Comparative Analysis of Phytohormone Biosynthesis Genes Responses to Long-Term High Light in Tolerant and Sensitive Wheat Cultivars
Abstract
:1. Introduction
2. Results
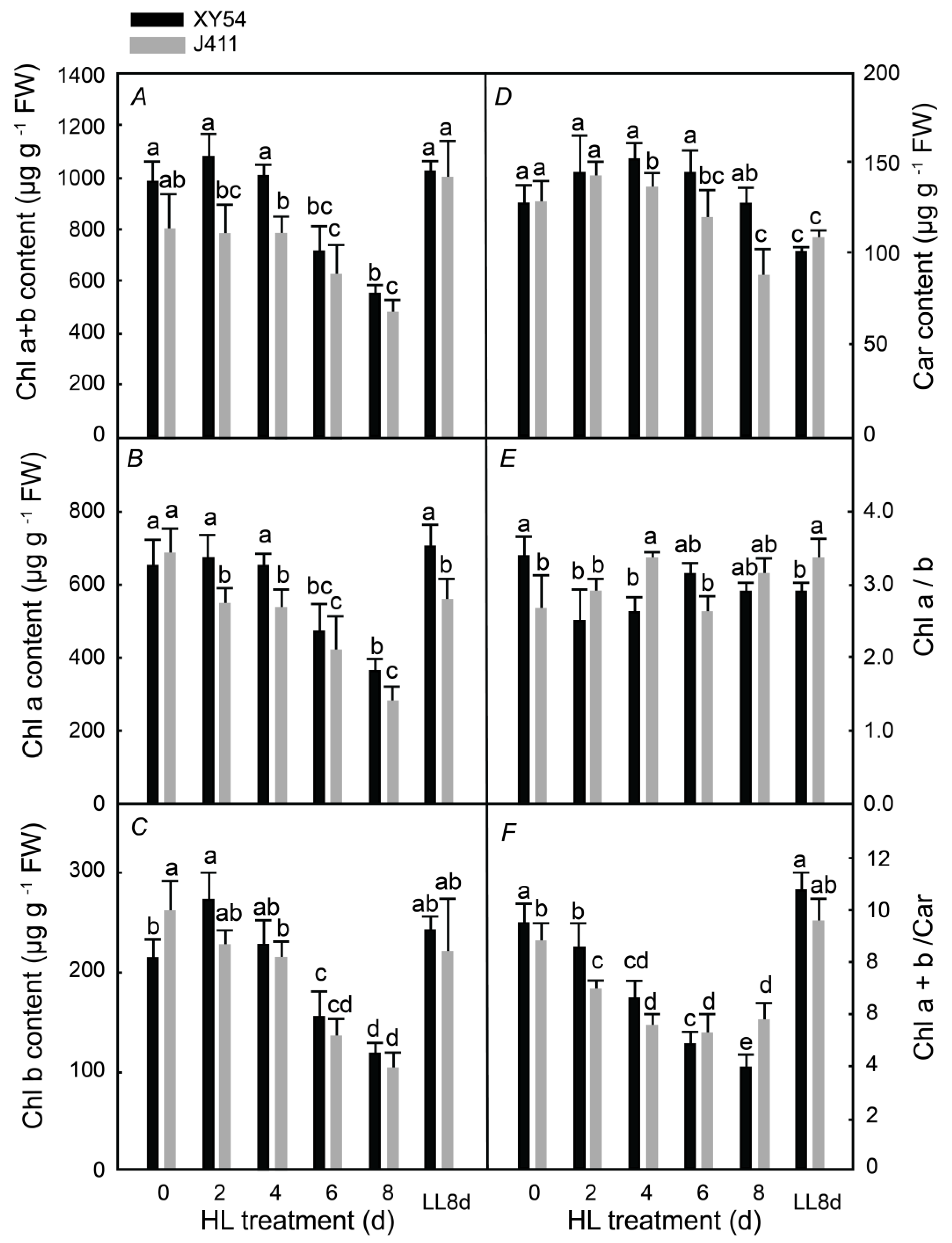
2.1. Photosynthetic Pigment Content under Long-Term HL Stress
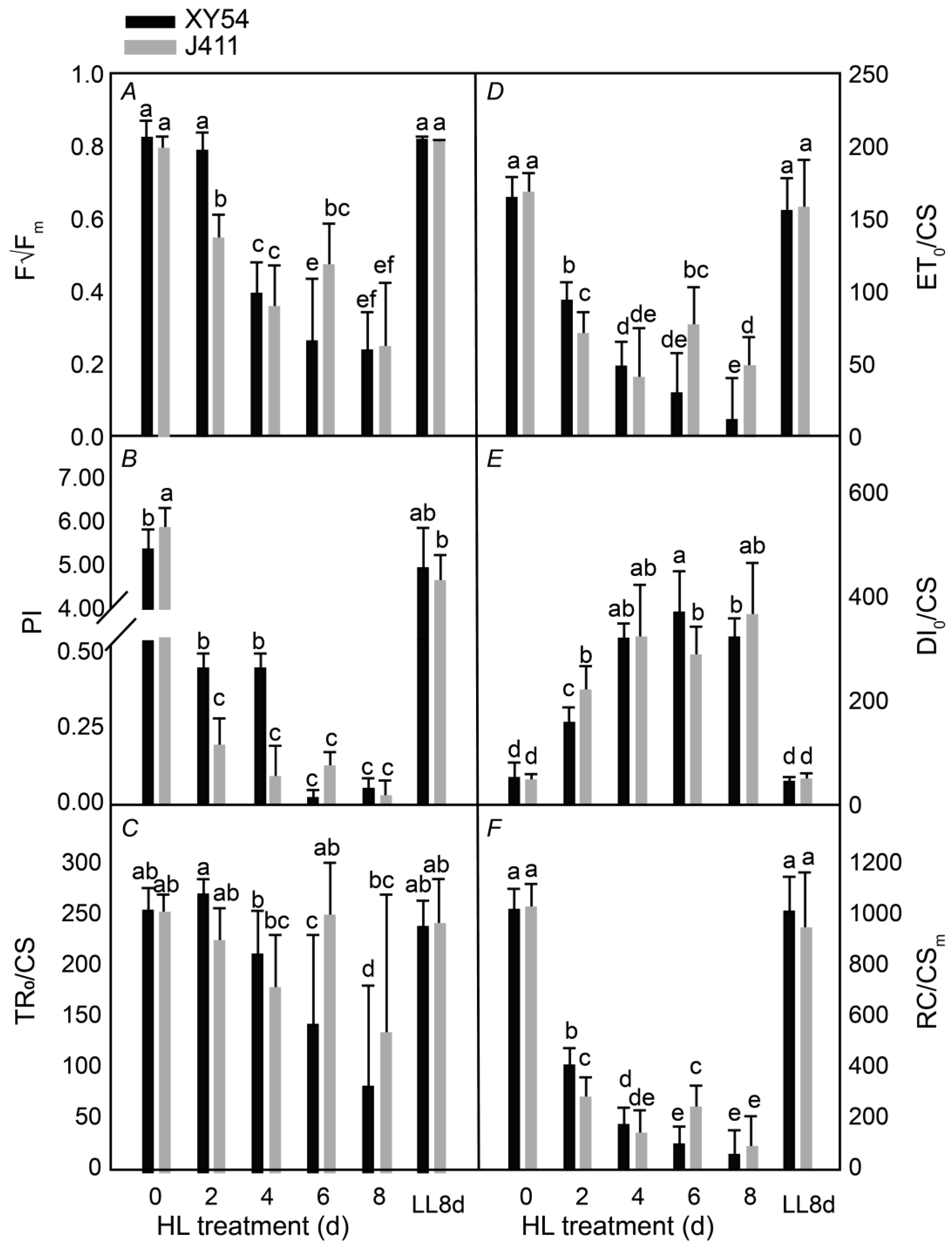
2.2. Assessment of Photosynthesis Performance under Long-Term HL Stress
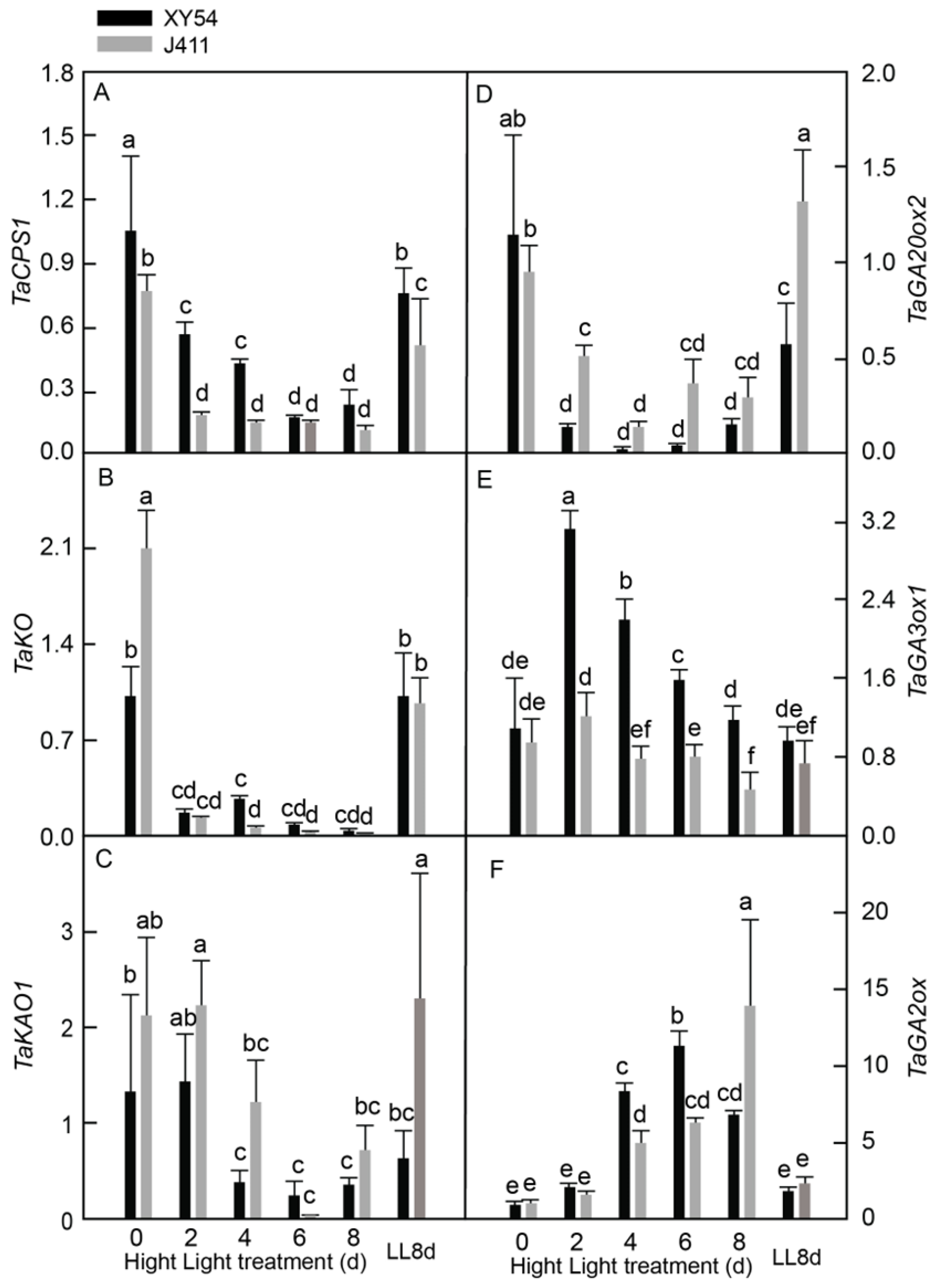
2.3. Effects of Long-Term HL Stress on GA Biosynthesis Pathway Genes and GA Content
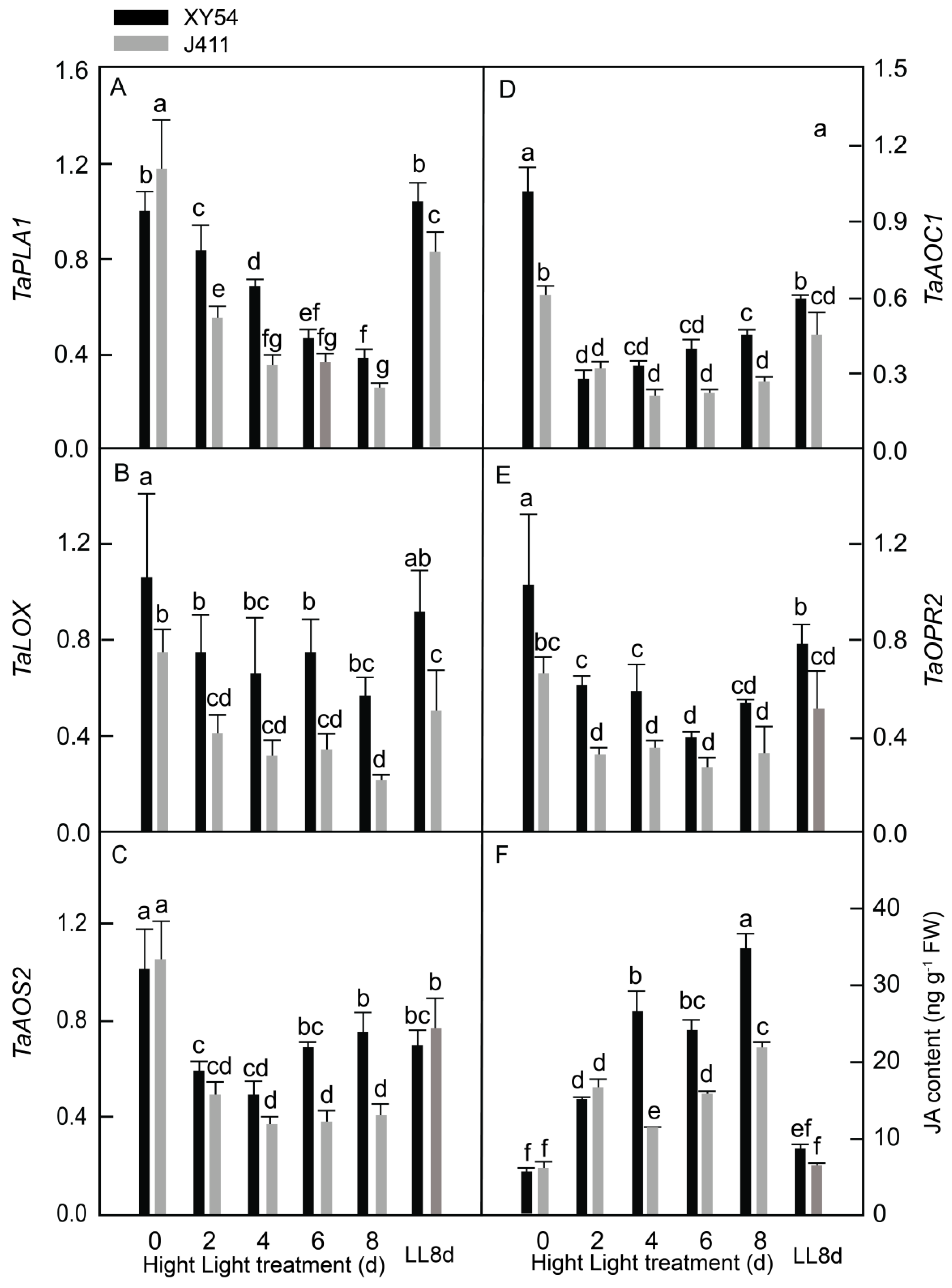
2.4. JA Biosynthesis Gene Expression and Accumulation under Long-Term HL Stress
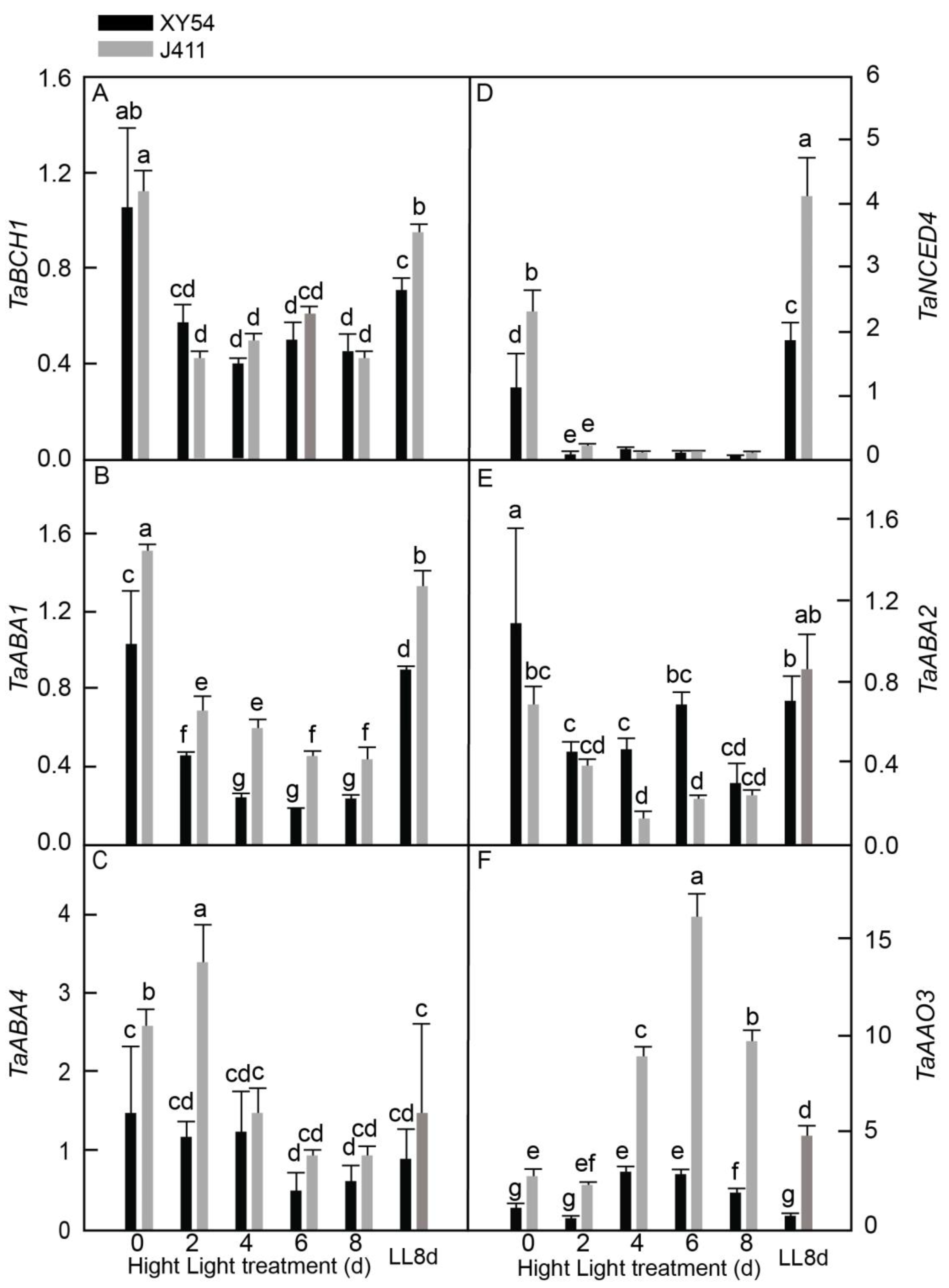
2.5. Expression and Accumulation of ABA Biosynthesis Pathway Genes under Long-Term HL Stress
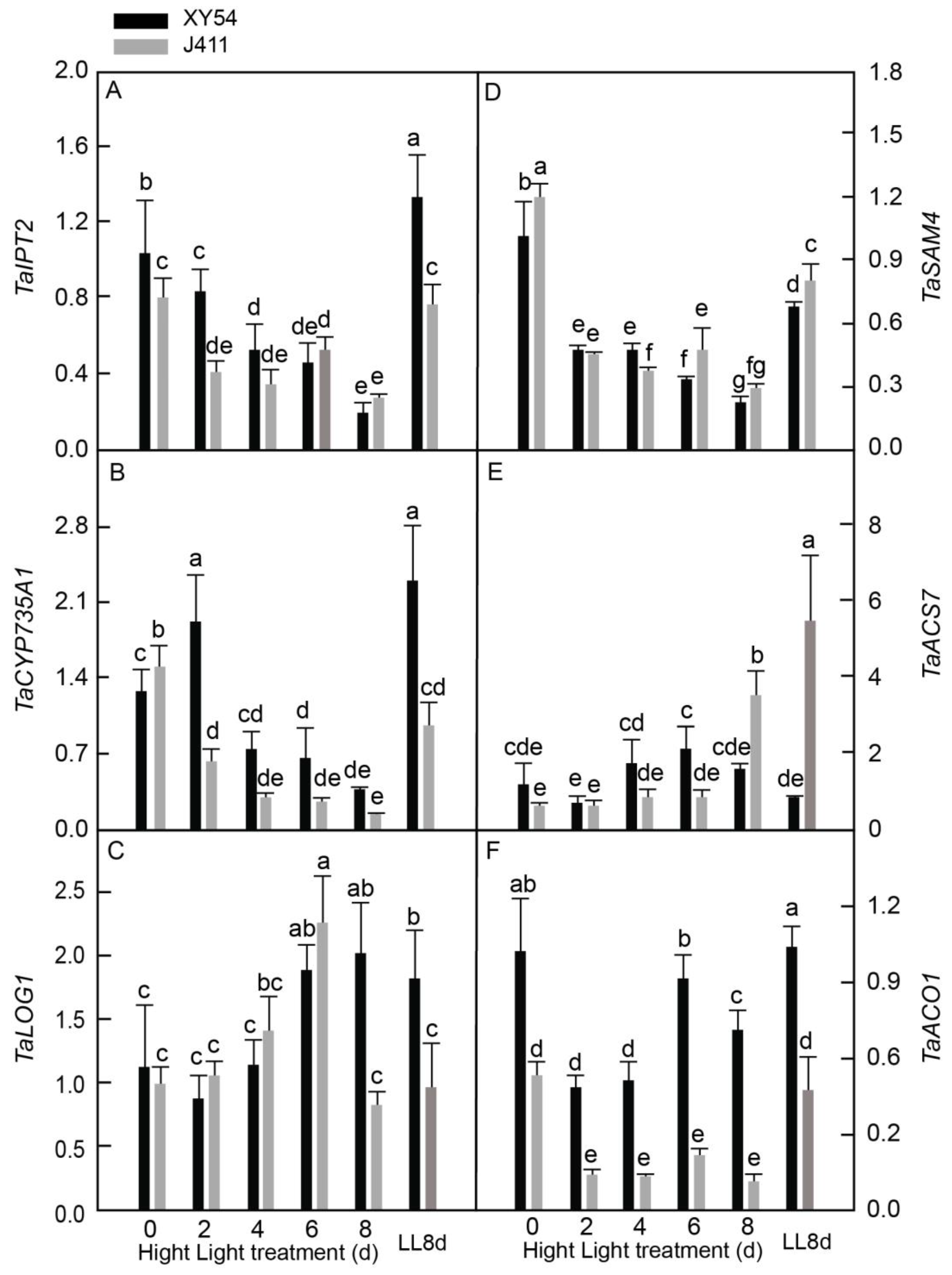
2.6. Differential Expressional Responses of CK and ET Biosynthesis Pathway Genes under Long-Term HL Stress
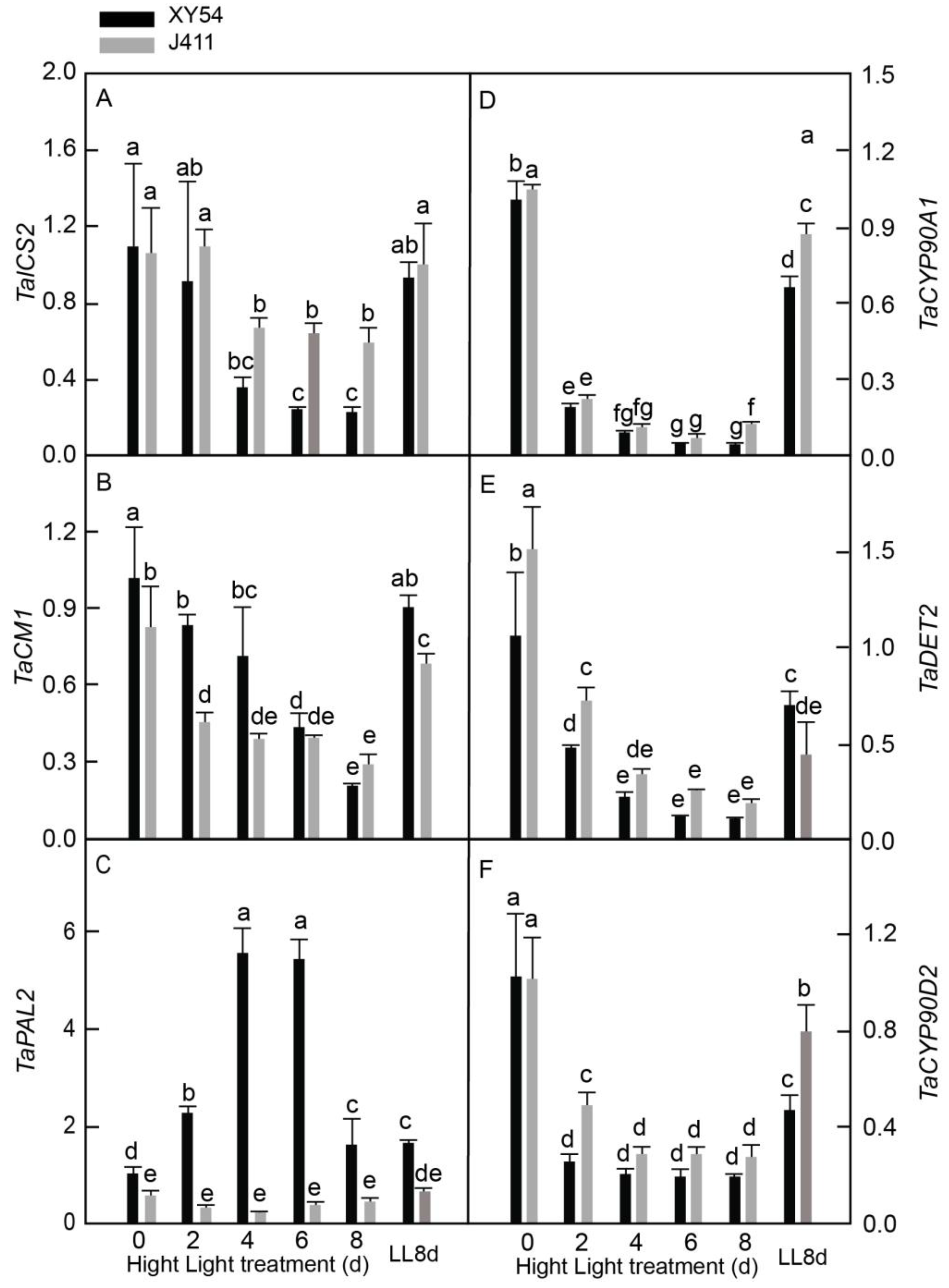
2.7. Expressional Dynamics of SA and BR Biosynthesis Pathway Genes under Long-Term HL Stress
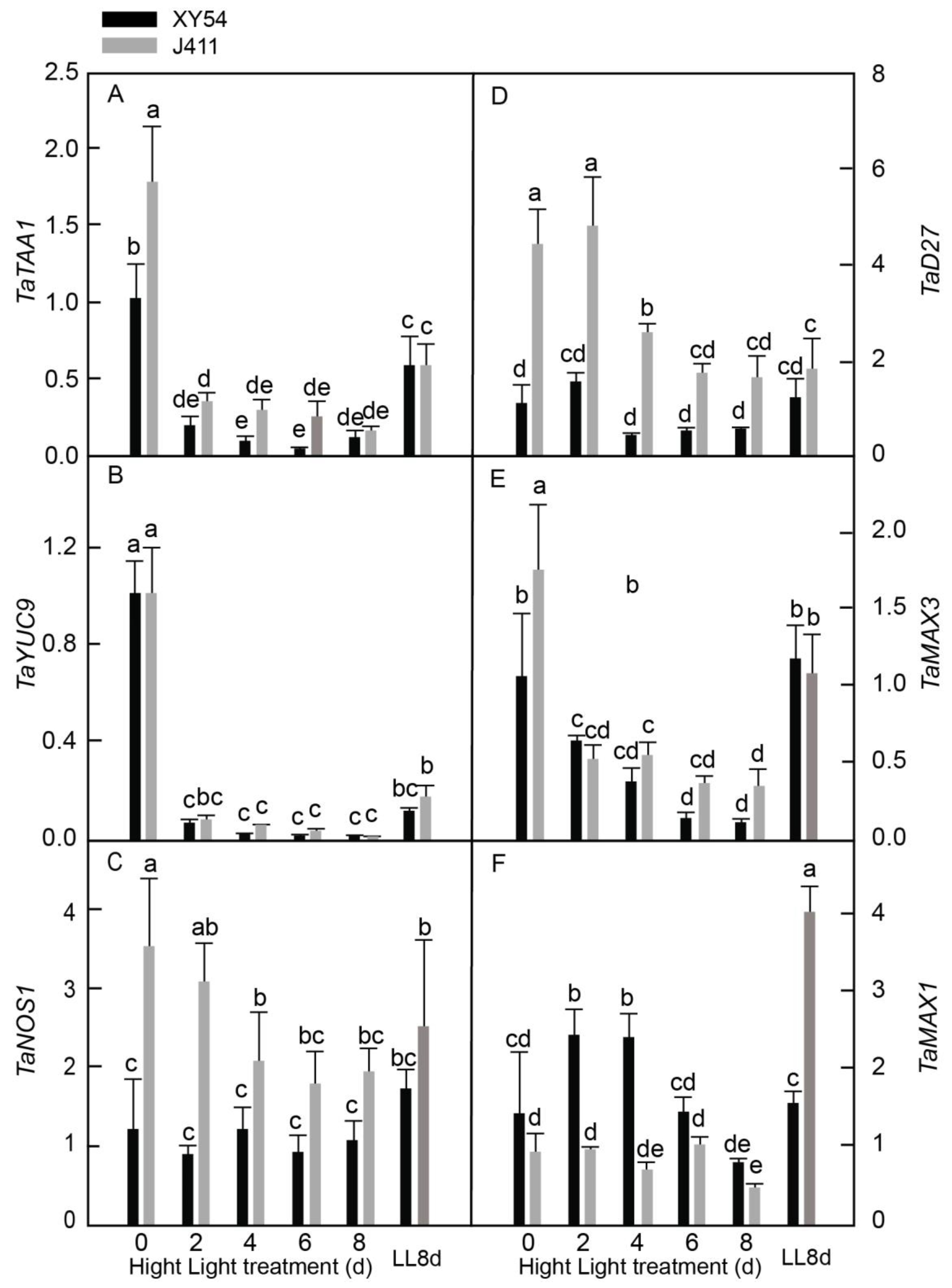
2.8. Expressional Responses of Auxin, Nitric Oxide, and Strigolactone Biosynthesis Pathway Genes under Long-Term HL Stress
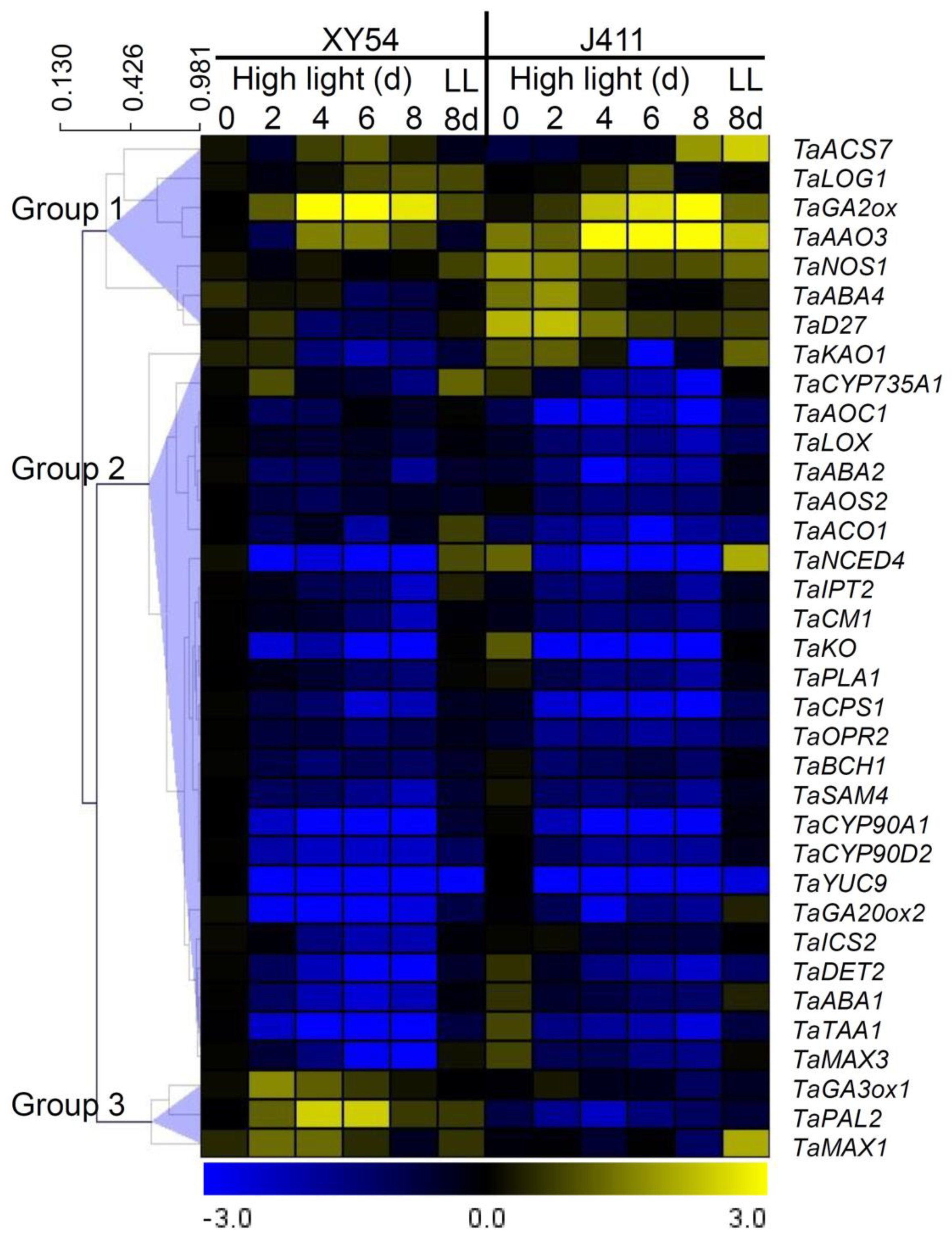
2.9. Hierarchical Clustering Analysis of Phytohormone Biosynthesis Gene Expression Patterns under Long-Term HL Stress
2.10. Correlation Analysis of Phytohormone Biosynthesis, Metabolic Changes, and Leaf Senescence under Long-Term HL Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Determination of Chlorophyll and Carotenoid Contents
4.3. Chlorophyll Fluorescence Measurement
4.4. RNA Isolation and First-Strand cDNA Synthesis
4.5. Quantification of Gene Expression
4.6. Jasmine Acid, Gibberellins, and Abscisic Acid, Quantification
4.7. Hierarchical Clustering Analysis
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shi, Y.; Ke, X.; Yang, X.; Liu, Y.; Hou, X. Plants response to light stress. J. Genet. Genom. 2022, 49, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Mudiyanselage, S.A. Interactive Response of Temperature and Light on Vegetative and Reproductive Growth of Grapevine (Vitis vinifera cv. Shiraz) under Field and Controlled Environment Conditions. Ph.D. Thesis, Charles Sturt University, Bathurst, Australia, 2016. [Google Scholar]
- Hajiboland, R. Reactive oxygen species and photosynthesis. In Oxidative Damage to Plants; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1–63. [Google Scholar]
- Ruban, A.V. Evolution under the sun: Optimizing light harvesting in photosynthesis. J. Exp. Bot. 2015, 66, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta (BBA)-Bioenerg. 2007, 1767, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Juvany, M.; Müller, M.; Munné-Bosch, S. Photo-oxidative stress in emerging and senescing leaves: A mirror image? J. Exp. Bot. 2013, 64, 3087–3098. [Google Scholar] [CrossRef]
- Wang, N.; Fang, W.; Han, H.; Sui, N.; Li, B.; Meng, Q.W. Overexpression of zeaxanthin epoxidase gene enhances the sensitivity of tomato PSII photoinhibition to high light and chilling stress. Physiol. Plant. 2008, 132, 384–396. [Google Scholar] [CrossRef]
- Wang, N.; Li, B.; Feng, H.-L.; Zhang, Q.-Y.; Yang, X.-H.; Meng, Q.-W. Antisense-mediated suppression of tomato zeaxanthin epoxidase alleviates photoinhibition of PSII and PSI during chilling stress under low irradiance. Photosynthetica 2010, 48, 409–416. [Google Scholar] [CrossRef]
- Galvez-Valdivieso, G.; Fryer, M.J.; Lawson, T.; Slattery, K.; Truman, W.; Smirnoff, N.; Asami, T.; Davies, W.J.; Jones, A.M.; Baker, N.R. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 2009, 21, 2143–2162. [Google Scholar] [CrossRef]
- Cortleven, A.; Nitschke, S.; Klaumünzer, M.; AbdElgawad, H.; Asard, H.; Grimm, B.; Riefler, M.; Schmülling, T. A novel protective function for cytokinin in the light stress response is mediated by the Arabidopsis histidine kinase2 and Arabidopsis histidine kinase3 receptors. Plant Physiol. 2014, 164, 1470–1483. [Google Scholar] [CrossRef]
- Ogweno, J.O.; Hu, W.H.; Song, X.S.; Shi, K.; Mao, W.H.; Zhou, Y.H.; Yu, J.Q. Photoinhibition-induced reduction in photosynthesis is alleviated by abscisic acid, cytokinin and brassinosteroid in detached tomato leaves. Plant Growth Regul. 2010, 60, 175–182. [Google Scholar] [CrossRef]
- Zhao, H.-J.; Zhao, X.-J.; Ma, P.-F.; Wang, Y.-X.; Hu, W.-W.; Li, L.-H.; Zhao, Y.-D. Effects of salicylic acid on protein kinase activity and chloroplast D1 protein degradation in wheat leaves subjected to heat and high light stress. Acta Ecol. Sin. 2011, 31, 259–263. [Google Scholar] [CrossRef]
- Parrine, D.; Wu, B.-S.; Muhammad, B.; Rivera, K.; Pappin, D.; Zhao, X.; Lefsrud, M. Proteome modifications on tomato under extreme high light induced-stress. Proteome Sci. 2018, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Zhao, M.; Tian, Y.; Wang, Y.; Han, C.; Fan, M.; Guo, H.; Bai, M.Y. Interaction between BZR1 and EIN3 mediates signalling crosstalk between brassinosteroids and ethylene. New Phytol. 2021, 232, 2308–2323. [Google Scholar] [CrossRef] [PubMed]
- Fahad, M.; Altaf, M.T.; Jamil, A.; Basit, A.; Aslam, M.M.; Liaqat, W.; Shah, M.N.; Ullah, I.; Mohamed, H.I. Functional characterization of transcriptional activator gene SIARRI in tomato reveals its role in fruit growth and ripening. Transgenic Res. 2023, 32, 77–93. [Google Scholar] [CrossRef]
- Balfagón, D.; Sengupta, S.; Gómez-Cadenas, A.; Fritschi, F.B.; Azad, R.K.; Mittler, R.; Zandalinas, S.I. Jasmonic acid is required for plant acclimation to a combination of high light and heat stress. Plant Physiol. 2019, 181, 1668–1682. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Gil Nam, H. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.-Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef]
- Jeong, Y.J.; Shang, Y.; Kim, B.H.; Kim, S.Y.; Song, J.H.; Lee, J.S.; Lee, M.M.; Li, J.; Nam, K.H. BAK7 displays unequal genetic redundancy with BAK1 in brassinosteroid signaling and early senescence in Arabidopsis. Mol. Cells 2010, 29, 259–266. [Google Scholar] [CrossRef]
- de Assis-Gomes, M.d.M.; Pinheiro, D.T.; Bressan-Smith, R.; Campostrini, E. Exogenous brassinosteroid application delays senescence and promotes hyponasty in Carica papaya L. leaves. Theor. Exp. Plant Physiol. 2018, 30, 193–201. [Google Scholar] [CrossRef]
- Sağlam-Çağ, S. The effect of epibrassinolide on senescence in wheat leaves. Biotechnol. Biotechnol. Equip. 2007, 21, 63–65. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef]
- Li, H.; Tong, Y.; Li, B.; Jing, R.; Lu, C.; Li, Z. Genetic analysis of tolerance to photo-oxidative stress induced by high light in winter wheat (Triticum aestivum L.). J. Genet. Genom. 2010, 37, 399–412. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Q.; Li, W.; Yang, X.; Zheng, Q.; Li, B.; Li, Z.; Li, H. Long-term high light stress induces leaf senescence in wheat (Triticum aestivum L.). Photosynthetica 2019, 57, 830–840. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Liu, M.; Zheng, Q.; Li, B.; Li, Z.; Li, H. Sugar accumulation is associated with leaf senescence induced by long-term high light in wheat. Plant Sci. 2019, 287, 110169. [Google Scholar] [CrossRef]
- Yuan, K.; Wysocka-Diller, J. Phytohormone signalling pathways interact with sugars during seed germination and seedling development. J. Exp. Bot. 2006, 57, 3359–3367. [Google Scholar] [CrossRef] [PubMed]
- Su-Wei, W.; Chang-Cheng, X.; Ke-Zhi, B.; Qi-De, Z.; Liang-Bi, L.; Ting-Yun, K.; Ji-Yun, L.; Zhen-Sheng, L. Comparative study on photoinhibition between two wheat genotypes. J. Integr. Plant Biol. 2000, 42, 1300. [Google Scholar]
- Chen, X.; Li, W.; Lu, Q.; Wen, X.; Li, H.; Kuang, T.; Li, Z.; Lu, C. The xanthophyll cycle and antioxidative defense system are enhanced in the wheat hybrid subjected to high light stress. J. Plant Physiol. 2011, 168, 1828–1836. [Google Scholar] [CrossRef]
- Li, H.; Wang, G.; Zheng, Q.; Li, B.; Jing, R.; Li, Z. Genetic analysis of biomass and photosynthetic parameters in wheat grown in different light intensities. J. Integr. Plant Biol. 2014, 56, 594–604. [Google Scholar] [CrossRef]
- Horton, P. Prospects for crop improvement through the genetic manipulation of photosynthesis: Morphological and biochemical aspects of light capture. J. Exp. Bot. 2000, 51, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.; Reynolds, M.; Salvucci, M.E.; Raines, C.; Andralojc, P.J.; Zhu, X.-G.; Price, G.D.; Condon, A.G.; Furbank, R.T. Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J. Exp. Bot. 2011, 62, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Altmann, M.; Altmann, S.; Rodriguez, P.A.; Weller, B.; Elorduy Vergara, L.; Palme, J.; Marín-De la Rosa, N.; Sauer, M.; Wenig, M.; Villaécija-Aguilar, J.A. Extensive signal integration by the phytohormone protein network. Nature 2020, 583, 271–276. [Google Scholar] [CrossRef]
- Li, J.; Yu, K.; Wei, J.; Ma, Q.; Wang, B.; Yu, D. Gibberellin retards chlorophyll degradation during senescence of Paris polyphylla. Biol. Plant. 2010, 54, 395–399. [Google Scholar] [CrossRef]
- Fletcher, R.; Osborne, D.J. Regulation of protein and nucleic acid synthesis by gibberellin during leaf senescence. Nature 1965, 207, 1176–1177. [Google Scholar] [CrossRef]
- Chen, M.; Maodzeka, A.; Zhou, L.; Ali, E.; Wang, Z.; Jiang, L. Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Sci. 2014, 219, 26–34. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z.; Wang, X.; Wang, J.; Fan, K.; Li, Z.; Lin, W. DELLA proteins negatively regulate dark-induced senescence and chlorophyll degradation in Arabidopsis through interaction with the transcription factor WRKY6. Plant Cell Rep. 2018, 37, 981–992. [Google Scholar] [CrossRef]
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flugge, U.-I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef]
- Radi, A.; Lange, T.; Niki, T.; Koshioka, M.; Lange, M.J.P. Ectopic expression of pumpkin gibberellin oxidases alters gibberellin biosynthesis and development of transgenic Arabidopsis plants. Plant Physiol. 2006, 140, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.G.; Yang, M.; Liu, P.; Yang, G.D.; Wu, C.A.; Zheng, C.C. GhDREB1 enhances abiotic stress tolerance, delays GA-mediated development and represses cytokinin signalling in transgenic Arabidopsis. Plant Cell Environ. 2009, 32, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef] [PubMed]
- Zarepour, M.; Simon, K.; Wilch, M.; Nieländer, U.; Koshiba, T.; Seo, M.; Lindel, T.; Bittner, F. Identification of superoxide production by Arabidopsis thaliana aldehyde oxidases AAO1 and AAO3. Plant Mol. Biol. 2012, 80, 659–671. [Google Scholar] [CrossRef]
- Raab, S.; Drechsel, G.; Zarepour, M.; Hartung, W.; Koshiba, T.; Bittner, F.; Hoth, S. Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 2009, 59, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Worley, E.; Udvardi, M. A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, X.; Yang, Y.; Kim, H.S.; Jia, X.; Yu, H.; Kwak, S.-S. Sweetpotato bZIP transcription factor IbABF4 confers tolerance to multiple abiotic stresses. Front. Plant Sci. 2019, 10, 630. [Google Scholar] [CrossRef]
- Smart, C.M.; Scofield, S.R.; Bevan, M.W.; Dyer, T.A. Delayed leaf senescence in tobacco plants transformed with tmr, a gene for cytokinin production in Agrobacterium. Plant Cell 1991, 3, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef]
- Sun, G.; Mei, Y.; Deng, D.; Xiong, L.; Sun, L.; Zhang, X.; Wen, Z.; Liu, S.; You, X.; Nasrullah; et al. N-terminus-mediated degradation of ACS7 is negatively regulated by senescence signaling to allow optimal ethylene production during leaf development in Arabidopsis. Front. Plant Sci. 2017, 8, 2066. [Google Scholar] [CrossRef] [PubMed]
- Gawroński, P.; Górecka, M.; Bederska, M.; Rusaczonek, A.; Ślesak, I.; Kruk, J.; Karpiński, S. Isochorismate synthase 1 is required for thylakoid organization, optimal plastoquinone redox status, and state transitions in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 3669–3679. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.-y.; Hsu, S.-Y.; Huei Kao, C. Nitric oxide counteracts the senescence of detached rice leaves induced by dehydration and polyethylene glycol but not by sorbitol. Plant Growth Regul. 2002, 38, 265–272. [Google Scholar] [CrossRef]
- Hung, K.T.; Kao, C.H. Nitric oxide counteracts the senescence of rice leaves induced by abscisic acid. J. Plant Physiol. 2003, 160, 871–879. [Google Scholar] [CrossRef]
- Niu, Y.H.; Guo, F.Q. Nitric Oxide Regulates Dark-Induced Leaf Senescence Through EIN2 in Arabidopsis F. J. Integr. Plant Biol. 2012, 54, 516–525. [Google Scholar] [CrossRef]
- Yamada, Y.; Furusawa, S.; Nagasaka, S.; Shimomura, K.; Yamaguchi, S.; Umehara, M. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 2014, 240, 399–408. [Google Scholar] [CrossRef]
- Ueda, H.; Kusaba, M. Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 2015, 169, 138–147. [Google Scholar] [CrossRef]
- Tian, M.-Q.; Jiang, K.; Takahashi, I.; Li, G.-D. Strigolactone-induced senescence of a bamboo leaf in the dark is alleviated by exogenous sugar. J. Pestic. Sci. 2018, 43, 173–179. [Google Scholar] [CrossRef]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar]
- Li, H.; Zheng, Q.; Zhang, J.; Li, B.; Li, Z. The analysis of determining factors and evaluation of tolerance to photoinhibition in wheat (Triticum aestivum L.). Photosynthetica 2017, 55, 69–76. [Google Scholar]
- Kitajima, M.; Butler, W. Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim. Biophys. Acta (BBA)-Bioenerg. 1975, 376, 105–115. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Schreiber, U.; Schliwa, U.; Bilger, W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 1986, 10, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Saeed, A.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
| Phytohormones | Genes | Fructose | Sucrose | Starch | Chlorophyll | TaAGP |
|---|---|---|---|---|---|---|
| Gibberellins | TaCPS1 | −0.821 ** | −0.817 ** | −0.786 ** | 0.678 ** | 0.727 ** |
| TaKO | −0.832 ** | −0.789 ** | −0.775 ** | 0.721 ** | 0.853 ** | |
| TaKAO1 | −0.461 ** | −0.450 ** | −0.510 ** | 0.673 ** | 0.591 ** | |
| TaGA20ox2 | −0.764 ** | −0.679 ** | −0.696 ** | 0.655 ** | 0.823 ** | |
| TaGA2ox | 0.679 ** | 0.657 ** | 0.758 ** | −0.766 ** | −0.721 ** | |
| GA content | - | 0.859 ** | 0.759 ** | 0.794 ** | −0.691 ** | −0.822 ** |
| Cytokinins | TaIPT2 | −0.795 ** | −0.782 ** | −0.779 ** | 0.611 ** | 0.684 ** |
| TaCYP735A1 | −0.664 ** | −0.698 ** | −0.699 ** | 0.646 ** | 0.584 ** | |
| Ethylene | TaSAM4 | −0.841 ** | −0.801 ** | −0.818 ** | 0.764 ** | 0.894 ** |
| Salicylic acid | TaICS2 | −0.599 ** | −0.544 ** | −0.636 ** | 0.691 ** | 0.719 ** |
| TaCM1 | −0.788 ** | −0.834 ** | −0.816 ** | 0.739 ** | 0.675 ** | |
| Abscisic acid | TaBCH1 | −0.788 ** | −0.720 ** | −0.719 ** | 0.625 ** | 0.829 ** |
| TaABA1 | −0.837 ** | −0.759 ** | −0.789 ** | 0.785 ** | 0.935 ** | |
| TaNCED4 | −0.794 ** | −0.734 ** | −0.718 ** | 0.690 ** | 0.740 ** | |
| TaABA2 | −0.750 ** | −0.778 ** | −0.698 ** | 0.552 ** | 0.616 ** | |
| TaAAO3 | 0.469 ** | 0.609 ** | 0.538 ** | −0.497 ** | −0.288 * | |
| ABA content | - | 0.816 ** | 0.788 ** | 0.688 ** | −0.767 ** | −0.798 ** |
| Brassinosteroids | TaCYP90A1 | −0.936 ** | −0.873 ** | −0.867 ** | 0.784 ** | 0.921 ** |
| TaDET2 | −0.703 ** | −0.669 ** | −0.723 ** | 0.746 ** | 0.860 ** | |
| TaCYP90D2 | −0.794 ** | −0.731 ** | −0.753 ** | 0.699 ** | 0.875 ** | |
| Indole-3-acetic acid | TaTAA1 | −0.706 ** | −0.647 ** | −0.673 ** | 0.656 ** | 0.863 ** |
| TaYUC9 | −0.660 ** | −0.623 ** | −0.628 ** | 0.565 ** | 0.744 ** | |
| Strigolactones | TaMAX3 | −0.820 ** | −0.778 ** | −0.788 ** | 0.782 ** | 0.904 ** |
| Jasmonic acid (JA) | TaLPLA1 | −0.878 ** | −0.895 ** | −0.881 ** | 0.822 ** | 0.827 ** |
| TaLOX | −0.579 ** | −0.650 ** | −0.603 ** | 0.422 ** | 0.430 ** | |
| TaAOS2 | −0.701 ** | −0.721 ** | −0.639 ** | 0.523 ** | 0.655 ** | |
| TaAOC1 | −0.559 ** | −0.567 ** | −0.498 ** | 0.305 * | 0.327 * | |
| TaOPR2 | −0.673 ** | −0.688 ** | −0.635 ** | 0.463 ** | 0.527 ** | |
| JA content | - | 0.759 ** | 0.694 ** | 0.777 ** | −0.747 ** | −0.806 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.-A.; Fahad, M.; Li, W.-C.; Tariq, L.; Liu, M.-M.; Liu, Y.-N.; Wang, T.-X. Comparative Analysis of Phytohormone Biosynthesis Genes Responses to Long-Term High Light in Tolerant and Sensitive Wheat Cultivars. Plants 2024, 13, 2628. https://doi.org/10.3390/plants13182628
Li Z-A, Fahad M, Li W-C, Tariq L, Liu M-M, Liu Y-N, Wang T-X. Comparative Analysis of Phytohormone Biosynthesis Genes Responses to Long-Term High Light in Tolerant and Sensitive Wheat Cultivars. Plants. 2024; 13(18):2628. https://doi.org/10.3390/plants13182628
Chicago/Turabian StyleLi, Zhi-Ang, Muhammad Fahad, Wan-Chang Li, Leeza Tariq, Miao-Miao Liu, Ya-Nan Liu, and Tai-Xia Wang. 2024. "Comparative Analysis of Phytohormone Biosynthesis Genes Responses to Long-Term High Light in Tolerant and Sensitive Wheat Cultivars" Plants 13, no. 18: 2628. https://doi.org/10.3390/plants13182628
APA StyleLi, Z.-A., Fahad, M., Li, W.-C., Tariq, L., Liu, M.-M., Liu, Y.-N., & Wang, T.-X. (2024). Comparative Analysis of Phytohormone Biosynthesis Genes Responses to Long-Term High Light in Tolerant and Sensitive Wheat Cultivars. Plants, 13(18), 2628. https://doi.org/10.3390/plants13182628





