Characterization of a Drought-Induced Betaine Aldehyde Dehydrogenase Gene SgBADH from Suaeda glauca
Abstract
:1. Introduction
2. Results
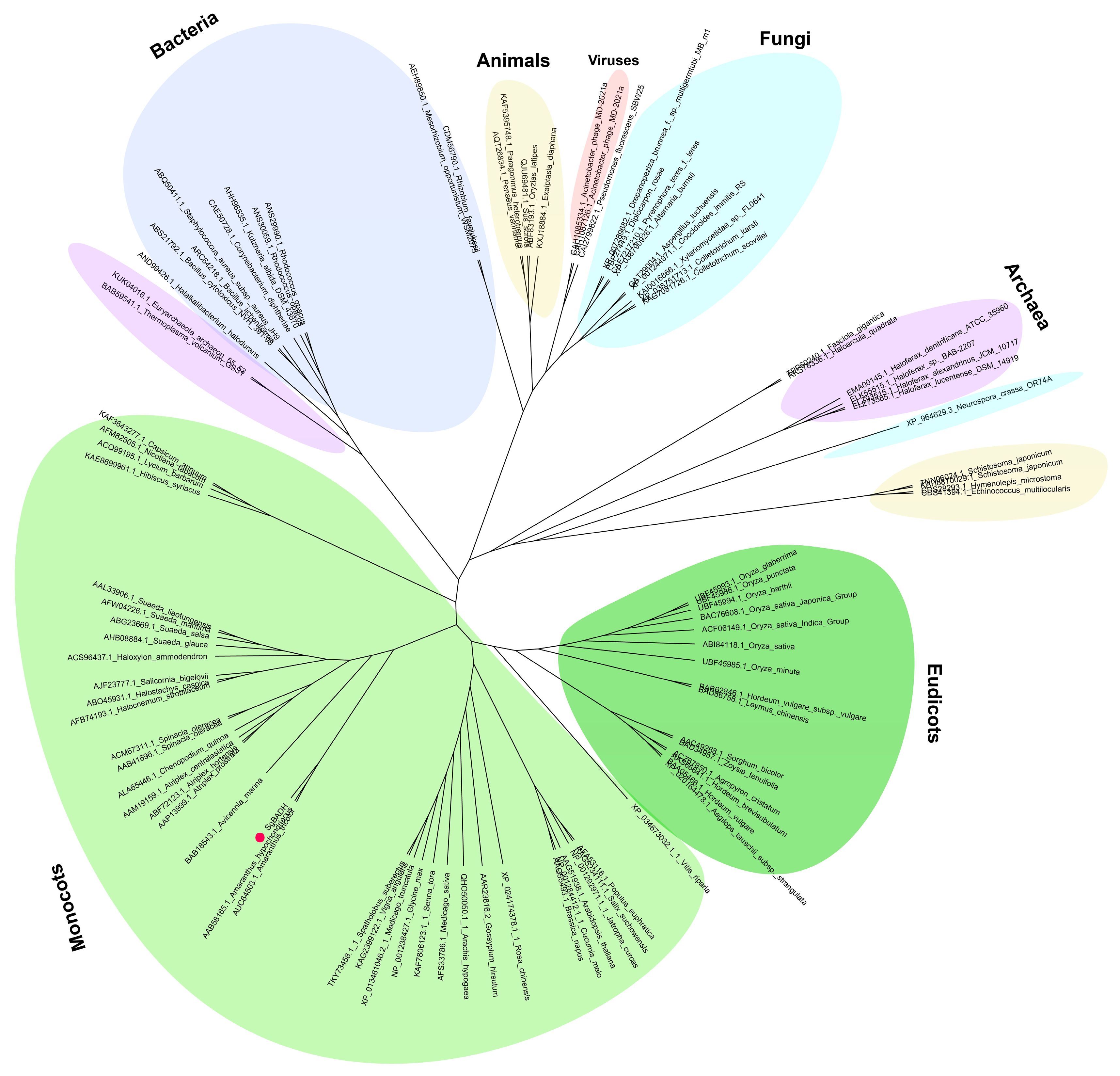
2.1. Evolutionary Analysis of SgBADH
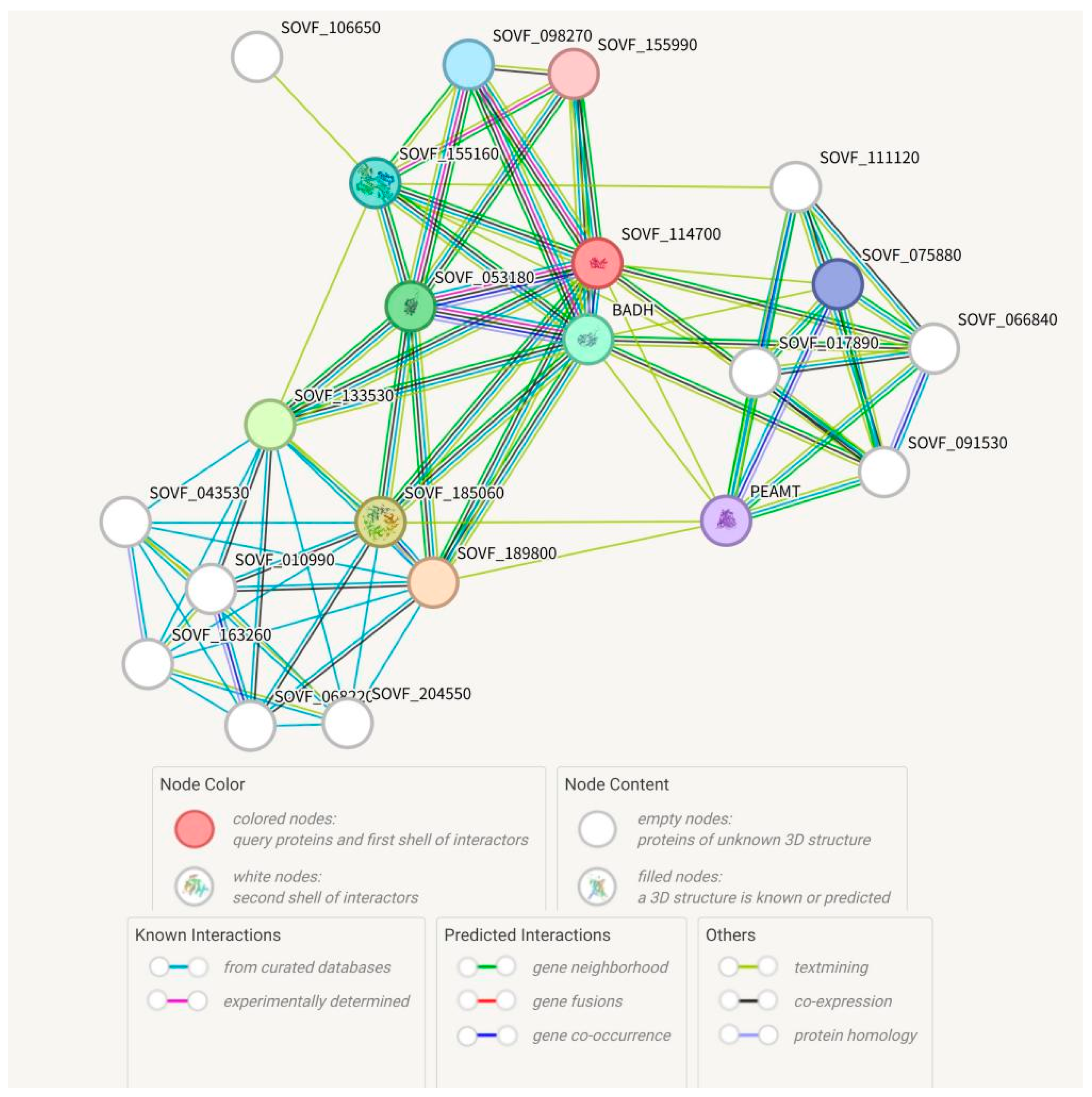
2.2. Analysis of the SgBADH Protein Interaction Network Collaborator
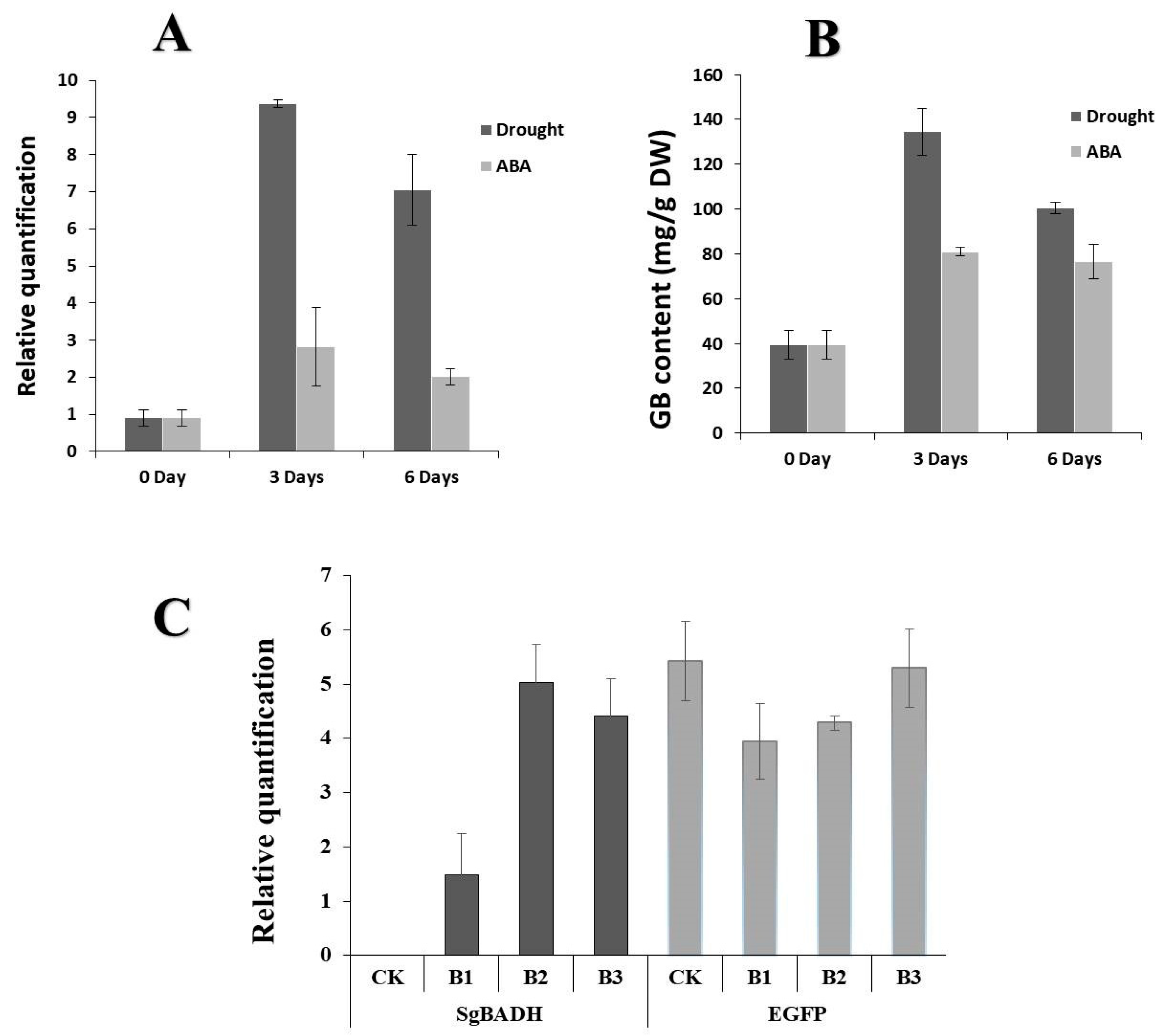
2.3. SgBADH Gene Is Stress-Responsive in Suaeda glauca
2.4. Expression of SgBADH in Transgenic Arabidopsis
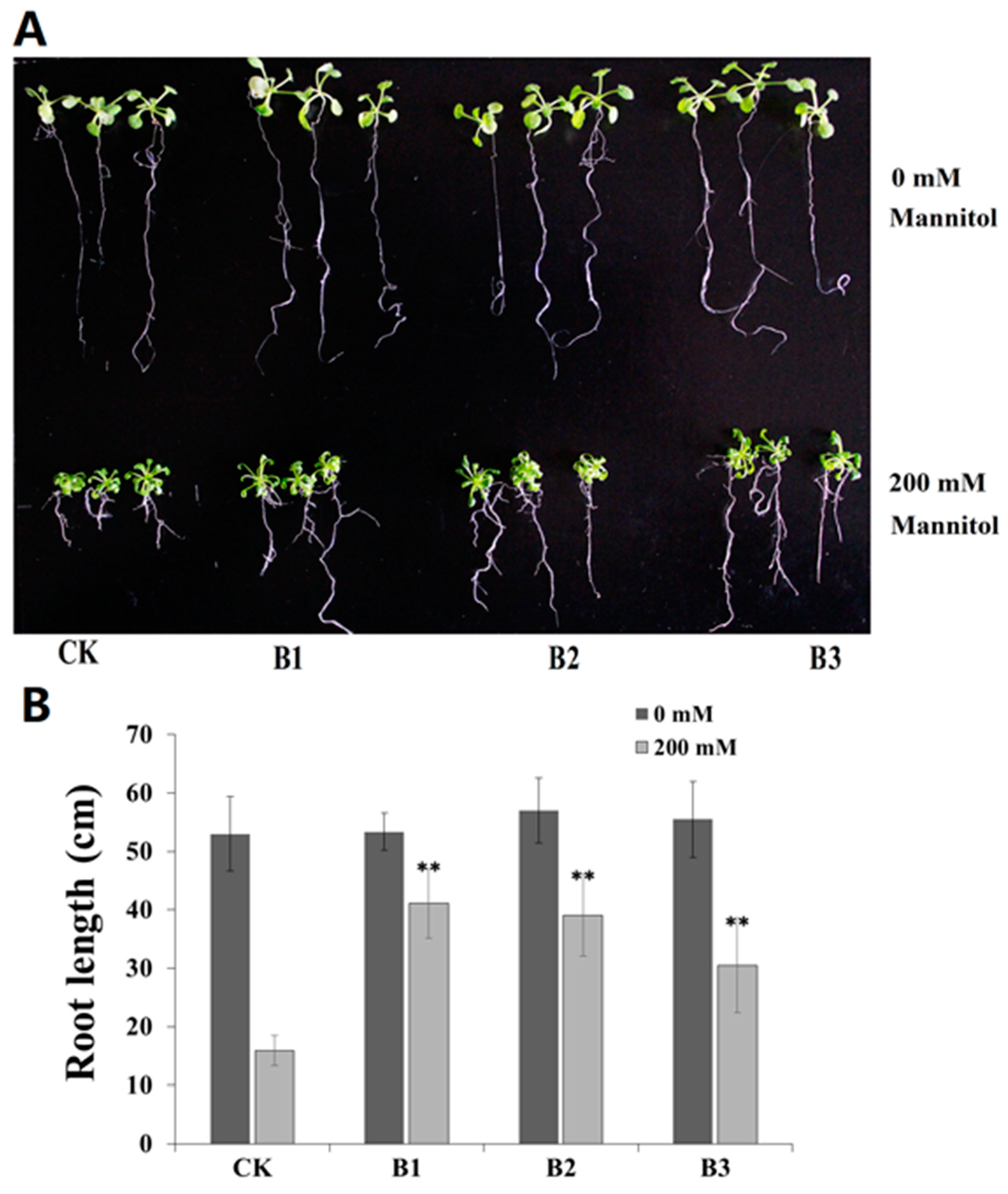
2.5. Overexpression of SgBADH Enhances Drought Tolerance in Transgenic Arabidopsis Seedlings
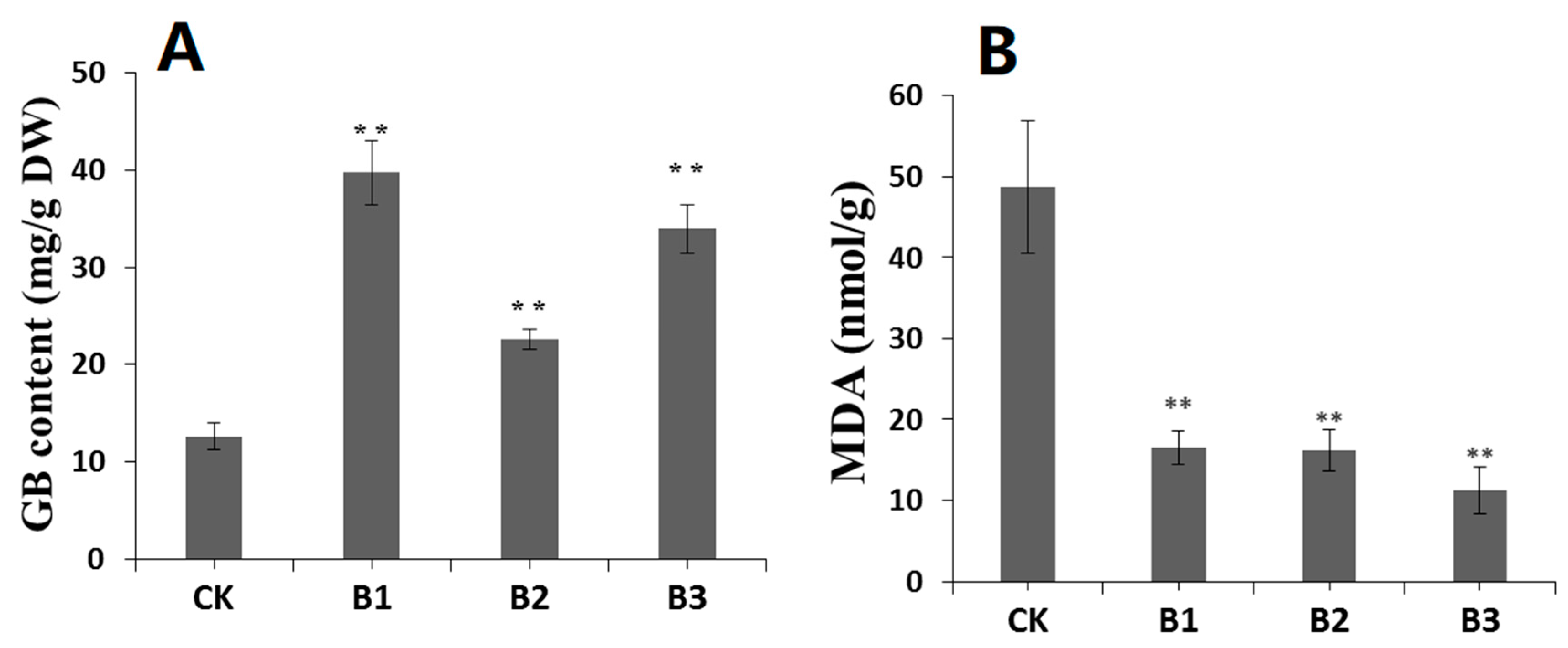
2.6. SgBADH-Overexpressing Arabidopsis Exhibits Increased GB Accumulation and Decreased MDA under Drought Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Salt Stress Treatment
4.2. RNA Extraction, cDNA Synthesis, and Cloning of SgBADH cDNAs
4.3. qRT-PCR Analysis
4.4. Phylogenetic Tree and Protein–Protein Interaction Analysis
4.5. Plasmid Construction, Arabidopsis Transformation
4.6. Verification of SgBADH-Overexpressing Transgenic Arabidopsis
4.7. Analysis of the Drought Tolerance in SgBADH-Overexpressing Transgenic Arabidopsis
4.8. Lipid Peroxidation Assay
4.9. Measurement of GB Content
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, C.; Zhou, Y.; Fan, J.; Fu, Y.; Shen, L.; Yao, Y.; Li, R.; Fu, S.; Duan, R.; Hu, X.; et al. SpBADH of the halophyte Sesuvium portulacastrum strongly confers drought tolerance through ROS scavenging in transgenic Arabidopsis. Plant Physiol. Biochem. 2015, 96, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Mitsuya, S.; Tsuchiya, A.; Kono-Ozaki, K.; Fujiwara, T.; Takabe, T.; Takabe, T. Functional and expression analyses of two kinds of betaine aldehyde dehydrogenases in a glycinebetaine-hyperaccumulating graminaceous halophyte, Leymus chinensis. SpringerPlus 2015, 4, 202. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Gaytan, M.F.; Hernandez-Palomares, M.L.; Sonanez-Organis, J.G.; Muhlia-Almazan, A.; Sanchez-Paz, A.; Stephens-Camacho, N.A.; Valenzuela-Soto, E.M.; Rosas-Rodriguez, J.A. Molecular characterization and organ-specific expression of the gene that encodes betaine aldehyde dehydrogenase from the white shrimp Litopenaeus vannamei in response to osmotic stress. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 189, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Q.; Wang, Y.G.; Yong, T.M.; She, Y.H.; Fu, F.L.; Li, W.C. Heterologous expression of betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus confers high salt and heat tolerance to Escherichia coli. Gene 2014, 549, 77–84. [Google Scholar] [CrossRef]
- Munoz-Clares, R.A.; Riveros-Rosas, H.; Garza-Ramos, G.; Gonzalez-Segura, L.; Mujica-Jimenez, C.; Julian-Sanchez, A. Exploring the evolutionary route of the acquisition of betaine aldehyde dehydrogenase activity by plant ALDH10 enzymes: Implications for the synthesis of the osmoprotectant glycine betaine. BMC Plant Biol. 2014, 14, 149. [Google Scholar] [CrossRef]
- Lambou, K.; Pennati, A.; Valsecchi, I.; Tada, R.; Sherman, S.; Sato, H.; Beau, R.; Gadda, G.; Latge, J.P. Pathway of glycine betaine biosynthesis in Aspergillus fumigatus. Eukaryot. Cell 2013, 12, 853–863. [Google Scholar] [CrossRef]
- Brocker, C.; Vasiliou, M.; Carpenter, S.; Carpenter, C.; Zhang, Y.; Wang, X.; Kotchoni, S.O.; Wood, A.J.; Kirch, H.H.; Kopecny, D.; et al. Aldehyde dehydrogenase (ALDH) superfamily in plants: Gene nomenclature and comparative genomics. Planta 2013, 237, 189–210. [Google Scholar] [CrossRef]
- Jackson, B.; Brocker, C.; Thompson, D.C.; Black, W.; Vasiliou, K.; Nebert, D.W.; Vasiliou, V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genom. 2011, 5, 283–303. [Google Scholar] [CrossRef]
- Weretilnyk, E.A.; Hanson, A.D. Molecular cloning of a plant betaine-aldehyde dehydrogenase, an enzyme implicated in adaptation to salinity and drought. Proc. Natl. Acad. Sci. USA 1990, 87, 2745–2749. [Google Scholar] [CrossRef]
- Ishitani, M.; Nakamura, T.; Han, S.Y.; Takabe, T. Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid. Plant Mol. Biol. 1995, 27, 307–315. [Google Scholar] [CrossRef]
- Shu, W. Genomic cloning and sequencing of betaine aldehyde dehydrogenase gene in spinach. Chin. Sci. Bull. 1998, 4, 325–330. [Google Scholar]
- Livingstone, J.R.; Maruo, T.; Yoshida, I.; Tarui, Y.; Hirooka, K.; Yamamoto, Y.; Tsutui, N.; Hirasawa, E. Purification and properties of betaine aldehyde dehydrogenase from Avena sativa. J. Plant Res. 2003, 116, 133–140. [Google Scholar] [CrossRef]
- Ye, C.; Wu, S.; Yang, Q.; Ma, C.; Yang, G.; Wang, B. Cloning, sequencing and salt induced expression of PEAMT and BADH in oilseed rape (Brassica napus). DNA Seq. J. DNA Seq. Mapp. 2005, 16, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, L.M.; Gillies, S.A.; Brushett, D.J.; Waters, D.L.; Henry, R.J. Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol. Biol. 2008, 68, 439–449. [Google Scholar] [CrossRef]
- Nakamura, T.; Yokota, S.; Muramoto, Y.; Tsutsui, K.; Oguri, Y.; Fukui, K.; Takabe, T. Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J. Cell Mol. Biol. 1997, 11, 1115–1120. [Google Scholar] [CrossRef]
- Missihoun, T.D.; Schmitz, J.; Klug, R.; Kirch, H.H.; Bartels, D. Betaine aldehyde dehydrogenase genes from Arabidopsis with different sub-cellular localization affect stress responses. Planta 2011, 233, 369–382. [Google Scholar] [CrossRef]
- Gao, T.P.; Wang, C.Y.; Xu, H.W.; Sun, H.L.; Zhang, M.; Zhang, Q.; Shi, X.N. Molecular Cloning and Expression Analysis of Betaine Aldehyde Dehydrogenase Gene from the Halophyte Halocnermum strobilaceum(Pall.)(Chenopodiaceae). Bull. Bot. Res. 2013, 33, 317–324. [Google Scholar]
- Chen, X.; Wang, G.; Xu, L.; Zhang, L.; Liu, Q.; Cui, H.; Zhang, X. Cloning and Expression Analysis of Betaine Aldehyde Dehydrogenase BADH Gene from Hazelnut (Corylus heterophylla Fisch.) during Overwintering Period. Shandong Agric. Sci. 2013, 45, 6–12. [Google Scholar]
- Wood, A.J.; Saneoka, H.; Rhodes, D.; Joly, R.J.; Goldsbrough, P.B. Betaine aldehyde dehydrogenase in sorghum. Plant Physiol. 1996, 110, 1301–1308. [Google Scholar] [CrossRef]
- Guo, P.; Baum, M.; Grando, S.; Ceccarelli, S.; Bai, G.; Li, R.; von Korff, M.; Varshney, R.K.; Graner, A.; Valkoun, J. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp. Bot. 2009, 60, 3531–3544. [Google Scholar] [CrossRef]
- Fan, W.; Zhang, M.; Zhang, H.; Zhang, P. Improved tolerance to various abiotic stresses in transgenic sweet potato (Ipomoea batatas) expressing spinach betaine aldehyde dehydrogenase. PLoS ONE 2012, 7, e37344. [Google Scholar] [CrossRef] [PubMed]
- Jacques, F.; Zhao, Y.; Kopecna, M.; Koncitikova, R.; Kopecny, D.; Rippa, S.; Perrin, Y. Roles for ALDH10 enzymes in gamma-butyrobetaine synthesis, seed development, germination, and salt tolerance in Arabidopsis. J. Exp. Bot. 2020, 71, 7088–7102. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Sun, J.; Liu, J.; Liu, F.; Yan, J.; Gou, X.; Lu, B.R.; Liu, Y. RNAi-directed downregulation of betaine aldehyde dehydrogenase 1 (OsBADH1) results in decreased stress tolerance and increased oxidative markers without affecting glycine betaine biosynthesis in rice (Oryza sativa). Plant Mol. Biol. 2014, 86, 443–454. [Google Scholar] [CrossRef] [PubMed]
- An, R.B.; Sohn, D.H.; Jeong, G.S.; Kim, Y.C. In vitro hepatoprotective compounds from Suaeda glauca. Arch. Pharm. Res. 2008, 31, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.X.; Zhong, R.Z.; Liu, H.W.; Wang, M.L.; Sun, J.Y.; Zhou, D.W. Meat quality, fatty acid composition of tissue and gastrointestinal content, and antioxidant status of lamb fed seed of a halophyte (Suaeda glauca). Meat Sci. 2015, 100, 10–16. [Google Scholar] [CrossRef]
- Yang, C.; Shi, D.; Wang, D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Reg. 2008, 56, 179–190. [Google Scholar] [CrossRef]
- Wang, F.W.; Wang, M.L.; Guo, C.; Wang, N.; Li, X.W.; Chen, H.; Dong, Y.Y.; Chen, X.F.; Wang, Z.M.; Li, H.Y. Cloning and characterization of a novel betaine aldehyde dehydrogenase gene from Suaeda corniculata. Genet. Mol. Res. 2016, 15, 15027848. [Google Scholar] [CrossRef]
- Li, Q.L.; Gao, X.R.; Yu, X.H.; Wang, X.Z.; An, L.J. Molecular cloning and characterization of betaine aldehyde dehydrogenase gene from Suaeda liaotungensis and its use in improved tolerance to salinity in transgenic tobacco. Biotechnol. Lett. 2003, 25, 1431. [Google Scholar]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 2010, 21, 535–553. [Google Scholar] [CrossRef]
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Jiaxu, L. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.G.; Du, B.X.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. AhCMO, regulated by stresses in Atriplex hortensis, can improve drought tolerance in transgenic tobacco. Theor. Appl. Genet. 2002, 105, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhao, R.; Li, F.; Tang, W.; Han, L. Simultaneous Expression of Spinacia oleracea Chloroplast Choline Monooxygenase (CMO) and Betaine Aldehyde Dehydrogenase (BADH) Genes Contribute to Dwarfism in Transgenic Lolium perenne. Plant Mol. Biol. Report. 2010, 29, 379–388. [Google Scholar] [CrossRef]
- Weretilnyk, E.A.; Hanson, A.D. Betaine aldehyde dehydrogenase polymorphism in spinach: Genetic and biochemical characterization. Biochem. Genet. 1988, 26, 143–151. [Google Scholar] [CrossRef]
- Ling, P.; Zhongfu, Y.; Jianping, W.; Pengxi, W.; Xiao, M.; Meiliang, Z.; Ji, L.; Nie, G.; Guangyan, F.; Junming, Z. Comparative proteomic analyses reveal the proteome response to short-term drought in Italian ryegrass (Lolium multiflorum). PLoS ONE 2017, 12, e0184289. [Google Scholar]
- Parida, A.K.; Dagaonkar, V.S.; Phalak, M.S.; Aurangabadkar, L.P. Differential responses of the enzymes involved in proline biosynthesis and degradation in drought tolerant and sensitive cotton genotypes during drought stress and recovery. Acta Physiol. Plant. 2008, 30, 619–627. [Google Scholar] [CrossRef]
- Quan, R.; Shang, M.; Zhang, H.; Zhao, Y.; Zhang, J. Engineering of enhanced glycine betaine synthesis improves drought tolerance in maize. Plant Biotechnol. J. 2004, 2, 477–486. [Google Scholar] [CrossRef]
- Yuan, X.; Li, Y.; Liu, S.; Xia, F.; Li, X.; Qi, B. Accumulation of eicosapolyenoic acids enhances sensitivity to abscisic acid and mitigates the effects of drought in transgenic Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 1637–1649. [Google Scholar] [CrossRef]
- Hong-Bo, S.; Xiao-Yan, C.; Li-Ye, C.; Xi-Ning, Z.; Gang, W.; Yong-Bing, Y.; Chang-Xing, Z.; Zan-Min, H. Investigation on the relationship of proline with wheat anti-drought under soil water deficits. Colloids Surf. B Biointerfaces 2006, 53, 113–119. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.; Zhang, W.; Li, S.; Gao, Y.; Ai, X.; Zhang, D.; Liu, B.; Li, Q. Metabolomics analysis reveals that elevated atmospheric CO2 alleviates drought stress in cucumber seedling leaves. Anal. Biochem. 2018, 559, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hu, C.; Yang, X.; Tan, Q.; Yao, S.; Zhou, Y.; Wang, X.; Sun, X. Molybdenum induces alterations in the glycerolipidome that confer drought tolerance in wheat. J. Exp. Bot. 2020, 71, 5074–5086. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.P.; Zhang, X.Y.; Li, F.; Luo, Y.; Wang, W. Overaccumulation of glycine betaine enhances tolerance of the photosynthetic apparatus to drought and heat stress in wheat. Photosynthetica 2010, 48, 117–126. [Google Scholar] [CrossRef]
- He, Q.; Yu, J.; Kim, T.S.; Cho, Y.H.; Lee, Y.S.; Park, Y.J. Resequencing Reveals Different Domestication Rate for BADH1 and BADH2 in Rice (Oryza sativa). PLoS ONE 2015, 10, e0134801. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Hori, K.; Ozaki, K.; Yokota, Y.; Mitsuya, S.; Ichiyanagi, T.; Hattori, T.; Takabe, T. Enzymatic characterization of peroxisomal and cytosolic betaine aldehyde dehydrogenases in barley. Physiol. Plant. 2010, 134, 22–30. [Google Scholar] [CrossRef]
- Nakamura, T.; Nomura, M.; Mori, H.; Jagendorf, A.T.; Ueda, A.; Takabe, T. An Isozyme of Betaine Aldehyde Dehydrogenase in Barley. Plant Cell Physiol. 2001, 42, 1088–1092. [Google Scholar] [CrossRef]
- Hasthanasombut, S.; Supaibulwatana, K.; Mii, M.; Nakamura, I. Genetic manipulation of Japonica rice using the OsBADH1 gene from Indica rice to improve salinity tolerance. Plant Cell Tiss. Organ. Cult. 2011, 104, 79–89. [Google Scholar] [CrossRef]
- Li, M.; Guo, S.; Xu, Y.; Meng, Q.; Li, G.; Yang, X. Glycine betaine-mediated potentiation of HSP gene expression involves calcium signaling pathways in tobacco exposed to NaCl stress. Physiol. Plant. 2014, 150, 63–75. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Chen, C.; Chen, T.; Lo, K.; Chiu, C. Effects of proline on copper transport in rice seedlings under excess copper stress. Plant Sci. 2004, 166, 103–111. [Google Scholar] [CrossRef]
- Dhindsa, R.; Matowe, W. Drought tolerance in two mosses correlated with enzymatic defence against lipid peroxidation. J. Exp. Bot. 1981, 32, 79–91. [Google Scholar] [CrossRef]
- Hong, Z.; Lakkineni, K.; Zhang, Z.; Verma, D. Removal of feedback inhibition of Delta 1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 2000, 122, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Martinez, S.E.V. Simultaneous determination of choline and betaine in some fish materials. Analyst 1983, 108, 1114–1119. [Google Scholar] [CrossRef]
| Gene | Primer Sequences |
|---|---|
| SgBADHF | ATGKCGWTCCCWATWCCTKCTCG |
| SgBADHR | TYAAGGAGACTTGTACCAYCCC |
| SgBADH-F | ATGGCGTTCCCAATTCCTGC |
| SgBADH-R | TCAAGGCGACTTGTACCATC |
| SgBADH192-F | AAATATGGAAGGAGGAAGT |
| SgBADH192-R | GTTGTGAGCAATTAACCC |
| AtActin124-F | ATCGCTGACCGTATGAG |
| AtActin124-R | TGAGGGAAGCAAGAATG |
| SgActin-F | CCGCAAAGATTACATACC |
| SgActin-R | TCACCGAAAGTGCTTCTA |
| EGFP-F | ACCCTCGTGACCACCCTGAC |
| EGFP-R | TGTAGTTGCCGTCGTCCTTGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, H.; Tang, M.; Zhu, L.; Yu, X.; Yang, Q.; Fu, X. Characterization of a Drought-Induced Betaine Aldehyde Dehydrogenase Gene SgBADH from Suaeda glauca. Plants 2024, 13, 2716. https://doi.org/10.3390/plants13192716
Jin H, Tang M, Zhu L, Yu X, Yang Q, Fu X. Characterization of a Drought-Induced Betaine Aldehyde Dehydrogenase Gene SgBADH from Suaeda glauca. Plants. 2024; 13(19):2716. https://doi.org/10.3390/plants13192716
Chicago/Turabian StyleJin, Hangxia, Min Tang, Longmin Zhu, Xiaomin Yu, Qinghua Yang, and Xujun Fu. 2024. "Characterization of a Drought-Induced Betaine Aldehyde Dehydrogenase Gene SgBADH from Suaeda glauca" Plants 13, no. 19: 2716. https://doi.org/10.3390/plants13192716





