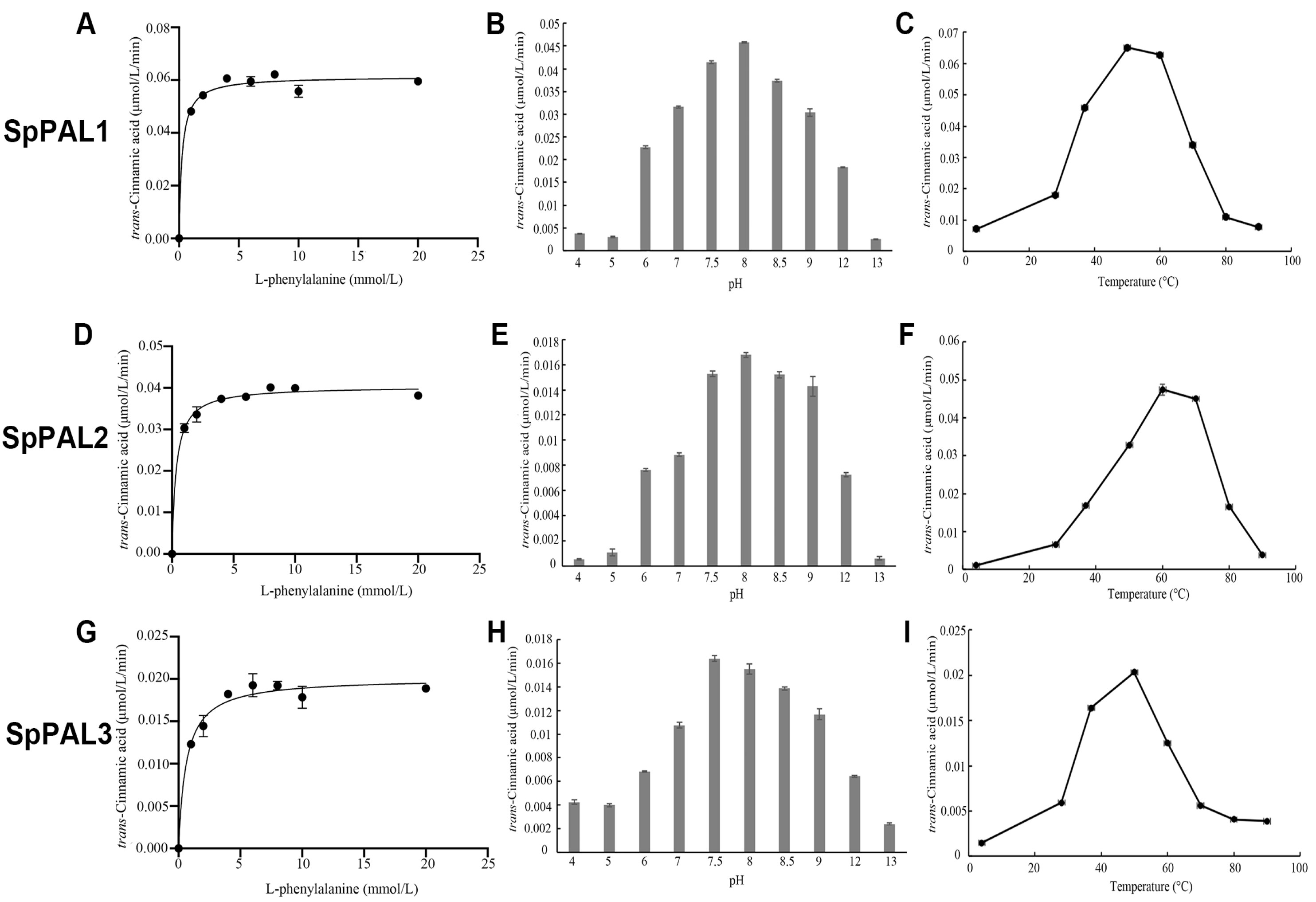
2.4. Functional Characterization of Recombinant SpPALs
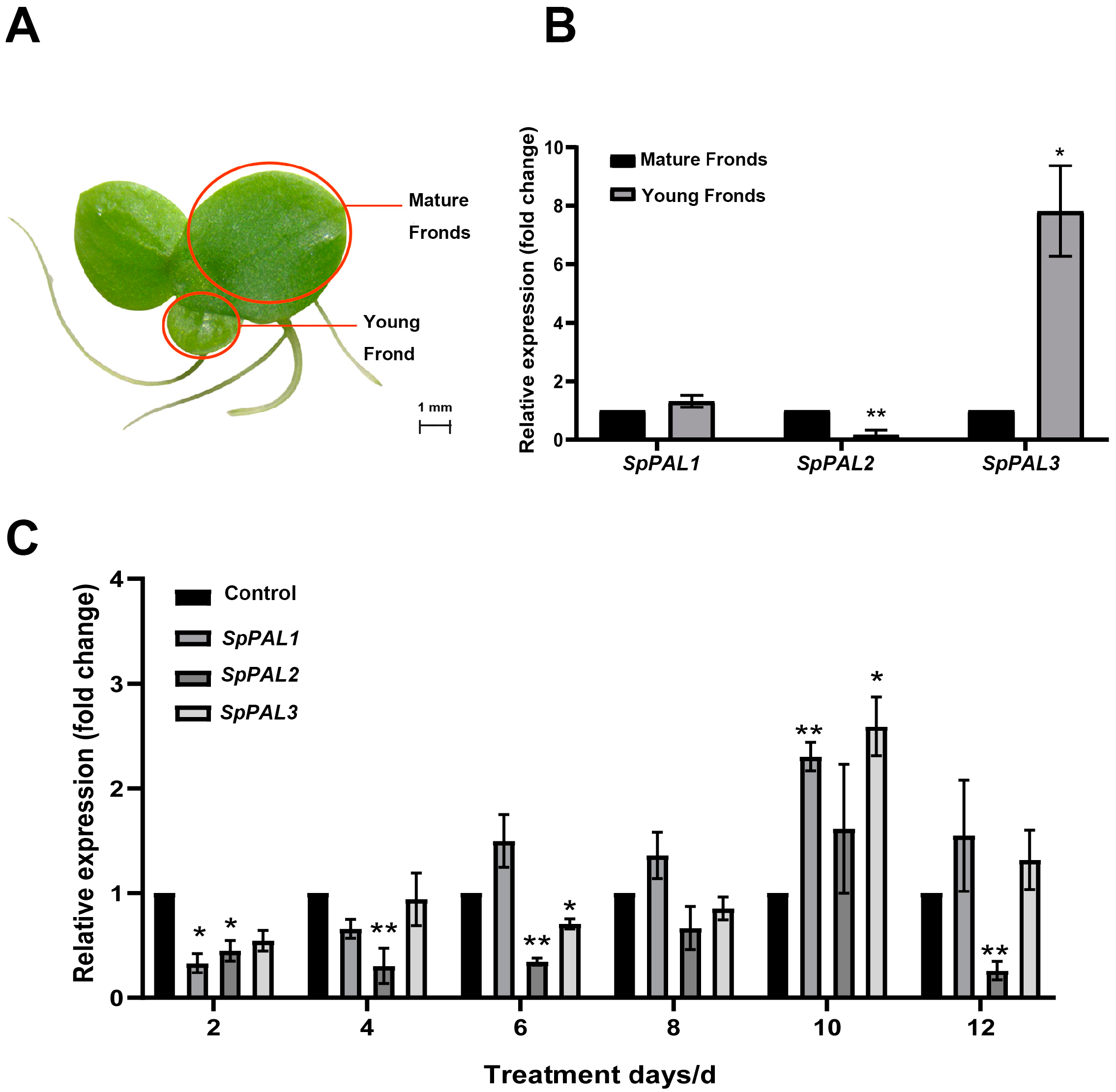
2.5. Transcription Profile of SpPALs in Different Tissues and under MeJA Treatment
2.6. Overexpression of SpPAL3 in Lemna turionifera
3. Discussion
4. Materials and Methods
4.1. Cultivation of Plant Materials and Application of MeJA Treatment
4.2. Bioinformatics Analysis
4.3. Cloning of SpPAL1, SpPAL2, and SpPAL3
4.4. Expression and Purification of Recombinant SpPALs in E. coli
4.5. Enzyme Activity Assays
4.6. Expression Analysis of SpPALs by qRT-PCR
4.7. Generating Transgenic SpPAL3 Duckweed Plants
4.8. Analysis of Fresh Weight, Soluble Protein Content, Pigment Content, and Phenolic Content
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, S.; Wang, Z.; Shen, L.; Xiao, H. Synthetic biology: A new frontier in food production. Trends Biotechnol. 2022, 40, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Stewart, C.N., Jr. Plant synthetic biology. Trends Plant Sci. 2015, 20, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, P.; Chavez, B.G.; Leite Dias, S.; D’Auria, J.C. Plant synthetic biology: From inspiration to augmentation. Curr. Opin. Biotechnol. 2023, 79, 102857. [Google Scholar] [CrossRef]
- Yang, Y.; Chaffin, T.A.; Ahkami, A.H.; Blumwald, E.; Stewart, C.N. Plant synthetic biology innovations for biofuels and bioproducts. Trends Biotechnol. 2022, 40, 1454–1468. [Google Scholar] [CrossRef]
- South, P.F.; Cavanagh, A.P.; Liu, H.W.; Ort, D.R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 2019, 363, eaat9077. [Google Scholar] [CrossRef]
- Ziegler, P.; Appenroth, K.J.; Sree, K.S. Survival Strategies of Duckweeds, the World’s Smallest Angiosperms. Plants 2023, 12, 2215. [Google Scholar] [CrossRef]
- Tao, X.; Fang, Y.; Huang, M.-J.; Xiao, Y.; Liu, Y.; Ma, X.-R.; Zhao, H. High flavonoid accompanied with high starch accu mulation triggered by nutrient starvation in bioenergy crop duckweed (Landoltia punctata). BMC Genom. 2017, 18, 166. [Google Scholar] [CrossRef]
- Yang, G.L.; Feng, D.; Liu, Y.T.; Lv, S.M.; Zheng, M.M.; Tan, A.J. Research Progress of a Potential Bioreactor: Duckweed. Biomolecules 2021, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhao, X.; Qi, G.; Bai, Z.; Wang, Y.; Wang, S.; Ma, Y.; Liu, Q.; Hu, R.; Zhou, G. Integrated analysis of transcript tome and metabolites reveals an essential role of metabolic flux in starch accumulation under nitrogen starvation in duckweed. Biotechnol. Biofuels 2017, 10, 167. [Google Scholar] [CrossRef]
- Qiao, X.; He, W.; Xiang, C.; Han, J.; Wu, L.; Guo, D.; Ye, M. Qualitative and Quantitative Analyses of Flavonoids in Spirodela polyrrhiza by High-performance Liquid Chromatography Coupled with Mass Spectrometry. Phytochem. Anal. 2011, 22, 475–483. [Google Scholar] [CrossRef]
- Hao, H.; Lin, L.; Liu, S.; Kang, Y.; Wang, Y.; Huang, J.; Weng, W. Deep Eutectic Solvent-Based Microwave-Assisted Extraction for the Chromatographic Analysis of Bioactive Flavonoids in Spirodela polyrrhiza. J. Chromatogr. Sci. 2022, 60, 501–510. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Xu, S.; Tang, X.; Zhao, J.; Yu, C.; He, G.; Xu, H.; Wang, S.; Tang, Y.; et al. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing in Lemna aequinoctialis. Plant Biotechnol. J. 2019, 17, 2143–2152. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Yu, X.H.; Anaokar, S.; Shi, H.; Dahl, W.B.; Cai, Y.; Luo, G.; Chai, J.; Cai, Y.; Molla-Morales, A.; et al. Engineering triacylglycerol accumulation in duckweed (Lemna japonica). Plant Biotechnol. J. 2023, 21, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Dixon, R.A. Plant Phenylalanine/Tyrosine Ammonia-lyases. Trends Plant Sci. 2020, 25, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C.-J. Multifaceted Regulations of Gateway Enzyme Phenylalanine Ammonia-Lyase in the Biosynthesis of Phe nylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Haberer, G.; Gundlach, H.; Gläßer, C.; Nussbaumer, T.; Luo, M.C.; Lomsadze, A.; Borodovsky, M.; Kerstetter, R.A.; Shanklin, J.; et al. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 2014, 5, 3311. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Y.; Yu, Y.; Ma, P.; Jia, Z.; Guo, X.; Xie, Y.; Bian, X. Overexpression of IbPAL1 promotes chlorogenic acid biosynthesis in sweetpotato. Crop J. 2021, 9, 204–215. [Google Scholar] [CrossRef]
- Zhan, C.; Li, Y.; Li, H.; Wang, M.; Gong, S.; Ma, D.; Li, Y. Phylogenomic analysis of phenylalanine ammonia-lyase (PAL) multigene family and their differential expression analysis in wheat (Triticum aestivum L.) suggested their roles during different stress responses. Front. Plant Sci. 2022, 13, 982457. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, Y.; Mu, K.; Cai, W.; Zhao, Y.; Shen, H.; Wang, X.; Ma, H. Phenylalanine Ammonia Lyase GmPAL1.1 Promotes Seed Vigor under High-Temperature and -Humidity Stress and Enhances Seed Germination under Salt and Drought Stress in Transgenic Arabidopsis. Plants 2022, 11, 3239. [Google Scholar] [CrossRef]
- Rahmatabadi, S.S.; Sadeghian, I.; Ghasemi, Y.; Sakhteman, A.; Hemmati, S. Identification and characterization of a steri cally robust phenylalanine ammonia-lyase among 481 natural isoforms through association of in silico and in vitro studies. Enzym. Microb. Technol. 2019, 122, 36–54. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Ma, G.J.; Yang, C.C.; Lee, P.D. Cloning, expression, site-directed mutagenesis and immunolocalization of phe nylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 2010, 71, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.M.; Wang, X.C.; Peng, Z.H.; Zheng, B.; Liu, Q. Characterization and primary functional analysis of phenylalanine ammonia-lyase gene from Phyllostachys edulis. Plant Cell Rep. 2012, 31, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Jiang, T.; Cong, Y.; Zheng, Z.; Ouyang, J. Molecular Characterization of a Recombinant Zea mays Phenylalanine Ammonia-Lyase (ZmPAL2) and Its Application in trans-Cinnamic Acid Production from L-Phenylalanine. Appl. Biochem. Biotechnol. 2015, 176, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Luo, W.; Tanaka, G.; Konishi, Y.; Matsuura, H.; Takahashi, K. Wounding stress induces phenylalanine ammonia lyases, leading to the accumulation of phenylpropanoids in the model liverwort Marchantia polymorpha. Phytochemistry 2018, 155, 30–36. [Google Scholar] [CrossRef]
- Chang, A.; Lim, M.-H.; Lee, S.-W.; Robb, E.J.; Nazar, R.N. Tomato Phenylalanine Ammonia-Lyase Gene Family, Highly Redundant but Strongly Underutilized. J. Biol. Chem. 2008, 283, 33591–33601. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, W.; Li, Y.; Dai, X.; Ma, G.; Xing, D.; Zhu, M.; Gao, L.; Xia, T. Six phenylalanine ammonia-lyases from Camellia sinensis: Evolution, expression, and kinetics. Plant Physiol. Biochem. 2017, 118, 413–421. [Google Scholar] [CrossRef]
- Hyun, M.W.; Yun, Y.H.; Kim, J.Y.; Kim, S.H. Fungal and Plant Phenylalanine Ammonia-lyase. Mycobiology 2011, 39, 257–265. [Google Scholar] [CrossRef]
- Fan, L.; Shi, G.; Yang, J.; Liu, G.; Niu, Z.; Ye, W.; Wu, S.; Wang, L.; Guan, Q. A Protective Role of Phenylalanine Am monia-Lyase from Astragalus membranaceus against Saline-Alkali Stress. Int. J. Mol. Sci. 2022, 23, 15686. [Google Scholar] [CrossRef]
- Li, W.; Yang, Y.; Qiao, C.; Zhang, G.; Luo, Y. Functional characterization of phenylalanine ammonia-lyase- and cinnamate 4-hydroxylase-encoding genes from Lycoris radiata, a galanthamine-producing plant. Int. J. Biol. Macromol. 2018, 117, 1264–1279. [Google Scholar] [CrossRef]
- Qin, Y.; Li, Q.; An, Q.; Li, D.; Huang, S.; Zhao, Y.; Chen, W.; Zhou, J.; Liao, H. A phenylalanine ammonia lyase from Fritillaria unibracteata promotes drought tolerance by regulating lignin biosynthesis and SA signaling pathway. Int. J. Biol. Macromol. 2022, 213, 574–588. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Luo, J.; Yao, R.; Huang, C.; Zhao, Y.; Kong, L. Functional characterization and correlation analysis of phenylala nine ammonia-lyase (PAL) in coumarin biosynthesis from Peucedanum praeruptorum Dunn. Phytochemistry 2019, 158, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xia, B.; Liang, L.; Li, X.; Xu, S.; Peng, F.; Wang, R. Molecular and analysis of a phenylalanine ammonia-lyase gene (LrPAL2) from Lycoris radiata. Mol. Biol. Rep. 2013, 40, 2293–2300. [Google Scholar] [CrossRef]
- Liu, A.; Zhu, Y.; Wang, Y.; Wang, T.; Zhao, S.; Feng, K.; Li, L.; Wu, P. Molecular identification of phenylalanine ammo nia lyase-encoding genes EfPALs and EfPAL2-interacting transcription factors in Euryale ferox. Front. Plant Sci. 2023, 14, 1114345. [Google Scholar] [CrossRef]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, S.; Cai, F.; Zheng, X.; Lin, N.; Qin, X.; Ou, Y.; Gu, X.; Zhu, X.; Xu, Y.; et al. Characterization, and expression profile of a phenylalanine ammonia lyase gene from Jatropha curcas L. Mol. Biol. Rep. 2012, 39, 3443–3452. [Google Scholar] [CrossRef]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.E.; Davis, K.R. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar] [CrossRef]
- Liu, R.; Xu, S.; Li, J.; Hu, Y.; Lin, Z. Expression profile of a PAL gene from Astragalus membranaceus var. Mongholicus and its crucial role in flux into flavonoid biosynthesis. Plant Cell Rep. 2006, 25, 705–710. [Google Scholar] [PubMed]
- Liu, Y.; Liu, L.; Yang, S.; Zeng, Q.; He, Z.; Liu, Y. Cloning, Characterization and Expression of the Phenylalanine Ammo nia-Lyase Gene (PaPAL) from Spruce Picea asperata. Forests 2019, 10, 613. [Google Scholar] [CrossRef]
- Habibollahi, M.; Kavousi, H.R.; Lohrasbi-Nejad, A.; Rahpeyma, S.A. Cloning, characterization and expression of a phenyl alanine ammonia-lyase gene (CcPAL) from cumin (Cuminum cyminum L.). J. Appl. Res. Med. Aromat. Plants 2020, 18, 100253. [Google Scholar]
- de Jong, F.; Hanley, S.J.; Beale, M.H.; Karp, A. Characterisation of the willow phenylalanine ammonia-lyase (PAL) gene family reveals expression differences compared with poplar. Phytochemistry 2015, 117, 90–97. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhong, X.; Jiang, X.; Cong, H.; Sun, H.; Qiao, F. Characterisation, expression and functional analysis of PAL gene family in Cephalotaxus hainanensis. Plant Physiol. Biochem. 2020, 156, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Wu, Q.; Li, J.; Huang, S.; Cai, L.; Mao, L.; Luo, Z.; Li, L.; Ying, T. Exogenous methyl jasmonate regulates phe nolic compounds biosynthesis during postharvest tomato ripening. Postharvest Biol. Technol. 2022, 184, 111760. [Google Scholar] [CrossRef]
- Mo, F.; Li, L.; Zhang, C.; Yang, C.; Chen, G.; Niu, Y.; Si, J.; Liu, T.; Sun, X.; Wang, S.; et al. Genome-Wide Analysis and Expression Profiling of the Phenylalanine Ammonia-Lyase Gene Family in Solanum tuberosum. Int. J. Mol. Sci. 2022, 23, 6833. [Google Scholar] [CrossRef]
- Abdollahi, M.R.; Kianersi, F.; Moosavi, S.S.; Dastan, D.; Asadi, S. Identification and Expression Analysis of Two Genes Involved in the Biosynthesis of t-Anethole in Fennel (Foeniculum vulgare Mill.) and Their Up-Regulation in Leaves in Response to Methyl Jasmonate Treatments. J. Plant Growth Regul. 2022, 42, 759–770. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, H.; Liu, Y.; Ji, Q.; Xie, J.; Zhang, R.; Huang, L.; Mei, K.; Wang, J.; Gao, W. Engineering of triterpene metabolism and overexpression of the lignin biosynthesis gene PAL promotes ginsenoside Rg(3) accumulation in ginseng plant chassis. J. Integr. Plant Biol. 2022, 64, 1739–1754. [Google Scholar] [CrossRef]
- Abramson, B.W.; Novotny, M.; Hartwick, N.T.; Colt, K.; Aevermann, B.D.; Scheuermann, R.H.; Michael, T.P. The genome and preliminary single-nuclei transcriptome of Lemna minuta reveals mechanisms of invasiveness. Plant Physiol. 2022, 188, 879–897. [Google Scholar] [CrossRef] [PubMed]
- Xiang, B.; Li, X.; Wang, Y.; Tian, X.; Yang, Z.; Ma, L.; Liu, X.; Wang, Y. Cloning and Characterization of Two Iridoid Synthase Homologs from Swertia Mussotii. Molecules 2017, 22, 1387. [Google Scholar] [CrossRef]
- Ma, W.; Wu, M.; Wu, Y.; Ren, Z.; Zhong, Y. Cloning and characterisation of a phenylalanine ammonia-lyase gene from Rhus chinensis. Plant Cell Rep. 2013, 32, 1179–1190. [Google Scholar] [CrossRef]
- Yang, L.; Han, Y.; Wu, D.; Yong, W.; Liu, M.; Wang, S.; Liu, W.; Lu, M.; Wei, Y.; Sun, J. Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat. Toxicol. 2017, 192, 127–135. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, X.; Gao, X.; Sun, J.; Ji, X.; Feng, G.; Shen, G.; Xiang, B.; Wang, Y. Molecular mechanism underlying the effect of maleic hydrazide treatment on starch accumulation in S. polyrrhiza 7498 fronds. Biotechnol. Biofuels 2021, 14, 99. [Google Scholar] [CrossRef] [PubMed]
- Dupont, P.Y.; Eaton, C.J.; Wargent, J.J.; Fechtner, S.; Solomon, P.; Schmid, J.; Day, R.C.; Scott, B.; Cox, M.P. Fungal endophyte infection of ryegrass reprograms host metabolism and alters development. New Phytol. 2015, 208, 1227–1240. [Google Scholar] [CrossRef] [PubMed]


| Gene | Gene Length (bp) | Protein Length (aa) | Molecular Weight (kDa) | pI | Subcellular Location | Transmembrane Helices | Signal Peptide |
|---|---|---|---|---|---|---|---|
| SpPAL1 | 2172 | 724 | 78.03 | 6.12 | Cytoplasmic (2.091) | No | No |
| SpPAL2 | 2277 | 759 | 83.59 | 6.01 | Cytoplasmic (1.892) | No | No |
| SpPAL3 | 2169 | 723 | 79.02 | 6.35 | Cytoplasmic (1.751) | No | No |
| Substrate | Enzyme | Km (μM) | Kcat (s−1) | Kcat/Km (s−1 M−1) | pH Optima | Temperature Optima (°C) |
|---|---|---|---|---|---|---|
| L-Phe | SpPAL1 | 252 | 9.83 | 39,008 | 8 | 50 |
| SpPAL2 | 345 | 7 | 20,290 | 8 | 60 | |
| SpPAL3 | 632 | 3.35 | 5301 | 7.5 | 50 | |
| L-Tyr | SpPAL1 | 1294 | 0.136 | 105 | NO | NO |
| SpPAL2 | 8718 | 0.313 | 36 | NO | NO | |
| SpPAL3 | ND | ND | ND | NO | NO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhang, Y.; Zhu, C.; Zheng, P.; Chen, C.; Zhang, N.; Ji, H.; Dong, C.; Yu, J.; Ren, J.; et al. Enzymatic Characterization of SpPAL Genes in S. polyrhiza and Overexpression of the SpPAL3. Plants 2024, 13, 2607. https://doi.org/10.3390/plants13182607
Li X, Zhang Y, Zhu C, Zheng P, Chen C, Zhang N, Ji H, Dong C, Yu J, Ren J, et al. Enzymatic Characterization of SpPAL Genes in S. polyrhiza and Overexpression of the SpPAL3. Plants. 2024; 13(18):2607. https://doi.org/10.3390/plants13182607
Chicago/Turabian StyleLi, Xiaoxue, Yinxing Zhang, Chunfeng Zhu, Pufan Zheng, Cunkun Chen, Na Zhang, Haipeng Ji, Chenghu Dong, Jinze Yu, Jie Ren, and et al. 2024. "Enzymatic Characterization of SpPAL Genes in S. polyrhiza and Overexpression of the SpPAL3" Plants 13, no. 18: 2607. https://doi.org/10.3390/plants13182607
APA StyleLi, X., Zhang, Y., Zhu, C., Zheng, P., Chen, C., Zhang, N., Ji, H., Dong, C., Yu, J., Ren, J., Zhu, Y., & Wang, Y. (2024). Enzymatic Characterization of SpPAL Genes in S. polyrhiza and Overexpression of the SpPAL3. Plants, 13(18), 2607. https://doi.org/10.3390/plants13182607







