Gene Expression Divergence in Eugenia uniflora Highlights Adaptation Across Contrasting Atlantic Forest Ecosystems
Abstract
:1. Introduction
2. Results
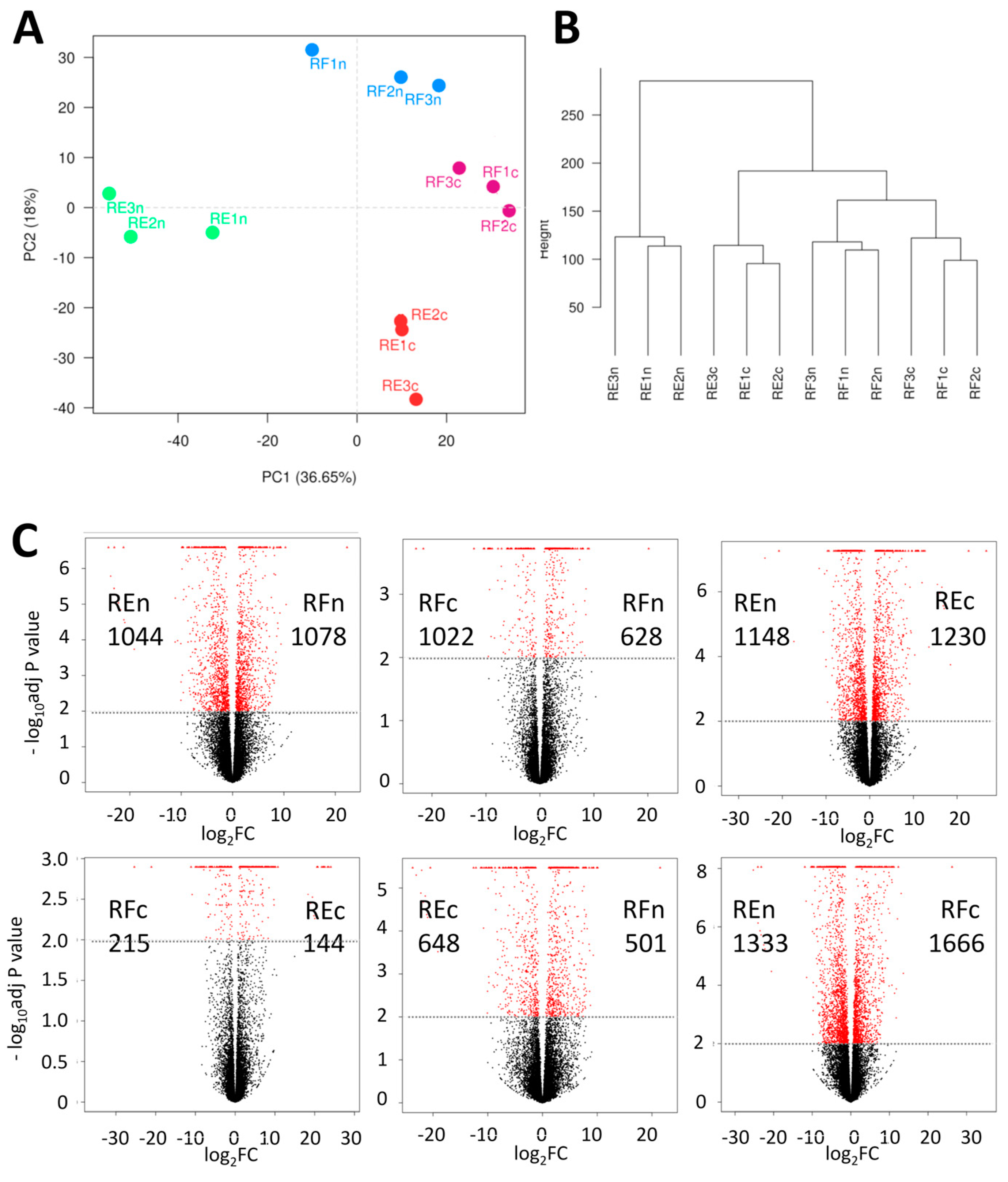
2.1. Comparing Samples from Different Environments
2.2. Patterns of Differentially Expressed Genes (DEGs)
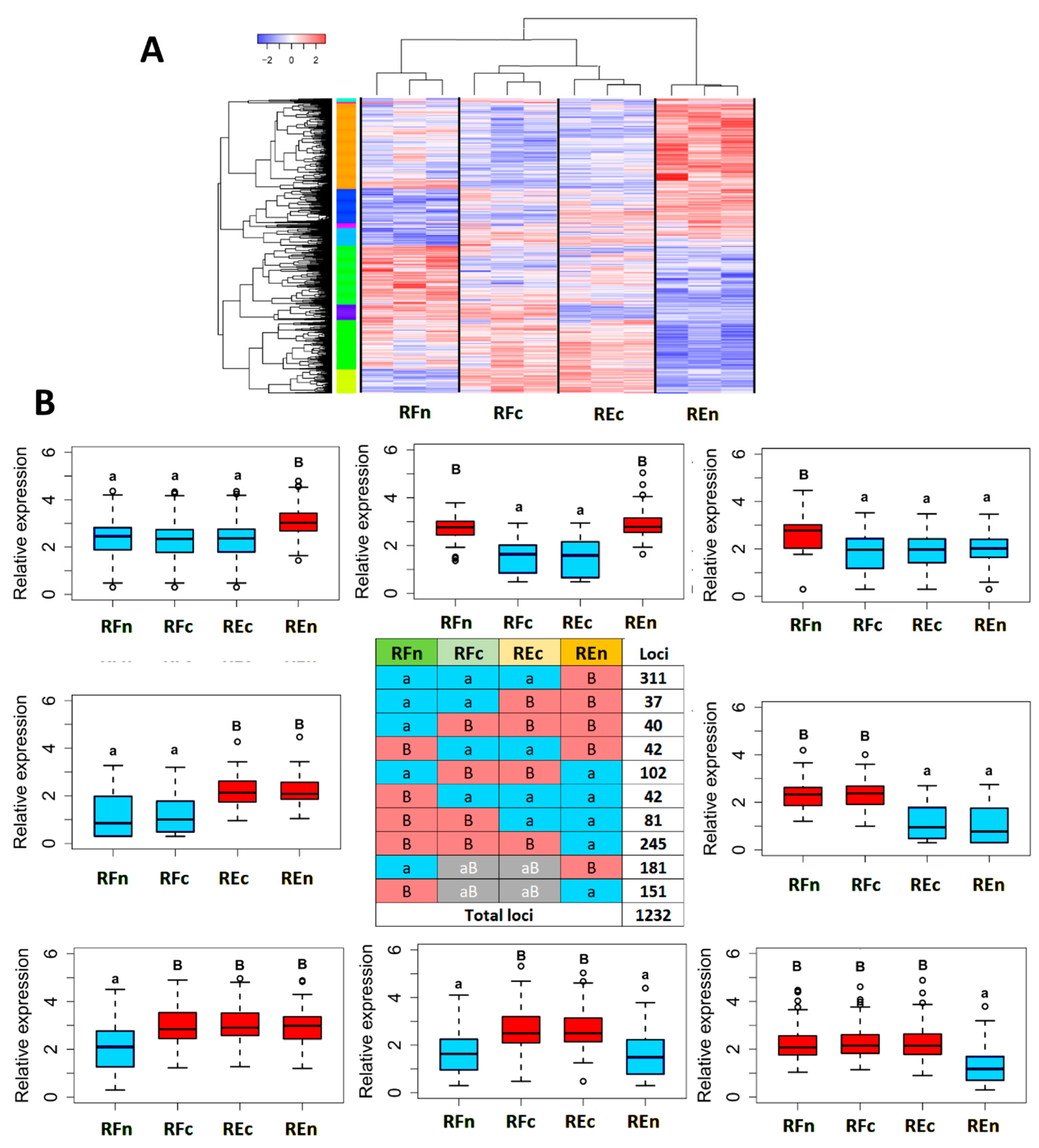
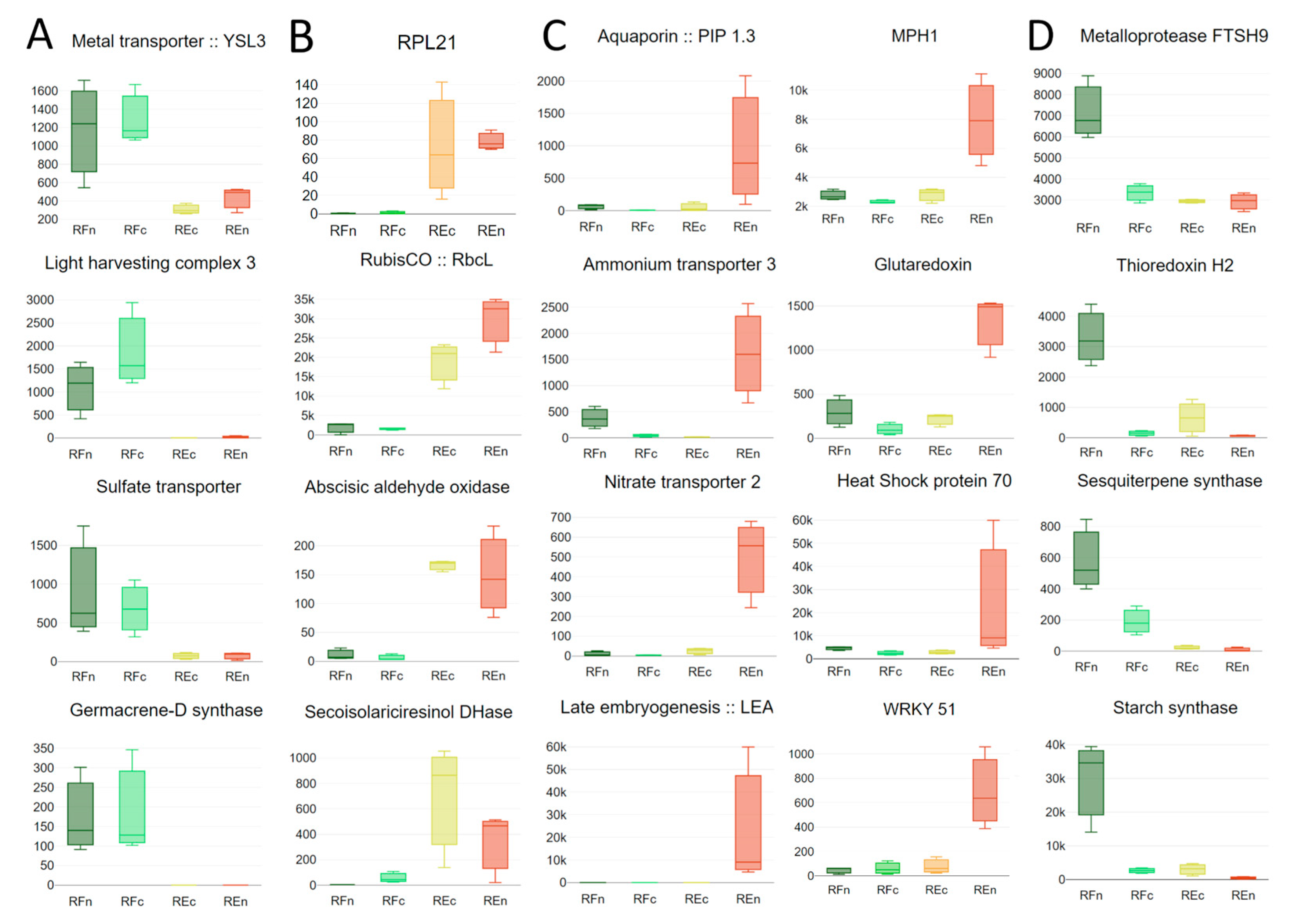
2.3. Genes Modulated by the Environment Compared to Those Exhibiting Constitutive Expression within Populations
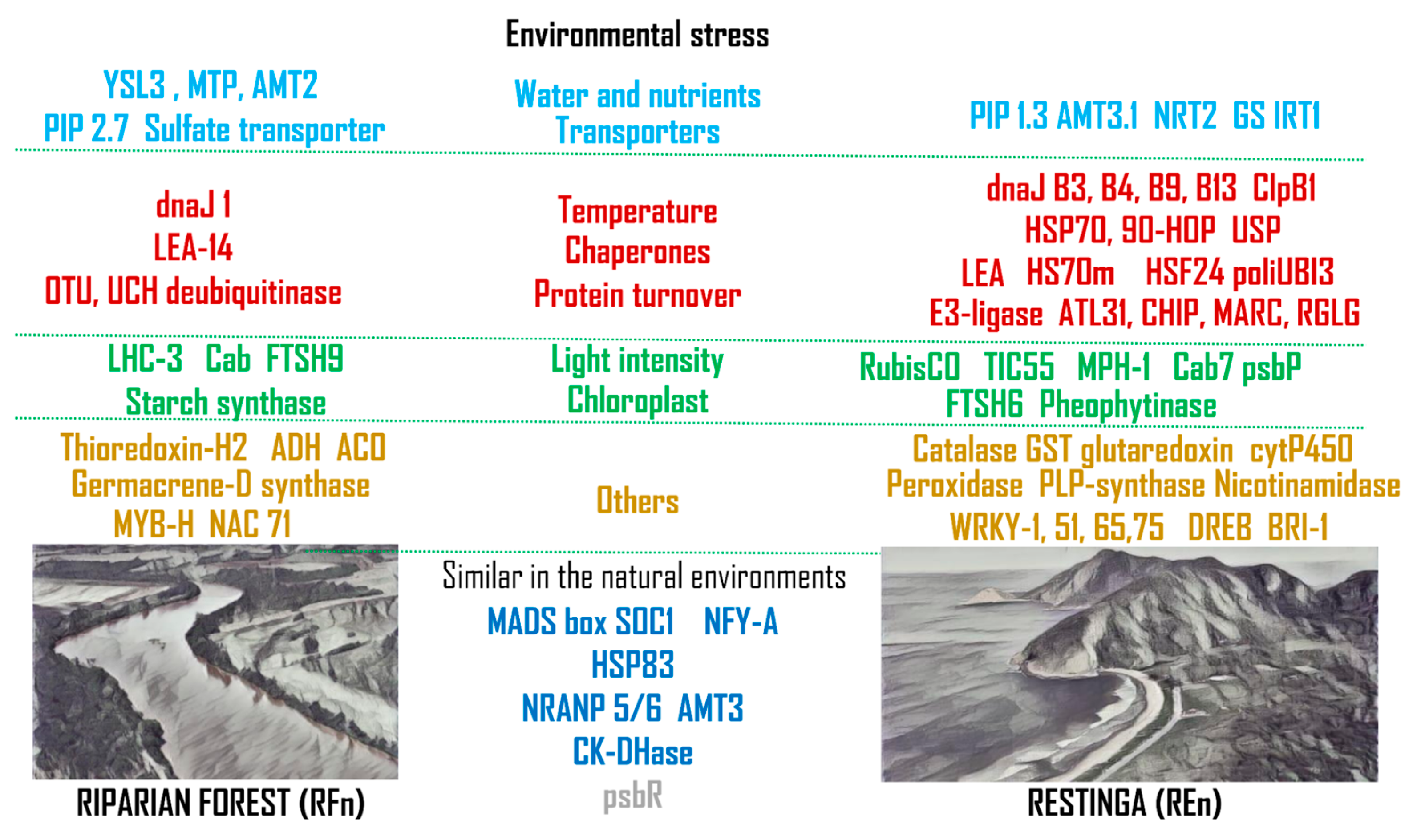
3. Discussion
3.1. Water and Nutrient Transport
3.2. Responses to High Temperatures
3.3. Managing Contrasting Light Intensities
3.4. Other Genes Modulated by Environmental Stresses
4. Materials and Methods
4.1. Study System and Experimental Design
4.2. RNA Extraction, Library Preparation, and Deep Sequencing
4.3. Read Trimming, De Novo Transcriptome Assembly, and Quality Assessment
4.4. Differential Expression and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fay, J.C.; Wittkopp, P.J. Evaluating the Role of Natural Selection in the Evolution of Gene Regulation. Heredity 2008, 100, 191–199. [Google Scholar] [CrossRef] [PubMed]
- King, M.-C.; Wilson, A.C. Evolution at Two Levels in Humans and Chimpanzees: Their Macromolecules Are so Alike That Regulatory Mutations May Account for Their Biological Differences. Science 1975, 188, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.C.; Maxson, L.R.; Sarich, V.M. Two Types of Molecular Evolution. Evidence from Studies of Interspecific Hybridization. Proc. Natl. Acad. Sci. USA. 1974, 71, 2843–2847. [Google Scholar] [CrossRef] [PubMed]
- Gilad, Y.; Oshlack, A.; Rifkin, S.A. Natural Selection on Gene Expression. Trends Genet. 2006, 22, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Necsulea, A.; Kaessmann, H. Evolutionary Dynamics of Coding and Non-Coding Transcriptomes. Nat. Rev. Genet. 2014, 15, 734–748. [Google Scholar] [CrossRef] [PubMed]
- Price, P.D.; Palmer Droguett, D.H.; Taylor, J.A.; Kim, D.W.; Place, E.S.; Rogers, T.F.; Mank, J.E.; Cooney, C.R.; Wright, A.E. Detecting Signatures of Selection on Gene Expression. Nat. Ecol. Evol. 2022, 6, 1035–1045. [Google Scholar] [CrossRef]
- Romero, I.G.; Ruvinsky, I.; Gilad, Y. Comparative Studies of Gene Expression and the Evolution of Gene Regulation. Nat. Rev. Genet. 2012, 13, 505–516. [Google Scholar] [CrossRef]
- Whitehead, A.; Crawford, D.L. Variation within and among Species in Gene Expression: Raw Material for Evolution. Mol. Ecol. 2006, 15, 1197–1211. [Google Scholar] [CrossRef]
- De Nadal, E.; Ammerer, G.; Posas, F. Controlling Gene Expression in Response to Stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef]
- Schlichting, C.D.; Smith, H. Phenotypic Plasticity: Linking Molecular Mechanisms with Evolutionary Outcomes. Evol. Ecol. 2002, 16, 189–211. [Google Scholar] [CrossRef]
- Lancaster, L.T.; Fuller, Z.L.; Berger, D.; Barbour, M.A.; Jentoft, S.; Wellenreuther, M. Understanding Climate Change Response in the Age of Genomics. J. Anim. Ecol. 2022, 91, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Schrey, A.W.; Richards, C.L. Ten Years of Transcriptomics in Wild Populations: What Have We Learned about Their Ecology and Evolution? Mol. Ecol. 2015, 24, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.L.; D’Agui, H.M.; Enright, N.J.; He, T. Characterization of Leaf Transcriptome in Banksia Hookeriana. Genom. Proteom. Bioinform. 2017, 15, 49–56. [Google Scholar] [CrossRef]
- Wos, G.; Požárová, D.; Kolář, F. Role of Phenotypic and Transcriptomic Plasticity in Alpine Adaptation of Arabidopsis Arenosa. Mol. Ecol. 2023, 32, 5771–5784. [Google Scholar] [CrossRef]
- Leal, B.S.S.; Brandão, M.M.; Palma-Silva, C.; Pinheiro, F. Differential Gene Expression Reveals Mechanisms Related to Habitat Divergence between Hybridizing Orchids from the Neotropical Coastal Plains. BMC Plant Biol. 2020, 20, 554. [Google Scholar] [CrossRef]
- Turchetto-Zolet, A.C.; Salgueiro, F.; Turchetto, C.; Cruz, F.; Veto, N.M.; Barros, M.J.F.; Segatto, A.L.A.; Freitas, L.B.; Margis, R. Phylogeography and Ecological Niche Modelling in Eugenia uniflora (Myrtaceae) Suggest Distinct Vegetational Responses to Climate Change between the Southern and the Northern Atlantic Forest. Bot. J. Linn. Soc. 2016, 182, 670–688. [Google Scholar] [CrossRef]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How Much Is Left, and How Is the Remaining Forest Distributed? Implications for Conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- De Mello Martins, F. Historical Biogeography of the Brazilian Atlantic Forest and the Carnaval-Moritz Model of Pleistocene Refugia: What Do Phylogeographical Studies Tell Us? Biol. J. Linn. Soc. 2011, 104, 499–509. [Google Scholar] [CrossRef]
- Vetö, N.M.; Postolache, D.; Guzman Escudero, F.L.; Vajana, E.; Burgo Braga, R.; Salgueiro, F.; Margis, R.; Vendramin, G.G.; Turchetto-Zolet, A.C. Population Structure and Signals of Local Adaptation in Eugenia uniflora L. (Myrtaceae), a Widely Distributed Species in the Atlantic Forest. Bot. J. Linn. Soc. 2023, 201, 100–113. [Google Scholar] [CrossRef]
- Lacerda, M.E.G. Heterogeneidade Nos Habitats de Eugenia Uniflora L. (Myrtaceae) Correlação Com as Variações No Tamanho e Forma Foliar. In Trabalhos de Conclusão de Curso de Graduação; Universidade Federal do Rio Grande do Sul: Porto Alegre, RS, Brazil, 2021. [Google Scholar]
- Anton, D.B.; Guzman, F.L.; Vetö, N.M.; Krause, F.A.; Kulcheski, F.R.; Coelho, A.P.D.; Duarte, G.L.; Margis, R.; Dillenburg, L.R.; Turchetto-Zolet, A.C. Characterization and Expression Analysis of P5CS (Δ1-Pyrroline-5-Carboxylate Synthase) Gene in Two Distinct Populations of the Atlantic Forest Native Species Eugenia Uniflora L. Mol. Biol. Rep. 2020, 47, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Filgueiras, J.P.C.; Da Silveira, T.D.; Kulcheski, F.R.; Turchetto-Zolet, A.C. Unraveling the Role of MYB Transcription Factors in Abiotic Stress Responses: An Integrative Approach in Eugenia uniflora L. Plant Mol. Biol. Report. 2024. [Google Scholar] [CrossRef]
- Waschburger, E.L.; Guzman, F.; Turchetto-Zolet, A.C. Genome-Wide Identification and Analysis of DOF Gene Family in Eugenia uniflora L. (Myrtaceae). Genes 2022, 13, 2235. [Google Scholar] [CrossRef]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological Consequences of Genetic Diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, F.; Chen, Z.; Wei, P.; Song, X.; Liu, C.; Liu, T.; Li, X.; Liu, X. Genetic Diversity and Gene Expression Diversity Shape the Adaptive Pattern of the Aquatic Plant Batrachium Bungei along an Altitudinal Gradient on the Qinghai–Tibet Plateau. Plant Mol. Biol. 2023, 111, 275–290. [Google Scholar] [CrossRef]
- Voelckel, C.; Gruenheit, N.; Lockhart, P. Evolutionary Transcriptomics and Proteomics: Insight into Plant Adaptation. Trends Plant Sci. 2017, 22, 462–471. [Google Scholar] [CrossRef]
- López-Serrano, L.; Martínez-Cuenca, M.; López-Galarza, S.; Calatayud, Á. Differential Gene Expression Patterns and Physiological Responses Improve Adaptation to High Salinity Concentration in Pepper Accessions. Physiol. Plant. 2023, 175, e14090. [Google Scholar] [CrossRef] [PubMed]
- Innes, P.A.; Goebl, A.M.; Smith, C.C.R.; Rosenberger, K.; Kane, N.C. Gene Expression and Alternative Splicing Contribute to Adaptive Divergence of Ecotypes. Heredity 2024, 132, 120–132. [Google Scholar] [CrossRef]
- Scarano, F.R.; Ceotto, P. Brazilian Atlantic Forest: Impact, Vulnerability, and Adaptation to Climate Change. Biodivers. Conserv. 2015, 24, 2319–2331. [Google Scholar] [CrossRef]
- Ribeiro, J.A. Flora e Vegetação Ribeirinha. Douro-Estud. Doc. 2000, 5, 39–45. [Google Scholar]
- Chrispeels, M.J.; Agre, P. Aquaporins: Water Channel Proteins of Plant and Animal Cells. Trends Biochem. Sci. 1994, 19, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Z.; Liu, F.; Sun, L.; Hao, F. Versatile Roles of Aquaporins in Plant Growth and Development. Int. J. Mol. Sci. 2020, 21, 9485. [Google Scholar] [CrossRef] [PubMed]
- Agre, P.; Sasaki, S.; Chrispeels, M.J. Aquaporins: A Family of Water Channel Proteins. Am. J. Physiol.-Ren. Physiol. 1993, 265, F461. [Google Scholar] [CrossRef]
- Heckwolf, M.; Pater, D.; Hanson, D.T.; Kaldenhoff, R. The Arabidopsis Thaliana Aquaporin AtPIP1;2 Is a Physiologically Relevant CO2 Transport Facilitator. Plant J. 2011, 67, 795–804. [Google Scholar] [CrossRef]
- Zwiazek, J.J.; Xu, H.; Tan, X.; Navarro-Ródenas, A.; Morte, A. Significance of Oxygen Transport through Aquaporins. Sci. Rep. 2017, 7, 40411. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Iwasaki, I.; Kitagawa, Y. Overexpression of a Lily PIP1 Gene in Tobacco Increased the Osmotic Water Permeability of Leaf Cells. Plant Cell Environ. 2004, 27, 177–186. [Google Scholar] [CrossRef]
- Cui, X.-H.; Hao, F.-S.; Chen, H.; Chen, J.; Wang, X.-C. Expression of the Vicia Faba VfPIP1 Gene in Arabidopsis Thaliana Plants Improves Their Drought Resistance. J. Plant Res. 2008, 121, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Israel, D.; Lee, S.H.; Robson, T.M.; Zwiazek, J.J. Plasma Membrane Aquaporins of the PIP1 and PIP2 Subfamilies Facilitate Hydrogen Peroxide Diffusion into Plant Roots. BMC Plant Biol. 2022, 22, 566. [Google Scholar] [CrossRef]
- Bots, M. PIP1 and PIP2 Aquaporins Are Differentially Expressed during Tobacco Anther and Stigma Development. J. Exp. Bot. 2004. [Google Scholar] [CrossRef]
- Patterson, K.; Cakmak, T.; Cooper, A.; Lager, I.; Rasmusson, A.G.; Escobar, M.A. Distinct Signalling Pathways and Transcriptome Response Signatures Differentiate Ammonium- and Nitrate-Supplied Plants: Transcriptome Signatures of Ammonium and Nitrate Responses. Plant Cell Environ. 2010, 33, 1486–1501. [Google Scholar] [CrossRef]
- Kiba, T.; Feria-Bourrellier, A.-B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Bréhaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis Nitrate Transporter NRT2.4 Plays a Double Role in Roots and Shoots of Nitrogen-Starved Plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Bu, Y.; Takano, T.; Liu, S. The Role of Ammonium Transporter (AMT) against Salt Stress in Plants. Plant Signal. Behav. 2019, 14, 1625696. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Fukushima, A.; Redestig, H.; Saito, K. Metabolomic Approaches toward Understanding Nitrogen Metabolism in Plants. J. Exp. Bot. 2011, 62, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Al-Saharin, R.; Hellmann, H.; Mooney, S. Plant E3 Ligases and Their Role in Abiotic Stress Response. Cells 2022, 11, 890. [Google Scholar] [CrossRef]
- Usman, M.G.; Rafii, M.Y.; Martini, M.Y.; Yusuff, O.A.; Ismail, M.R.; Miah, G. Molecular Analysis of Hsp70 Mechanisms in Plants and Their Function in Response to Stress. Biotechnol. Genet. Eng. Rev. 2017, 33, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Deswal, R. Ecophysiolomic Analysis of Stress Tolerant Himalayan Shrub Hipppophae Rhamnoides Shows Multifactorial Acclimation Strategies Induced by Diverse Environmental Conditions. Physiol. Plant. 2020, 168, 58–76. [Google Scholar] [CrossRef]
- Parankusam, S.; Bhatnagar-Mathur, P.; Sharma, K.K. Heat Responsive Proteome Changes Reveal Molecular Mechanisms Underlying Heat Tolerance in Chickpea. Environ. Exp. Bot. 2017, 141, 132–144. [Google Scholar] [CrossRef]
- Saux, M.; Ponnaiah, M.; Langlade, N.; Zanchetta, C.; Balliau, T.; El-Maarouf-Bouteau, H.; Bailly, C. A Multiscale Approach Reveals Regulatory Players of Water Stress Responses in Seeds during Germination. Plant Cell Environ. 2020, 43, 1300–1313. [Google Scholar] [CrossRef]
- Yadav, A.; Singh, J.; Ranjan, K.; Kumar, P.; Khanna, S.; Gupta, M.; Kumar, V.; Wani, S.H.; Sirohi, A. Heat Shock Proteins: Master Players for Heat-stress Tolerance in Plants during Climate Change. In Heat Stress Tolerance in Plants; Wani, S.H., Kumar, V., Eds.; Wiley: New York, NY, USA, 2020; pp. 189–211. ISBN 978-1-119-43236-4. [Google Scholar]
- Archana, P.R.; Aleena, J.; Pragna, P.; Vidya, M.K.; Nyas, A.P.A.; Bagath, M.; Krishnan, G.; Manimaran, A.; Beena, V.; Kurien, E.K.; et al. Role of Heat Shock Proteins in Livestock Adaptation to Heat Stress. J. Dairy Vet. Anim. Res. 2017, 5. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef]
- Wan, C.; Zhang, H.; Cheng, H.; Sowden, R.G.; Cai, W.; Jarvis, R.P.; Ling, Q. Selective Autophagy Regulates Chloroplast Protein Import and Promotes Plant Stress Tolerance. EMBO J. 2023, 42, e112534. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, A.; Sedlářová, M.; Pospíšil, P. Malondialdehyde Enhances PsbP Protein Release during Heat Stress in Arabidopsis. Plant Physiol. Biochem. 2023, 202, 107984. [Google Scholar] [CrossRef]
- Majsec, K.; Bhuiyan, N.H.; Sun, Q.; Kumari, S.; Kumar, V.; Ware, D.; Van Wijk, K.J. The Plastid and Mitochondrial Peptidase Network in Arabidopsis Thaliana: A Foundation for Testing Genetic Interactions and Functions in Organellar Proteostasis. Plant Cell 2017, 29, 2687–2710. [Google Scholar] [CrossRef] [PubMed]
- Sasi, S.; Venkatesh, J.; Daneshi, R.F.; Gururani, M.A. Photosystem II Extrinsic Proteins and Their Putative Role in Abiotic Stress Tolerance in Higher Plants. Plants 2018, 7, 100. [Google Scholar] [CrossRef]
- Abdulbaki, A.S.; Alsamadany, H.; Alzahrani, Y.; Olayinka, B.U. Rubisco and Abiotic Stresses in Plants: Current Assessment. Turk. J. Bot. 2022, 46, 541–552. [Google Scholar] [CrossRef]
- Hammel, A.; Sommer, F.; Zimmer, D.; Stitt, M.; Mühlhaus, T.; Schroda, M. Overexpression of Sedoheptulose-1,7-Bisphosphatase Enhances Photosynthesis in Chlamydomonas Reinhardtii and Has No Effect on the Abundance of Other Calvin-Benson Cycle Enzymes. Front. Plant Sci. 2020, 11, 868. [Google Scholar] [CrossRef] [PubMed]
- Gershenzon, J.; Dudareva, N. The Function of Terpene Natural Products in the Natural World. Nat. Chem. Biol. 2007, 3, 408–414. [Google Scholar] [CrossRef]
- Pazouki, L.; Niinemets, Ü. Multi-Substrate Terpene Synthases: Their Occurrence and Physiological Significance. Front. Plant Sci. 2016, 7, 1019. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.S.; Köllner, T.G.; Wiggins, G.; Grant, J.; Degenhardt, J.; Chen, F. Molecular and Genomic Basis of Volatile-mediated Indirect Defense against Insects in Rice. Plant J. 2008, 55, 491–503. [Google Scholar] [CrossRef]
- Chiriboga, X.; Campos-Herrera, R.; Jaffuel, G.; Röder, G.; Turlings, T.C.J. Diffusion of the Maize Root Signal (E)-β-Caryophyllene in Soils of Different Textures and the Effects on the Migration of the Entomopathogenic Nematode Heterorhabditis Megidis. Rhizosphere 2017, 3, 53–59. [Google Scholar] [CrossRef]
- Tamiru, A.; Bruce, T.J.A.; Richter, A.; Woodcock, C.M.; Midega, C.A.O.; Degenhardt, J.; Kelemu, S.; Pickett, J.A.; Khan, Z.R. A Maize Landrace That Emits Defense Volatiles in Response to Herbivore Eggs Possesses a Strongly Inducible Terpene Synthase Gene. Ecol. Evol. 2017, 7, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Visser, E.A.; Mangwanda, R.; Becker, J.V.W.; Külheim, C.; Foley, W.J.; Myburg, A.A.; Naidoo, S. Foliar Terpenoid Levels and Corresponding Gene Expression Are Systemically and Differentially Induced in Eucalyptus Grandis Clonal Genotypes in Response to Chrysoporthe Austroafricana Challenge. Plant Pathol. 2015, 64, 1320–1325. [Google Scholar] [CrossRef]
- Santonja, M.; Bousquet-Mélou, A.; Greff, S.; Ormeño, E.; Fernandez, C. Allelopathic Effects of Volatile Organic Compounds Released from Pinus Halepensis Needles and Roots. Ecol. Evol. 2019, 9, 8201–8213. [Google Scholar] [CrossRef]
- Lu, C.; Tian, Y.; Hou, X.; Hou, X.; Jia, Z.; Li, M.; Hao, M.; Jiang, Y.; Wang, Q.; Pu, Q.; et al. Multiple Forms of Vitamin B6 Regulate Salt Tolerance by Balancing ROS and Abscisic Acid Levels in Maize Root. Stress Biol. 2022, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, Q.; Zhang, Y. Overexpression of the Poplar WRKY51 Transcription Factor Enhances Salt Tolerance in Arabidopsis Thaliana. Forests 2023, 14, 191. [Google Scholar] [CrossRef]
- Javed, T.; Gao, S.-J. WRKY Transcription Factors in Plant Defense. Trends Genet. 2023, 39, 787–801. [Google Scholar] [CrossRef]
- Lee, D.-K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Choi, Y.D.; Kim, J.-K. The NF-YA Transcription Factor OsNF-YA7 Confers Drought Stress Tolerance of Rice in an Abscisic Acid Independent Manner. Plant Sci. 2015, 241, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, S.; Ren, T.; Niu, M.; Liu, X.; Liu, C.; Wang, H.; Yin, W.; Xia, X. Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants. Int. J. Mol. Sci. 2023, 24, 4426. [Google Scholar] [CrossRef]
- Guzman, F.; Kulcheski, F.R.; Turchetto-Zolet, A.C.; Margis, R. De Novo Assembly of Eugenia uniflora L. Transcriptome and Identification of Genes from the Terpenoid Biosynthesis Pathway. Plant Sci. 2014, 229, 238–246. [Google Scholar] [CrossRef]
- Eguiluz, M.; Rodrigues, N.F.; Guzman, F.; Yuyama, P.; Margis, R. The Chloroplast Genome Sequence from Eugenia uniflora, a Myrtaceae from Neotropics. Plant Syst. Evol. 2017, 303, 1199–1212. [Google Scholar] [CrossRef]
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and Effective Method for RNA Extraction from Different Tissues of Grapevine and Other Woody Plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2018. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 March 2021).
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turchetto-Zolet, A.C.; Salgueiro, F.; Guzman, F.; Vetö, N.M.; Rodrigues, N.F.; Balbinott, N.; Margis-Pinheiro, M.; Margis, R. Gene Expression Divergence in Eugenia uniflora Highlights Adaptation Across Contrasting Atlantic Forest Ecosystems. Plants 2024, 13, 2719. https://doi.org/10.3390/plants13192719
Turchetto-Zolet AC, Salgueiro F, Guzman F, Vetö NM, Rodrigues NF, Balbinott N, Margis-Pinheiro M, Margis R. Gene Expression Divergence in Eugenia uniflora Highlights Adaptation Across Contrasting Atlantic Forest Ecosystems. Plants. 2024; 13(19):2719. https://doi.org/10.3390/plants13192719
Chicago/Turabian StyleTurchetto-Zolet, Andreia C., Fabiano Salgueiro, Frank Guzman, Nicole M. Vetö, Nureyev F. Rodrigues, Natalia Balbinott, Marcia Margis-Pinheiro, and Rogerio Margis. 2024. "Gene Expression Divergence in Eugenia uniflora Highlights Adaptation Across Contrasting Atlantic Forest Ecosystems" Plants 13, no. 19: 2719. https://doi.org/10.3390/plants13192719






