Pomological and Molecular Characterization of Apple Cultivars in the German Fruit Genebank
Abstract
:1. Introduction
2. Results
2.1. True-to-Type Apple Cultivars across GFG Collection
2.2. Dataset Assessment and Power of SSR Markers
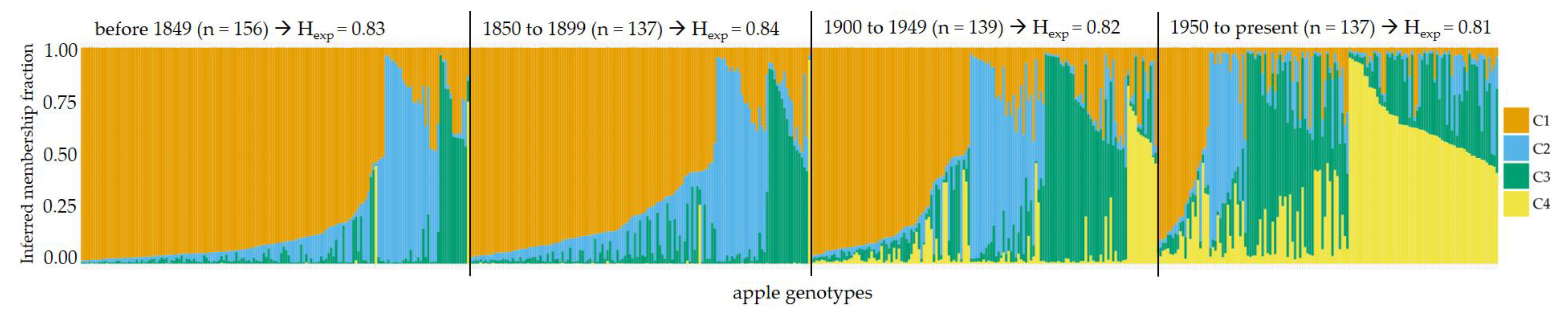
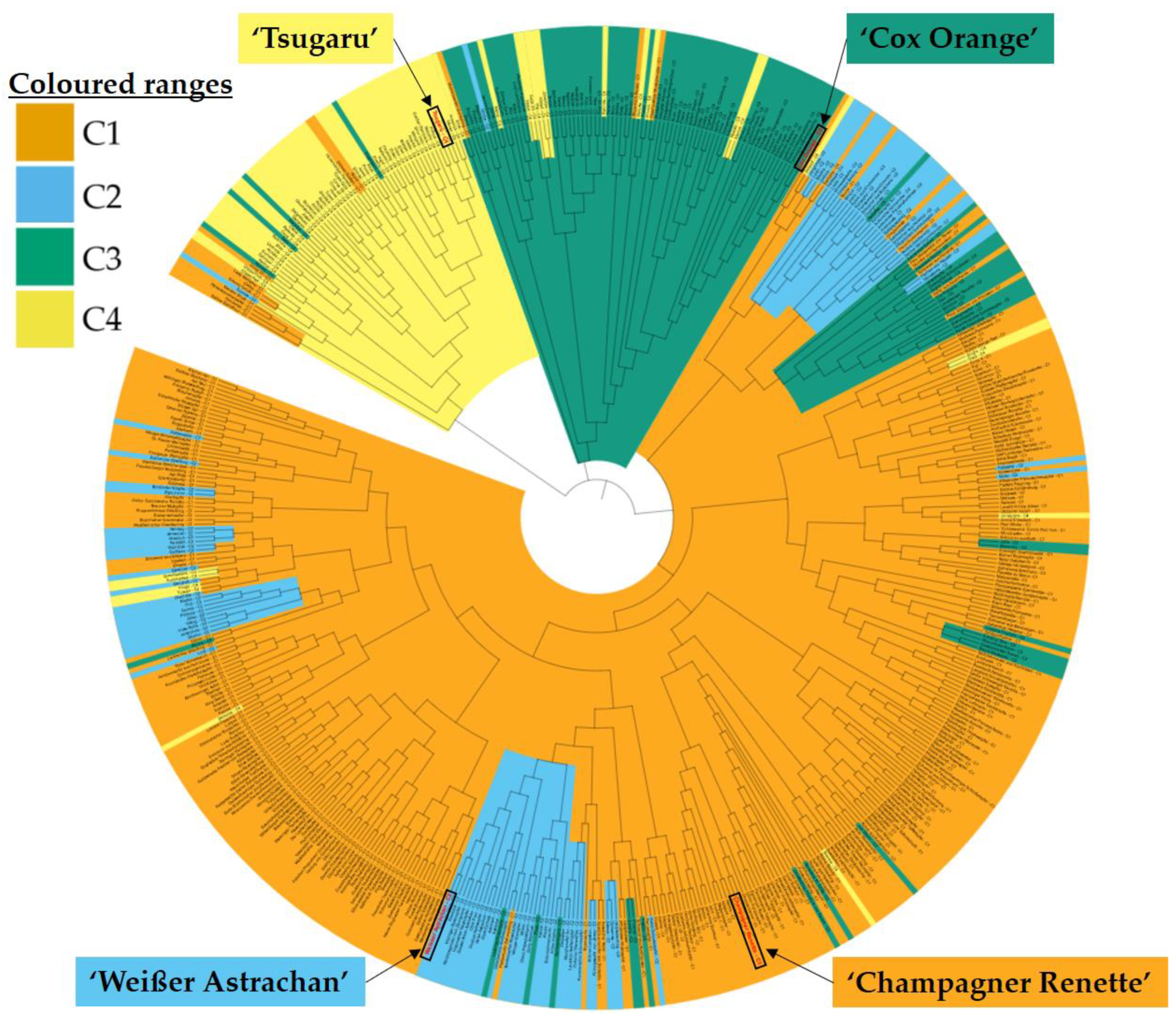
2.3. Genetic Structure of Apple Cultivars in Context of Age and Origin
2.4. Parent-Child-Relationships between Apple Cultivars
3. Discussion
3.1. The Importance of Cultivar Identification and the Cultivar Context
3.2. Genetic Diversity and Structure
3.3. Assumed Parentages Confirmed for Most Apple Cultivars
4. Conclusions
5. Materials and Methods
5.1. Trueness-to-Type Criterion
5.2. Probability of Identity Calculation and Diversity Parameters
5.3. STRUCTURE Analysis and Phylogenetic Tree
5.4. Parentage Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahbandeh, M. Global Production of Fruit by Variety Selected. 2021. Available online: https://www.statista.com/statistics/264001/worldwide-production-of-fruit-by-variety/ (accessed on 8 August 2023).
- BLE. Äpfel Produktinformation. Available online: https://www.ble-medienservice.de/simpledownloadable/freedownload/link/hash/509e27ccc3b1b81ce7b6405f8154c63a/ (accessed on 8 August 2023).
- BZL. Äpfel. Available online: https://www.landwirtschaft.de/landwirtschaftliche-produkte/wie-werden-unsere-lebensmittel-erzeugt/pflanzliche-produkte/aepfel (accessed on 23 February 2024).
- Ahrens, S. Sortenanteile der Wichtigsten Apfelsorten in Deutschland 2021/22. Available online: https://de.statista.com/statistik/daten/studie/29118/umfrage/bedeutende-apfelsorten-in-deutschland-nach-sortenanteil/ (accessed on 8 August 2023).
- Ahrens, S. Pro-Kopf-Konsum von Obst in Deutschland nach Art Bis 2021/22. Available online: https://de.statista.com/statistik/daten/studie/247425/umfrage/die-beliebtesten-obstsorten-der-deutschen/ (accessed on 8 August 2023).
- Laurens, F.; Aranzana, M.J.; Arus, P.; Bassi, D.; Bink, M.; Bonany, J.; Caprera, A.; Corelli-Grappadelli, L.; Costes, E.; Durel, C.E.; et al. An integrated approach for increasing breeding efficiency in apple and peach in Europe. Hortic. Res. 2018, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Mittler, R.; Blumwald, E.; Zandalinas, S.I. Developing climate-resilient crops: Improving plant tolerance to stress combination. Plant J. 2022, 109, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M. Zur Geschichte der Genbank Obst Dresden-Pillnitz. Vortr. Pflanzenzüchtg. 2003, 57, 5–6. [Google Scholar]
- Salgotra, R.K.; Chauhan, B.S. Genetic Diversity, Conservation, and Utilization of Plant Genetic Resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Fu, Y.B. Understanding crop genetic diversity under modern plant breeding. Theor. Appl. Genet. 2015, 128, 2131–2142. [Google Scholar] [CrossRef]
- Khoury, C.K.; Brush, S.; Costich, D.E.; Curry, H.A.; de Haan, S.; Engels, J.M.M.; Guarino, L.; Hoban, S.; Mercer, K.L.; Miller, A.J.; et al. Crop genetic erosion: Understanding and responding to loss of crop diversity. New Phytol. 2022, 233, 84–118. [Google Scholar] [CrossRef]
- Höfer, M.; Flachowsky, H. Preservation of fruit genetic resources in Germany. Acta Hortic. 2021, 1307, 163–170. [Google Scholar] [CrossRef]
- Broschewitz, L.; Bannier, H.-J.; Reim, S.; Flachowsky, H.; Höfer, M. Microsatellite/SSR dataset: Pomological and molecular characterization of apple cultivars (Malus × domestica Borkh.) of the German Fruit Genebank. OpenAgrar Repos. 2023. [Google Scholar] [CrossRef]
- Muranty, H.; Denancé, C.; Feugey, L.; Crépin, J.-L.; Barbier, Y.; Tartarini, S.; Ordidge, M.; Troggio, M.; Lateur, M.; Nybom, H.; et al. Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biology 2020, 20, 2. [Google Scholar] [CrossRef]
- Azizi, M.M.F.; Lau, H.Y.; Abu-Bakar, N. Integration of advanced technologies for plant variety and cultivar identification. J Biosci 2021, 46, 1–20. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R.; Chen, X.; Panaud, O.; Temnykh, S.; Xu, Y.; Cho, Y.G.; Huang, N.; Ishii, T.; Matthew Blair, M. Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol. Biol. 1997, 35, 89–99. [Google Scholar] [CrossRef]
- Frey, J.E.; Koller, B.; Frey, B.; Bünter, M. Identifikation von Obstsorten: Validierung einer Analysemethode. AGRARForschung Schweiz 2007, 14, 536–541. [Google Scholar]
- Testolin, R.; Messina, R.; Cipriani, G.; de Mori, G. SSR-based DNA fingerprinting of fruit crops. Crop Sci. 2023, 63, 390–459. [Google Scholar] [CrossRef]
- Evans, K.M.; Patocchi, A.; Rezzonico, F.; Mathis, F.; Durel, C.E.; Fernández-Fernández, F.; Boudichevskaia, A.; Dunemann, F.; Stankiewicz-Kosyl, M.; Gianfranceschi, L.; et al. Genotyping of pedigreed apple breeding material with a genome-covering set of SSRs: Trueness-to-type of cultivars and their parentages. Mol. Breed. 2010, 28, 535–547. [Google Scholar] [CrossRef]
- Denancé, C.; Muranty, H.; Durel, C.-E. MUNQ—Malus UNiQue genotype code for grouping apple accessions corresponding to a unique genotypic profile. Portail Data INRAE 2020, 1. [Google Scholar] [CrossRef]
- Durel, C.E.; Denancé, C.; Muranty, H.; Lateur, M.; Ordidge, M. MUNQ and PUN—A European and international apple and pear germplasm coding system. Acta Hortic. 2023, 1384, 471–476. [Google Scholar] [CrossRef]
- Julius Kühn-Institut—Federal Research Centre for Cultivated Plants. Deutsche Genbank Obst (German Fruit Genebank). Available online: https://www.deutsche-genbank-obst.de (accessed on 10 December 2023).
- Bannier, H.-J.; Schuricht, W. Pomologischer Abschlussbericht: Zweite pomologische Bestimmung der Apfelsorten der Deutschen Genbank Obst. Available online: https://www.deutsche-genbank-obst.de/uploads/3QsxZ01fFus116dtuGQlnQCxjRojJdiU.pdf (accessed on 10 September 2024).
- Morgan, J.; Richards, A.; Dowle, E. The Book of Apples; Ebury Press, Brogdale Horticulture Trust: London, UK, 1993. [Google Scholar]
- Reim, S.; Schiffler, J.; Braun-Lullemann, A.; Schuster, M.; Flachowsky, H.; Hofer, M. Genetic and Pomological Determination of the Trueness-to-Type of Sweet Cherry Cultivars in the German National Fruit Genebank. Plants 2023, 12, 205. [Google Scholar] [CrossRef]
- Sticca, E.L.; Belbin, G.M.; Gignoux, C.R. Current Developments in Detection of Identity-by-Descent Methods and Applications. Front. Genet. 2021, 12, 722602. [Google Scholar] [CrossRef]
- Lassois, L.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Hibrand-Saint-Oyant, L.; Poncet, C.; Lasserre-Zuber, P.; Feugey, L.; Durel, C.-E. Genetic Diversity, Population Structure, Parentage Analysis, and Construction of Core Collections in the French Apple Germplasm Based on SSR Markers. Plant Mol. Biol. Report. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Denance, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Poncet, C.; Lateur, M.; Houben, P.; Ordidge, M.; et al. Analysis of the genetic diversity and structure across a wide range of germplasm reveals prominent gene flow in apple at the European level. BMC Plant Biol. 2016, 16, 130. [Google Scholar] [CrossRef] [PubMed]
- Baric, S.; Storti, A.; Hofer, M.; Guerra, W.; Dalla Via, J. Molecular Genetic Identification of Apple Cultivars Based on Microsatellite DNA Analysis. I. The Database of 600 Validated Profiles. Erwerbs-Obstbau 2020, 62, 117–154. [Google Scholar] [CrossRef]
- Van Treuren, R.; Kemp, H.; Ernsting, G.; Jongejans, B.; Houtman, H.; Visser, L. Microsatellite genotyping of apple (Malus × domestica Borkh.) genetic resources in the Netherlands: Application in collection management and variety identification. Genet. Resour. Crop Evol. 2010, 57, 853–865. [Google Scholar] [CrossRef]
- Watkins, R. Apple and pear. In Evolution of Crop Plants, 2nd ed.; Smartt, J., Simmonds, N.W., Eds.; Wiley: Hoboken, NJ, USA, 1995; pp. 418–422. [Google Scholar]
- Janick, J. The Origins of Fruits, Fruit Growing, and Fruit Breeding. Plant Breed. Rev. 2005, 25, 255–321. [Google Scholar]
- Migicovsky, Z.; Gardner, K.M.; Richards, C.; Thomas Chao, C.; Schwaninger, H.R.; Fazio, G.; Zhong, G.-Y.; Myles, S. Genomic consequences of apple improvement. Hortic. Res. 2021, 8, 9. [Google Scholar] [CrossRef]
- Hokanson, S.C.; Lamboy, W.F.; Szewc-McFadden, A.K.; McFerson, J.R. Microsatellite (SSR) variation in a collection of Malus (apple) species and hybrids. Euphytica 2001, 118, 281–294. [Google Scholar] [CrossRef]
- Larsen, B.; van Dooijeweert, W.; Durel, C.-E.; Denancé, C.; Rutten, M.; Howard, N.P. SNP genotyping Dutch heritage apple cultivars allows for germplasm characterization, curation, and pedigree reconstruction using genotypic data from multiple collection sites across the world. Tree Genet. Genomes 2024, 20, 21. [Google Scholar] [CrossRef]
- Luby, J.J.; Howard, N.P.; Tillman, J.R.; Bedford, D.S. Extended Pedigrees of Apple Cultivars from the University of Minnesota Breeding Program Elucidated Using SNP Array Markers. HortScience 2022, 57, 472–477. [Google Scholar] [CrossRef]
- Khan, A.; Carey, S.B.; Serrano, A.; Zhang, H.; Hargarten, H.; Hale, H.; Harkess, A.; Honaas, L. A phased, chromosome-scale genome of ‘Honeycrisp’ apple (Malus domestica). Gigabyte 2022, 2022, gigabyte69. [Google Scholar] [CrossRef]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef]
- James, T.; Johnson, A.; Schaller, A.; Vanderzande, S.; Luo, F.; Sandefur, P.; Ru, S.; Peace, C. As It Stands: The Palouse Wild Cider Apple Breeding Program. Plants 2022, 11, 517. [Google Scholar] [CrossRef] [PubMed]
- Reim, S.; Flachowsky, H.; Hanke, M.V.; Peil, A. Verifying the parents of Pillnitzer apple cultivars. Acta Hortic. 2009, 814, 319–324. [Google Scholar] [CrossRef]
- Ecogenics GmbH c/o Microsynth AG. Molekularer Abschlussbericht: Zweite molekulargenetische Bestimmung der Apfelsorten der Deutschen Genbank Obst. Available online: https://www.deutsche-genbank-obst.de/uploads/c0aGk44pCMq-gu1805Ac7zhPs8D_FyzE.pdf (accessed on 10 September 2024).
- Parker, A.; Namuth-Covert, D. APPLE: Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability; TG/14/10; UPOV: Geneva, Switzerland, 2023; pp. 14–23. [Google Scholar]
- Broschewitz, L.; Reim, S.; Flachowsky, H.; Höfer, M. Microsatellite/SSR dataset: Characterization of apple cultivars of the German Fruit Genebank. OpenAgrar Repos. 2024, submitted.
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Kimura, M.; Crow, J.F. The Number of Alleles That Can Be Maintained in a Finite Population. Genetics 1964, 49, 725–738. [Google Scholar] [CrossRef]
- Tajima, F.; Tokunaga, T.; T. Miyashita, N. Statistical methods for estimating the effective number of alleles, expected heterozygosity and genetic distance in self-incompatibility locus. Jpn. J. Genet. 1994, 69, 287–295. [Google Scholar] [CrossRef]
- Microsoft Corporation. Microsoft Excel; Microsoft Corporation: Redmond, WA, USA, 2016. [Google Scholar]
- Microsoft Corporation. Microsoft PowerPoint; Microsoft Corporation: Redmond, WA, USA, 2016. [Google Scholar]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grunwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Posit Team. RStudio: Integrated Development Environment for R; 2024.4.1.748; Posit Software, PBC: Boston, MA, USA, 2024. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; 4.4.0 (2024-04-24 ucrt); R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Schauberger, P.; Walker, A. Openxlsx: Read, Write and Edit xlsx Files; R Package Version 4.2.5.2; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://CRAN.R-project.org/package=openxlsx (accessed on 17 September 2024).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation; R Package Version 1.1.4; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://CRAN.R-project.org/package=dplyr (accessed on 17 September 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Wickham, H.; Vaughan, D.; Girlich, M. Tidyr: Tidy Messy Data; R Package Version 1.3.1; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://CRAN.R-project.org/package=tidyr (accessed on 17 September 2024).
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: https://darwin.cirad.fr/ (accessed on 1 July 2024).
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]


| Locus | Assessed Genotypes | Na * | Ne * | Hobs * (Locus) | Hexp * (Locus) | PIC * | PID * | PIDsib * |
|---|---|---|---|---|---|---|---|---|
| Ch01h01 | 1085 | 22 | 7.04 | 0.86 | 0.86 | 0.84 | 0.04 | 0.33 |
| Ch01f07a | 1085 | 18 | 7.30 | 0.87 | 0.86 | 0.85 | 0.03 | 0.33 |
| CH03d07 | 1083 | 26 | 7.35 | 0.88 | 0.86 | 0.85 | 0.03 | 0.33 |
| CH04c07 | 1085 | 19 | 7.46 | 0.88 | 0.87 | 0.85 | 0.03 | 0.33 |
| CH05f06 | 1085 | 14 | 6.33 | 0.84 | 0.84 | 0.82 | 0.04 | 0.34 |
| CH01f03b | 1084 | 14 | 4.90 | 0.81 | 0.80 | 0.77 | 0.07 | 0.37 |
| GD12 | 1085 | 20 | 3.36 | 0.70 | 0.70 | 0.68 | 0.11 | 0.43 |
| CH02d08 | 1083 | 20 | 6.54 | 0.83 | 0.85 | 0.83 | 0.04 | 0.34 |
| CH05e03 | 1073 | 38 | 9.01 | 0.65 | 0.89 | 0.88 | 0.02 | 0.31 |
| CH02c09 | 1083 | 17 | 6.62 | 0.86 | 0.85 | 0.83 | 0.04 | 0.34 |
| CH02g09 | 1085 | 26 | 9.01 | 0.89 | 0.89 | 0.88 | 0.02 | 0.31 |
| CH01f02 | 1085 | 27 | 8.55 | 0.88 | 0.88 | 0.87 | 0.03 | 0.32 |
| CH04f10 | 1050 | 37 | 8.00 | 0.56 | 0.88 | 0.87 | 0.03 | 0.32 |
| COL | 1083 | 17 | 5.08 | 0.78 | 0.80 | 0.78 | 0.07 | 0.37 |
| Hi02c07 | 1085 | 18 | 5.10 | 0.80 | 0.80 | 0.78 | 0.06 | 0.36 |
| CH04e05 | 1085 | 26 | 4.18 | 0.79 | 0.76 | 0.74 | 0.08 | 0.39 |
| GD147 | 1085 | 16 | 5.68 | 0.83 | 0.82 | 0.80 | 0.05 | 0.35 |
| Mean | 1081.71 | 22.06 | 6.56 | 0.81 | 0.84 | 0.82 | 0.05 | 0.34 |
| n * | Hexp * | Na | Pairwise Population Matrix of Nei Genetic Identity | ||||
|---|---|---|---|---|---|---|---|
| before 1849 | 1850 to 1899 | 1900 to 1949 | 1950 to present | ||||
| before 1849 | 156 | 0.83 | 15.65 | 1 | |||
| 1850 to 1899 | 137 | 0.84 | 15.53 | 0.98 | 1 | ||
| 1900 to 1949 | 139 | 0.82 | 13.00 | 0.962 | 0.965 | 1 | |
| 1950 to present | 137 | 0.81 | 12.24 | 0.894 | 0.899 | 0.941 | 1 |
| C1 (in %) | C2 (in %) | C3 (in %) | C4 (in %) | Total (in %) | |
|---|---|---|---|---|---|
| before 1849 | 78.2 | 14.1 | 7.1 | 0.6 | 100 |
| 1850 to 1899 | 72.3 | 15.3 | 11.7 | 0.7 | 100 |
| 1900 to 1949 | 46 | 21.6 | 23.7 | 8.6 | 100 |
| 1950 to present | 15.3 | 10.9 | 29.9 | 43.8 | 100 |
| Cultivar Name | Candidate Mother | Candidate Father |
|---|---|---|
| Akane | Jonathan | Worcester Parmäne |
| Alkmene | Geheimrat Dr. Oldenburg | Cox Orange |
| Allington Pepping | Cox Orange | Goldparmäne |
| Angold | Antonovka | Golden Delicious |
| Apollo | Cox Orange | Geheimrat Dr. Oldenburg |
| Arlet | Golden Delicious | Idared |
| Aroma | Ingrid Marie | Filippa |
| Astillisch | Roter Astrachan | Signe Tillisch |
| Auralia | Cox Orange | Schöner aus Nordhausen |
| Bancroft | Forest | McIntosh |
| Berlon | Goldrenette Freiherr von Berlepsch | Ontario |
| Charles Roß | Cox Orange | Peasgoods Sondergleichen |
| Charlotte | McIntosh | Greensleeves |
| Clivia | Geheimrat Dr. Oldenburg | Cox Orange |
| Cripps Pink | Lady Williams | Golden Delicious |
| Cripps Red | Lady Williams | Golden Delicious |
| Delcorf | Jongrimes | Golden Delicious |
| Delikates | James Grieve | Cortland |
| Delorina | Blushing Golden | Florina |
| Discovery | Worcester Parmäne | Schöner aus Bath |
| Early McIntosh | Weißer Klarapfel | McIntosh |
| Ecolette | Elstar | Coop 2 |
| Elan | Golden Delicious | James Grieve |
| Elektra | Cox Orange | Geheimrat Dr. Oldenburg |
| Elstar | Golden Delicious | Ingrid Marie |
| Empire | McIntosh | Delicious |
| Ernst Bosch | Manks Küchenapfel | Ananasrenette |
| Fantazja | McIntosh | Linda |
| Fiesta | Cox Orange | Idared |
| Freyberg | Cox Orange | Golden Delicious |
| Früher Victoriaapfel | Lord Grosvenor | Keswick’s Küchenapfel |
| Frureru | Red Winter | Rafzubin |
| Gala | Kidd’s Orange Red | Golden Delicious |
| Geheimrat Dr. Oldenburg | Ananasrenette | Kaiser Alexander |
| Gloria | Gloster | James Grieve |
| Gloster | Weißer Winterglockenapfel | Delicious |
| Goldjon | Golden Delicious | Jonathan |
| Greensleeves | Golden Delicious | James Grieve |
| Havelgold | Undine | Clivia |
| Holiday | Macoun | Jonathan |
| Idagold | Wagenerapfel | Esopus Spitzenburg |
| Idared | Jonathan | Wagenerapfel |
| Iduna | Golden Delicious | Weißer Winterglockenapfel |
| Ingol | Ingrid Marie | Golden Delicious |
| Ivette | Cox Orange | Golden Delicious |
| Jamba | Melba | James Grieve |
| Jan Steen | Rote Sternrenette | Cox Orange |
| Jester | Worcester Parmäne | Golden Delicious |
| Jonadel | Jonathan | Delicious |
| Jonamac | McIntosh | Jonathan |
| Julia | Quinte | Discovery |
| Katja | James Grieve | Worcester Parmäne |
| Kidd’s Orange Red | Cox Orange | Delicious |
| Laxtons Triumph | Goldparmäne | Cox Orange |
| Lonjon | London Pepping | Jonathan |
| Lord Lambourne | James Grieve | Worcester Parmäne |
| Maigold | Fraurotacher | Golden Delicious |
| Margol | Ingrid Marie | Golden Delicious |
| Marina | Kidd’s Orange Red | Idared |
| Melrose | Jonathan | Delicious |
| Merton Beauty | Ellisons Orangenpepping | Cox Orange |
| Merton Charm | McIntosh | Cox Orange |
| Millicent Barnes | Gascoynes Scharlachroter | Cox Orange |
| Milton | Weißer Klarapfel | McIntosh |
| Monarch | Peasgoods Sondergleichen | Dumelows Seedling |
| Monrö | Jonathan | Rome Beauty |
| Neujahrsapfel | London Pepping | McIntosh |
| Odin | Golden Delicious | Ingrid Marie |
| Ontario | Wagenerapfel | Northern Spy |
| Orangenburg | Cox Orange | Geheimrat Dr. Oldenburg |
| Orleans Renette | Karmeliter Renette | Französische Edelrenette |
| Pia | Idared | Helios |
| Pidi | Britemac | Coop 2 |
| Piflora | Idared | Golden Delicious |
| Pikant | Undine | Carola |
| Pikkolo | Clivia | Auralia |
| Pikosa | Pirella | Idared |
| Pilana | Pirella | Idared |
| Pilot | Clivia | Undine |
| Pimona | Clivia | Auralia |
| Pinett | Idared | Bancroft |
| Pingo | Idared | Bancroft |
| Pinova | Clivia | Golden Delicious |
| Pirella | Golden Delicious | Alkmene |
| Pirina | Alkmene | Geheimrat Dr. Oldenburg |
| Piros | Helios | Apollo |
| Pivita | Pinova | Idared |
| Pohorka | Cox Orange | Ontario |
| Prinzeßin Irene | Jonathan | Cox Orange |
| Quinte | Crimson Beauty | Melba |
| Rafzubin | Golden Delicious | Cox Orange |
| Rebella | Golden Delicious | Remo |
| Rekarda | Golden Delicious | Remo |
| Rival | Peasgoods Sondergleichen | Cox Orange |
| Rubin | Golden Delicious | Lord Lambourne |
| Rubinola | Coop 2 | Rubin |
| Sansa | Gala | Akane |
| Schöner aus Mlejew | Goldparmäne | McIntosh |
| Schweizer Orangenapfel | Ontario | Cox Orange |
| Shampion | Golden Delicious | Lord Lambourne |
| Shinsei | Golden Delicious | Early McIntosh |
| Slava Pobeditelaam | Weißer Klarapfel | McIntosh |
| Sommerregent | Anton Fischer | James Grieve |
| Spencer | McIntosh | Golden Delicious |
| Summerland | McIntosh | Golden Delicious |
| Telamon | McIntosh | Golden Delicious |
| Tuscan | McIntosh | Greensleeves |
| Tydemans Early | McIntosh | Worcester Parmäne |
| Tydemans Oktoberpepping | Cox Orange | Ellisons Orangenpepping |
| Weißer Winterglockenapfel | Boikenapfel | Prinzenapfel |
| n | Both Parents Confirmed | Mother Confirmed/Father Rejected | Father Confirmed/Mother Rejected | Both Parents Rejected | |
|---|---|---|---|---|---|
| Both parents suggested | 128 | 110 | 12 | 3 | 3 |
| Mother suggested | 100 | - | 82 | 18 | - |
| Father suggested | 25 | - | 7 | 18 | - |
| German Fruit Genebank Trueness-to-Type Criteria | Explanation | |
|---|---|---|
| 0 | deceased | tree died during project duration |
| 1 | true-to-type | pomologically and molecularly assessed |
| 2 | true-to-type (group/mutant) | pomologically and molecularly assessed |
| 3 | not determined | no references available, cultivar name unknown |
| 4 | not assessed | not assessed (pomologically) |
| pomologically assessed, not assessed molecularly | ||
| 5 | true-to-type with reservations | molecularly assessed for at least 3 accessions and pomologically assessed for at least 2 accessions |
| pomologically assessed with reservation, molecularly assessed | ||
| Category | Group Name | n * |
|---|---|---|
| Age | Before 1849 | 156 |
| 1850 to 1899 | 137 | |
| 1900 to 1949 | 139 | |
| 1950 to present | 137 | |
| Origin | Germany | 264 |
| Europe | 208 | |
| Other | 97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broschewitz, L.; Reim, S.; Flachowsky, H.; Höfer, M. Pomological and Molecular Characterization of Apple Cultivars in the German Fruit Genebank. Plants 2024, 13, 2699. https://doi.org/10.3390/plants13192699
Broschewitz L, Reim S, Flachowsky H, Höfer M. Pomological and Molecular Characterization of Apple Cultivars in the German Fruit Genebank. Plants. 2024; 13(19):2699. https://doi.org/10.3390/plants13192699
Chicago/Turabian StyleBroschewitz, Lea, Stefanie Reim, Henryk Flachowsky, and Monika Höfer. 2024. "Pomological and Molecular Characterization of Apple Cultivars in the German Fruit Genebank" Plants 13, no. 19: 2699. https://doi.org/10.3390/plants13192699
APA StyleBroschewitz, L., Reim, S., Flachowsky, H., & Höfer, M. (2024). Pomological and Molecular Characterization of Apple Cultivars in the German Fruit Genebank. Plants, 13(19), 2699. https://doi.org/10.3390/plants13192699






