GWAS and Meta-QTL Analysis of Kernel Quality-Related Traits in Maize
Abstract
1. Introduction
2. Results
2.1. Phenotypic Analysis of Quality-Related Traits
2.2. GWAS Analysis
2.3. Expression Analysis of Candidate Genes
2.4. Screening and Functional Analysis of Candidate Genes
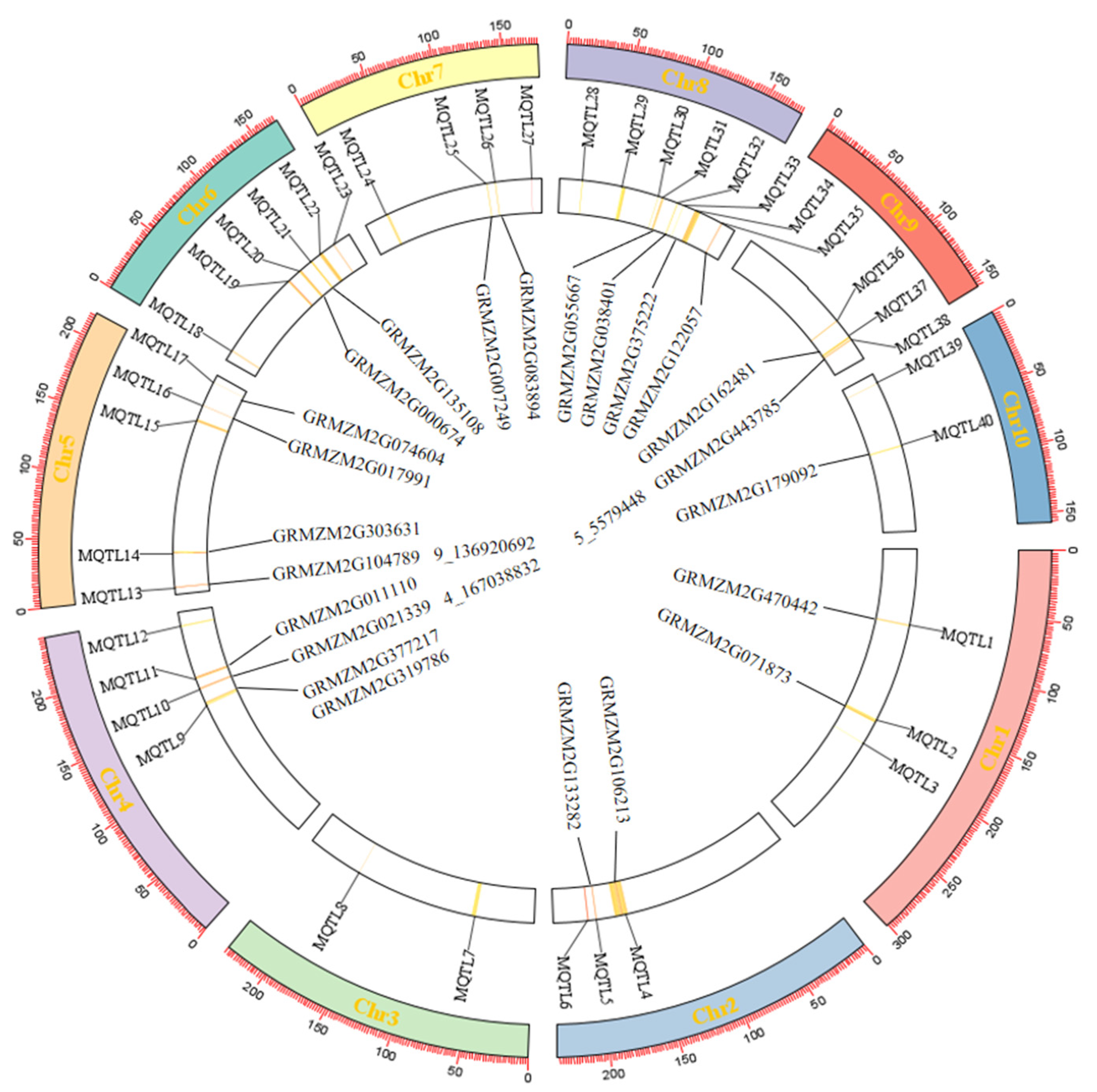
2.5. QTL Distribution of Corn Grain Quality Traits
2.6. Meta-QTL Analysis
3. Discussion
3.1. Genetic Basis of Kernel Quality-Related Traits
3.2. Function Prediction Analysis of Candidate Genes
3.3. Combination of GWAS and Meta-QTL to Analyze the Genetic Basis of Quality Traits
4. Materials and Methods
4.1. Materials and Experimental Design
4.2. Determination of Phenotypic Traits
4.3. Phenotypic Data Analysis
4.4. Genome-Wide Association Study
4.5. Candidate Gene Mining and Functional Analysis
4.6. The Collection of QTL Information Related to Corn Quality Traits
4.7. Integration of QTL Information
4.8. QTL Prediction and Meta-Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Reddappa, S.B.; Muthusamy, V.; Zunjare, R.U.; Chhabra, R.; Talukder, Z.A.; Maman, S.; Chand, G.; Pal, D.; Kumar, R.; Mehta, B.K. Composition of kernel-amylose and-resistant starch among subtropically adapted maize. J. Food Compos. Anal. 2023, 119, 105236. [Google Scholar] [CrossRef]
- Zhao, H.-L.; Yao, Q.; Xiao, Z.-Y.; Qin, S.; Gong, D.-M.; Qiu, F.-Z. Revealing the process of storage protein rebalancing in high quality protein maize by proteomic and transcriptomic. J. Integr. Agric. 2023, 22, 1308–1323. [Google Scholar] [CrossRef]
- Ogunyemi, A.M.; Otegbayo, B.O.; Fagbenro, J.A. Effects of NPK and biochar fertilized soil on the proximate composition and mineral evaluation of maize flour. Food Sci. Nutr. 2018, 6, 2308–2313. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; He, T. The Formation and Regulation of Maize Quality. Bot. Res. 2022, 11, 541. [Google Scholar]
- Mangolin, C.; De Souza, C.; Garcia, A.A.F.; Garcia, A.; Sibov, S.; De Souza, A. Mapping QTLs for kernel oil content in a tropical maize population. Euphytica 2004, 137, 251–259. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.; Zhao, Y. Difference analysis of kernel test weight and nutritional quality traits in maize (Zea mays L.) germplasm resources. J. Plant Genet. Res. 2016, 17, 832–839. [Google Scholar]
- Cook, J.P.; McMullen, M.D.; Holland, J.B.; Tian, F.; Bradbury, P.; Ross-Ibarra, J.; Buckler, E.S.; Flint-Garcia, S.A. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 2012, 158, 824–834. [Google Scholar] [CrossRef]
- Liu, N.; Xue, Y.; Guo, Z.; Li, W.; Tang, J. Genome-wide association study identifies candidate genes for starch content regulation in maize kernels. Front. Plant Sci. 2016, 7, 1046. [Google Scholar] [CrossRef]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Z.; Deng, D.; Ding, H.; Zhang, R.; Wang, S.; Bian, Y.; Yin, Z.; Xu, X. Meta-analysis combined with syntenic metaQTL mining dissects candidate loci for maize yield. Mol. Breed. 2013, 31, 601–614. [Google Scholar] [CrossRef]
- Goffinet, B.; Gerber, S. Quantitative trait loci: A meta-analysis. Genetics 2000, 155, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Arriagada, O.; Arévalo, B.; Cabeza, R.A.; Carrasco, B.; Schwember, A.R. Meta-QTL analysis for yield components in common bean (Phaseolus vulgaris L.). Plants 2022, 12, 117. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-wide association studies in maize: Praise and stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhou, B.; Xu, W. Transcriptome analysis revealed sh2 gene mutation leads reduced zein protein accumulation in maize endosperm. Genet. Resour. Crop Evol. 2023, 70, 1663–1676. [Google Scholar] [CrossRef]
- Sethi, M.; Singh, A.; Kaur, H.; Phagna, R.K.; Rakshit, S.; Chaudhary, D.P. Expression profile of protein fractions in the developing kernel of normal, Opaque-2 and quality protein maize. Sci. Rep. 2021, 11, 2469. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xiang, X.; Huang, Y.; Zhou, Y.; An, D.; Dong, J.; Zhao, C.; Liu, H.; Li, Y.; Wang, Q. Long-read sequencing reveals genomic structural variations that underlie creation of quality protein maize. Nat. Commun. 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, H.; Zhu, Y.; Huang, X.; Li, S.; Wu, X.; Zhao, Y.; Bao, Z.; Qin, L.; Jin, Y. THP9 enhances seed protein content and nitrogen-use efficiency in maize. Nature 2022, 612, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wang, M.; Zhang, X.; Chen, W.; Song, X.; Fu, X.; Fang, H.; Xu, J.; Xiao, Y.; Li, Y. Genetic basis of kernel starch content decoded in a maize multi-parent population. Plant Biotechnol. J. 2021, 19, 2192–2205. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef]
- Yang, G.; Dong, Y.; Li, Y.; Wang, Q.; Shi, Q.; Zhou, Q. QTL verification of grain protein content and its correlation with oil content by using connected RIL populations of high-oil maize. Genet. Mol. Res. 2014, 13, 881–894. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Li, Y.; Wei, M.; Li, X.; Fu, J. QTL detection for grain oil and starch content and their associations in two connected F 2: 3 populations in high-oil maize. Euphytica 2010, 174, 239–252. [Google Scholar] [CrossRef]
- Zheng, Y.; Yuan, F.; Huang, Y.; Zhao, Y.; Jia, X.; Zhu, L.; Guo, J. Genome-wide association studies of grain quality traits in maize. Sci. Rep. 2021, 11, 9797. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yang, X.; Chander, S.; Yan, J.; Zhang, J.; Song, T.; Li, J. Identification of unconditional and conditional QTL for oil, protein and starch content in maize. Crop J. 2013, 1, 34–42. [Google Scholar] [CrossRef]
- Varshney, V.; Hazra, A.; Rao, V.; Ghosh, S.; Kamble, N.U.; Achary, R.K.; Gautam, S.; Majee, M. The Arabidopsis F-box protein SKP1-INTERACTING PARTNER 31 modulates seed maturation and seed vigor by targeting JASMONATE ZIM DOMAIN proteins independently of jasmonic acid-isoleucine. Plant Cell 2023, 35, 3712–3738. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Z.; Zhang, Q.; Wang, R.; Xiao, L.; Ma, H.; Chong, K.; Xu, Y. SKP1 is involved in abscisic acid signalling to regulate seed germination, stomatal opening and root growth in Arabidopsis thaliana. Plant Cell Environ. 2012, 35, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, Y.; Wang, X.; Jiao, Z.; Zhang, W.; Long, Y. Characterization of Glycosyltransferase Family 1 (GT1) and Their Potential Roles in Anthocyanin Biosynthesis in Maize. Genes 2023, 14, 2099. [Google Scholar] [CrossRef]
- Ng, D.W.; Wang, T.; Chandrasekharan, M.B.; Aramayo, R.; Kertbundit, S.; Hall, T.C. Plant SET domain-containing proteins: Structure, function and regulation. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2007, 1769, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Gräfe, K.; Schmitt, L. The ABC transporter G subfamily in Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 92–106. [Google Scholar] [CrossRef]
- Ly, L.K.; Bui, T.P.; Van Thi Le, A.; Van Nguyen, P.; Ong, P.X.; Pham, N.B.; Zhang, Z.J.; Do, P.T.; Chu, H.H. Enhancing plant growth and biomass production by overexpression of GA20ox gene under control of a root preferential promoter. Transgenic Res. 2022, 31, 73–85. [Google Scholar] [CrossRef]
- Yue, K.; Lingling, L.; Xie, J.; Coulter, J.A.; Luo, Z. Synthesis and regulation of auxin and abscisic acid in maize. Plant Signal. Behav. 2021, 16, 1891756. [Google Scholar] [CrossRef] [PubMed]
- Decker, D.; Kleczkowski, L.A. UDP-sugar producing pyrophosphorylases: Distinct and essential enzymes with overlapping substrate specificities, providing de novo precursors for glycosylation reactions. Front. Plant Sci. 2019, 9, 1822. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Shi, H.; Wang, H.; Ji, K.; Zhang, L.; Wang, Y.; Dong, Y.; Li, Y. ZmMPK6, a mitogen-activated protein kinase, regulates maize kernel weight. J. Exp. Bot. 2024, 75, 3287–3299. [Google Scholar] [CrossRef] [PubMed]
- Welcker, C.; Sadok, W.; Dignat, G.; Renault, M.; Salvi, S.; Charcosset, A.; Tardieu, F. A common genetic determinism for sensitivities to soil water deficit and evaporative demand: Meta-analysis of quantitative trait loci and introgression lines of maize. Plant Physiol. 2011, 157, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Choudhary, M.; Singh, A.; Sheoran, S.; Singla, D.; Rakshit, S. Meta-QTL analysis for mining of candidate genes and constitutive gene network development for fungal disease resistance in maize (Zea mays L.). Crop J. 2023, 11, 511–522. [Google Scholar] [CrossRef]
- Camus-Kulandaivelu, L.; Veyrieras, J.-B.; Madur, D.; Combes, V.; Fourmann, M.; Barraud, S.; Dubreuil, P.; Gouesnard, B.; Manicacci, D.; Charcosset, A. Maize adaptation to temperate climate: Relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 2006, 172, 2449–2463. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, C.; Zhang, X.; Li, Y.; Zhang, D.; Shi, Y.; Song, Y.; Li, Y.; Yang, D.; Wang, T. Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci. 2020, 292, 110380. [Google Scholar] [CrossRef]
- Urrutia, M.; Blein-Nicolas, M.; Prigent, S.; Bernillon, S.; Deborde, C.; Balliau, T.; Maucourt, M.; Jacob, D.; Ballias, P.; Bénard, C. Maize metabolome and proteome responses to controlled cold stress partly mimic early-sowing effects in the field and differ from those of Arabidopsis. Plant Cell Environ. 2021, 44, 1504–1521. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 2016, 9, plantgenome2015.04.0025. [Google Scholar] [CrossRef]
- Sosnowski, O.; Charcosset, A.; Joets, J. BioMercator V3: An upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 2012, 28, 2082–2083. [Google Scholar] [CrossRef] [PubMed]
- Darvasi, A.; Soller, M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 1997, 27, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-M.; Shao, X.-Y.; Pei, Y.-H.; Guo, X.-M.; Li, J.; Song, X.-Y.; Zhao, M.-A. Genetic diversity and genome-wide association study of major ear quantitative traits using high-density SNPs in maize. Front. Plant Sci. 2018, 9, 966. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhang, Z.; Shi, Q.; Wang, Q.; Zhou, Q.; Li, Y. QTL identification and meta-analysis for kernel composition traits across three generations in popcorn. Euphytica 2015, 204, 649–660. [Google Scholar] [CrossRef]
- Lin, F.; Zhou, L.; He, B.; Zhang, X.; Dai, H.; Qian, Y.; Ruan, L.; Zhao, H. QTL mapping for maize starch content and candidate gene prediction combined with co-expression network analysis. Theor. Appl. Genet. 2019, 132, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Dong, Y.; Li, Y.; Wang, Q.; Shi, Q.; Zhou, Q. Verification of QTL for grain starch content and its genetic correlation with oil content using two connected RIL populations in high-oil maize. PLoS ONE 2013, 8, e53770. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, G.; Li, Y.; Wang, Q.; Zhou, Y.; Zhou, Q.; Shen, B.; Zhang, F.; Liang, X. Detection and integration of quantitative trait loci for grain yield components and oil content in two connected recombinant inbred line populations of high-oil maize. Mol. Breed. 2012, 29, 313–333. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, X.; Song, X.; Yan, J.; Song, T.; Dai, J.; Rocheford, T.; Li, J. Mapping quantitative trait loci for oil, starch, and protein concentrations in grain with high-oil maize by SSR markers. Euphytica 2008, 162, 335–344. [Google Scholar] [CrossRef]
- Wassom, J.J.; Wong, J.C.; Martinez, E.; King, J.J.; DeBaene, J.; Hotchkiss, J.R.; Mikkilineni, V.; Bohn, M.O.; Rocheford, T.R. QTL associated with maize kernel oil, protein, and starch concentrations; kernel mass; and grain yield in Illinois high oil× B73 backcross-derived lines. Crop Sci. 2008, 48, 243–252. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Wei, M.; Li, X.; Fu, J. QTL identification of grain protein concentration and its genetic correlation with starch concentration and grain weight using two populations in maize (Zea mays L.). J. Genet. 2009, 88, 61–67. [Google Scholar] [CrossRef]
- Yanyang, L.; Yongbin, D.; Suzhen, N.; Dangqun, C.; Yanzhao, W.; Mengguan, W.; Xuehui, L.; Jiafeng, F.; Zhongwei, Z.; Huanqing, C. QTL identification of kernel composition traits with popcorn using both F2: 3 and BC2F2 populations developed from the same cross. J. Cereal Sci. 2008, 48, 625–631. [Google Scholar] [CrossRef]
- Yang, X.; Ma, H.; Zhang, P.; Yan, J.; Guo, Y.; Song, T.; Li, J. Characterization of QTL for oil content in maize kernel. Theor. Appl. Genet. 2012, 125, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yu, X.; Yuan, J.; Cao, Y.; Sun, Q.; Yan, Z.; Ke, Y. QTL, Mapping of Nutritional Quality Traits in Maize. J. Sichuan Univ. 2009, 46, 1454–1460. [Google Scholar]
- Feng, X. QTL Analysis of the Kernel Nutritional Characters and Filling Speed in Different Filling Stages in Maize. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2008. [Google Scholar]
- Li, X.; Shen, S.; Li, Y. QTL Analysis of Protein in Content in High-Oil Maize Using Seed Trait QTL Explorer. Crops 2011, 27, 40–42. [Google Scholar]
- Li, Y.; Wang, Y.; Dong, Y. QTL Analysis of Protein in Content in Popcorn Kernels by Using the Trisomic Inheritance of the Endosperm Model. J. Maize Sci. 2006, 14, 13–16. [Google Scholar]
- Sun, H.; Cai, Y.; Wang, J.; Wang, G.; Dai, G.; Xu, D.; Sun, J. QTL Mapping for Nutritional Quality Traits in Maize. J. Agric. Biotechnol. 2011, 19, 616–623. [Google Scholar]
- Sun, H. QTL Mapping for Nutritional Quality Traits in Maize. Master’s Thesis, Southwest University, Chongqing, China, 2008. [Google Scholar]
- Xie, H.; Duan, L.; Wu, X.; Zhao, S.; Zhu, Z.; Cui, Z.; Fu, Z.; Tang, J. QTL Analysis of Protein Content in Maize Kernels at Different Developing Stages. J. Henan Agric. Univ. 2008, 42, 371–374. [Google Scholar]
- Yang, G. Construction of Genetic Map and QTL Analysis for Main Traits Using Two Connected RIL Populations in Maize. Ph.D. Thesis, Henan Agricultural University, Zhengzhou, China, 2011. [Google Scholar]
- Yang, M. QTL Analysisi for Maize Grain Filling Related and Kernel Nutritive Traits at Different Stages Use RIL Population. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2010. [Google Scholar]
- Zhao, S. QTLs Analysis of Quality Characters in Maize Kernels under Different Developing Stages. Doctoral Dissertation, Xinjiang Agricultural University, Urumqi, China, 2007. [Google Scholar]
- Zhang, Z. Construction of Normal Corn × Popcorn RIL Population and QTL Analysis for Main Traits. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2009. [Google Scholar]
- Pei, Y.; Li, Y.; Guo, X.; Song, X. QTL Mapping for Main Nutritional Quality Traits in Maize. J. Maize Sci. 2014, 22, 21–26. [Google Scholar]
- Zhao, D. QTL Analysis of Kernel Nutritional Quality Traits in Maize. Master’s Thesis, Sichuan Agricultural University, Chengdu, China, 2016. [Google Scholar]
- Lan, T.; Cui, T.; He, K.; Chang, L.; Liu, J. QTL Mapping of Kernel Quality Traits under Different Nitrogen Treatments in Maize. J. Maize Sci. 2017, 25, 6–11. [Google Scholar]
- Yang, Z.; Li, X.; Zhang, N.; Zhang, Y.; Jiang, H.; Gao, J.; Kuai, B.; Ding, Y.; Huang, X. Detection of quantitative trait loci for kernel oil and protein concentration in a B73 and Zheng58 maize cross. Genet. Mol. Res. 2016, 15, gmr.15038951. [Google Scholar] [CrossRef]
- Katral, A.; Hossain, F.; Zunjare, R.U.; Chhabra, R.; Vinutha, T.; Duo, H.; Kumar, B.; Karjagi, C.G.; Jacob, S.R.; Pandey, S. Multilocus functional characterization of indigenous and exotic inbreds for dgat1-2, fatb, ge2 and wri1a genes affecting kernel oil and fatty acid profile in maize. Gene 2024, 895, 148001. [Google Scholar] [CrossRef] [PubMed]
- Varalakshmi, S.; Sahoo, S.; Singh, N.K.; Pareek, N.; Garkoti, P.; Senthilkumar, V.; Kashyap, S.; Jaiswal, J.P.; Jacob, S.R.; Nankar, A.N. Marker–Trait Association for Protein Content among Maize Wild Accessions and Coix Using SSR Markers. Agronomy 2023, 13, 2138. [Google Scholar] [CrossRef]
- Niu, L.; Liu, L.; Zhang, J.; Scali, M.; Wang, W.; Hu, X.; Wu, X. Genetic Engineering of Starch Biosynthesis in Maize Seeds for Efficient Enzymatic Digestion of Starch during Bioethanol Production. Int. J. Mol. Sci. 2023, 24, 3927. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Sun, X.; Chen, Z.; Wang, Q.; Zhang, C.; Liu, X.; Wu, W.; Yin, Y. Exploring the roles of ZmARM gene family in maize development and abiotic stress response. PeerJ 2023, 11, e16254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, J.; Zhang, Y.; Li, J.; Liu, M.; Li, L.; Li, S.; Wang, T.; Shaw, R.K.; Jiang, F. Hotspot regions of Quantitative Trait Loci and candidate genes for ear-related traits in maize: A literature review. Genes 2023, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Maghraby, A.; Alzalaty, M. Genome-wide identification and evolutionary analysis of the AP2/EREBP, COX and LTP genes in Zea mays L. under drought stress. Sci. Rep. 2024, 14, 7610. [Google Scholar] [CrossRef]
- Luo, M.; Lu, B.; Shi, Y.; Zhao, Y.; Liu, J.; Zhang, C.; Wang, Y.; Liu, H.; Shi, Y.; Fan, Y. Genetic basis of the oil biosynthesis in ultra-high-oil maize grains with an oil content exceeding 20%. Front. Plant Sci. 2023, 14, 1168216. [Google Scholar] [CrossRef]
- Cheng, C.; An, L.; Li, F.; Ahmad, W.; Aslam, M.; Ul Haq, M.Z.; Yan, Y.; Ahmad, R.M. Wide-range portrayal of AP2/ERF transcription factor family in maize (Zea mays L.) development and stress responses. Genes 2023, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, X.; Fu, J.; Wang, Q. Functional identification of maize transcription factor ZmMYB12 to enhance drought resistance and low phosphorus tolerance in plants. Acta Agron. Sin. 2024, 50, 76–88. [Google Scholar]
- Jiang, Y.; Yang, L.; Xie, H.; Qin, L.; Wang, L.; Xie, X.; Zhou, H.; Tan, X.; Zhou, J.; Cheng, W. Metabolomics and transcriptomics strategies to reveal the mechanism of diversity of maize kernel color and quality. BMC Genom. 2023, 24, 194. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Zhu, L.; Wang, L.; Zhang, H.; Zhang, X.; Xu, S.; Xue, J. Identification of the maize LEA gene family and its relationship with kernel dehydration. Plants 2023, 12, 3674. [Google Scholar] [CrossRef]
- Sharma, J.; Sharma, S.; Karnatam, K.S.; Raigar, O.P.; Lahkar, C.; Saini, D.K.; Kumar, S.; Singh, A.; Das, A.K.; Sharma, P. Surveying the genomic landscape of silage-quality traits in maize (Zea mays L.). Crop J. 2023, 11, 1893–1901. [Google Scholar] [CrossRef]
- Chen, R.; Wei, Q.; Liu, Y.; Li, J.; Du, X.; Chen, Y.; Wang, J.; Liu, Y. The pentatricopeptide repeat protein EMP601 functions in maize seed development by affecting RNA editing of mitochondrial transcript ccmC. Crop J. 2023, 11, 1368–1379. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Y.; Tang, C.; Wu, J.; Fu, J.; Wang, Q. Genome-wide identification of ZmMYC2 binding sites and target genes in maize. BMC Genom. 2024, 25, 397. [Google Scholar]
- Xie, S.; Luo, H.; Huang, W.; Jin, W.; Dong, Z. Striking a growth–defense balance: Stress regulators that function in maize development. J. Integr. Plant Biol. 2024, 66, 424–442. [Google Scholar] [CrossRef] [PubMed]
- Okunlola, G.; Badu-Apraku, B.; Ariyo, O.; Agre, P.; Offernedo, Q.; Ayo-Vaughan, M. Genome-wide association studies of Striga resistance in extra-early maturing quality protein maize inbred lines. G3 2023, 13, jkac237. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Hou, Q.; Zhao, J.; Fang, C.; An, X.; Wan, X. Fatty acid de novo biosynthesis in plastids: Key enzymes and their critical roles for male reproduction and other processes in plants. Plant Physiol. Biochem. 2024, 210, 108654. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Wang, Y.; Bao, J.; Li, Y.; Yin, Z.; Long, Y.; Wan, X. The genetic structures and molecular mechanisms underlying ear traits in maize (Zea mays L.). Cells 2023, 12, 1900. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yang, D.; Wang, D.; Liang, C.; Wang, J.; Lisch, D.; Zhao, M. Heritable changes of epialleles near genes in maize can be triggered in the absence of CHH methylation. Plant Physiol. 2024, 194, 2511–2532. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.; Lin, Y.; Dang, K.; Wan, J.; Meng, S.; Qiu, X.; Wang, Q.; Mu, L.; Ding, D. Comparative transcriptomics analysis at the key stage of maize ear development dissect heterosis. Plant Genome 2023, 16, e20293. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hou, Y.; Cao, Y.; Wei, B.; Gu, L. A Comprehensive Identification and Expression Analysis of the WUSCHEL Homeobox-Containing Protein Family Reveals Their Special Role in Development and Abiotic Stress Response in Zea mays L. Int. J. Mol. Sci. 2023, 25, 441. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Su, S.; Chen, H.; Li, S.; Shan, X.; Li, H.; Liu, H.; Dong, H.; Yuan, Y. Transcriptome analysis of drought-responsive and drought-tolerant mechanisms in maize leaves under drought stress. Physiol. Plant. 2023, 175, e13875. [Google Scholar] [CrossRef] [PubMed]
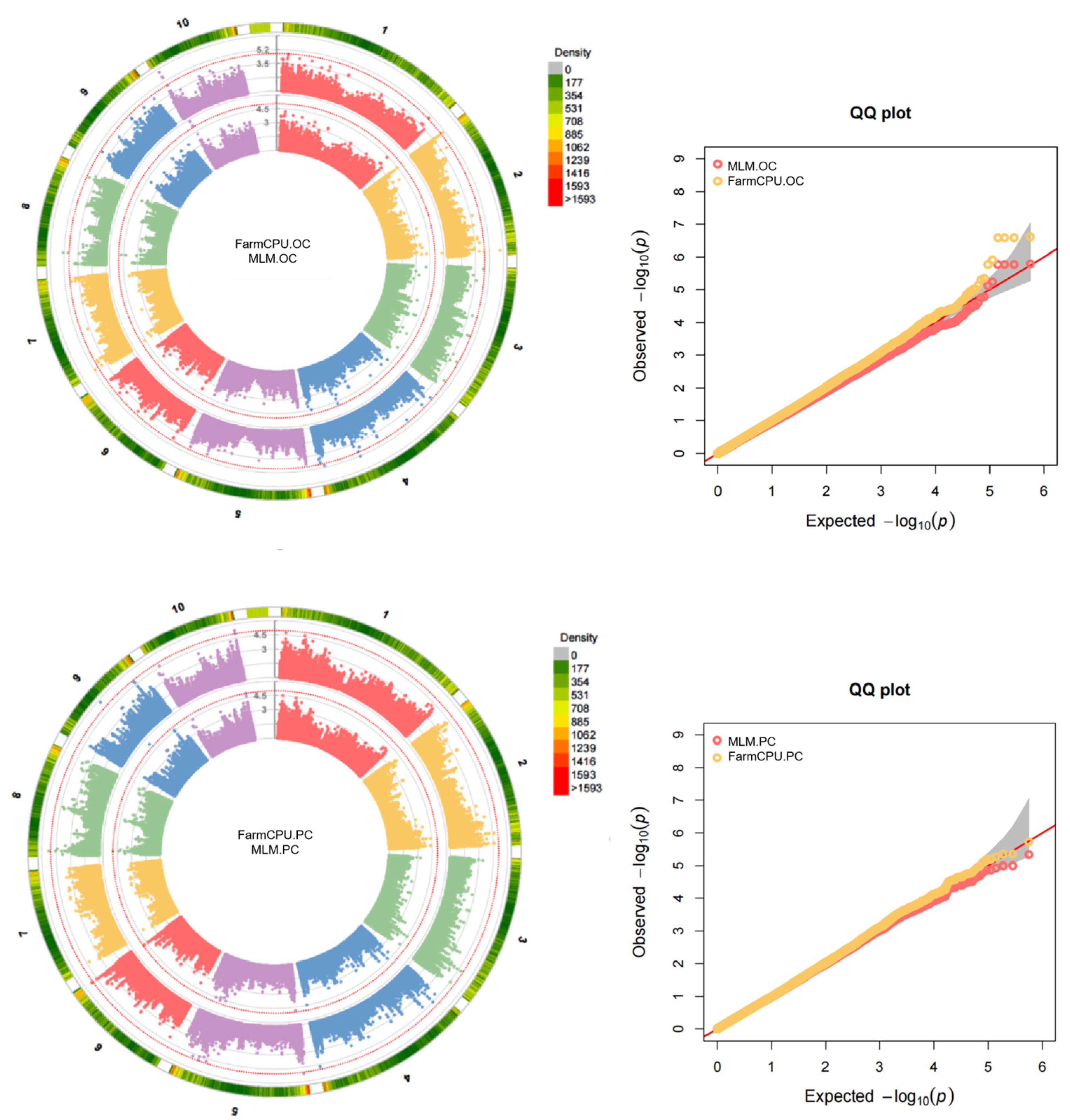
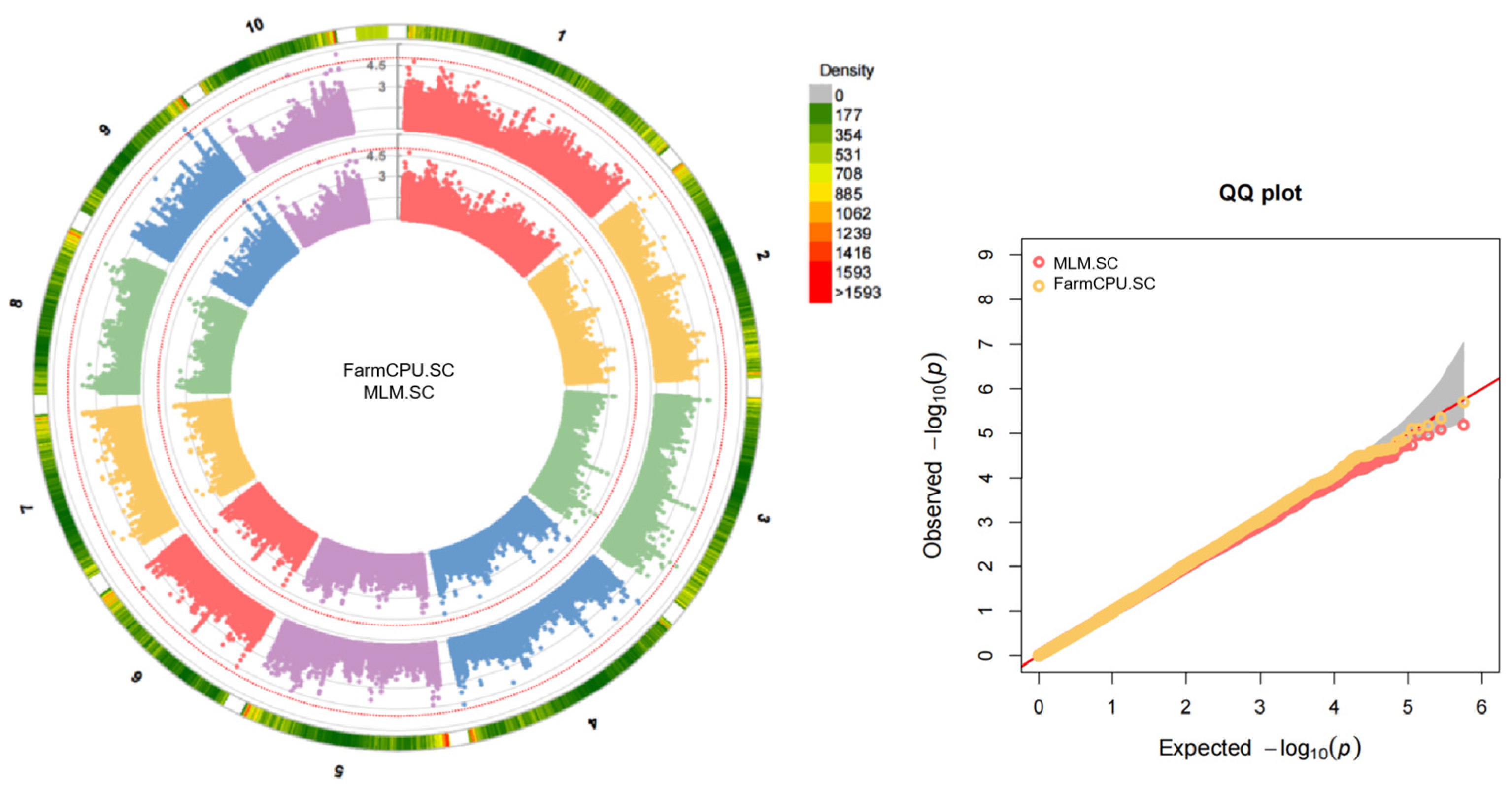
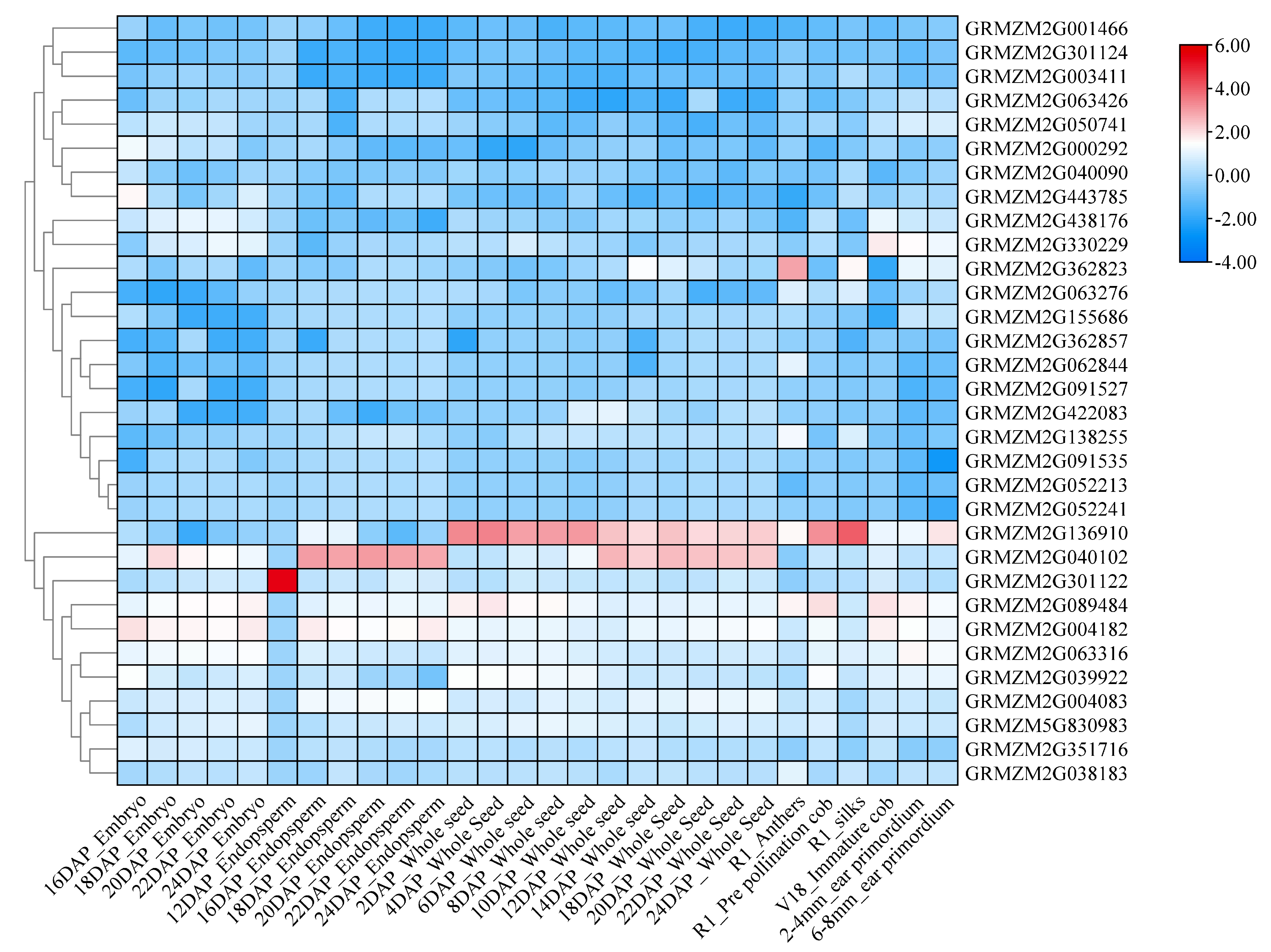
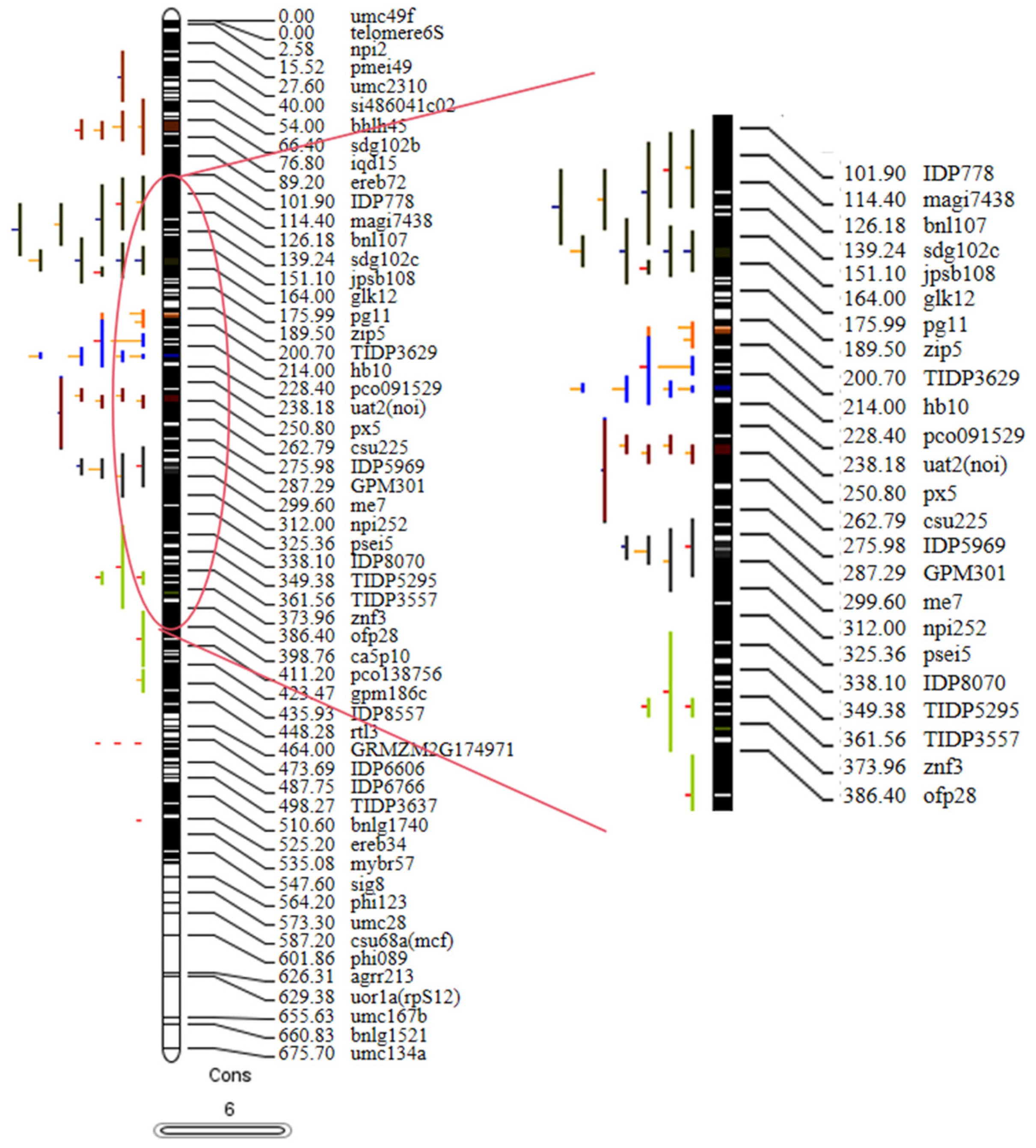
| Character | Mean | Range | CV/% | Skewness | Kurtosis |
|---|---|---|---|---|---|
| Protein content | 11.22 ± 1.14 | 7.74–14.58 | 10.00 | 0.16 | 0.22 |
| Oil content | 3.29 ± 0.72 | 1.79–4.97 | 18.00 | 0.51 | 0.92 |
| Starch content | 61.06 ± 1.83 | 55.38–66.04 | 3.00 | −0.48 | 0.21 |
| Trait | SNPs | Gene | Annotation |
|---|---|---|---|
| OC | 2_229132132 | GRMZM2G052213 | SKP1-like protein 1 |
| OC | 10_8849369 | GRMZM2G301122 | GT1 superfamily protein |
| OC | 4_167038832 | GRMZM2G063316 | SET domain containing protein |
| OC | 4_231963888 | GRMZM2G003411 | ABC transporter G family member 39 |
| OC | 7_4959916 | GRMZM2G039922 | Probable receptor-like protein kinase At1g30570 |
| OC | 10_8849424 | GRMZM2G136910 | Abscisic stress protein homolog |
| SC | 9_136920692 | GRMZM5G830983 | Probable UDP-arabinose 4-epimerase 3-like |
| SC | 10_88685468 | GRMZM2G422083 | Putative MYB DNA-binding domain superfamily protein |
| SC | 10_88685442 | GRMZM2G089484 | MAP kinase6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, R.; Zhuang, Z.; Bian, J.; Ren, Z.; Ta, W.; Peng, Y. GWAS and Meta-QTL Analysis of Kernel Quality-Related Traits in Maize. Plants 2024, 13, 2730. https://doi.org/10.3390/plants13192730
Tang R, Zhuang Z, Bian J, Ren Z, Ta W, Peng Y. GWAS and Meta-QTL Analysis of Kernel Quality-Related Traits in Maize. Plants. 2024; 13(19):2730. https://doi.org/10.3390/plants13192730
Chicago/Turabian StyleTang, Rui, Zelong Zhuang, Jianwen Bian, Zhenping Ren, Wanling Ta, and Yunling Peng. 2024. "GWAS and Meta-QTL Analysis of Kernel Quality-Related Traits in Maize" Plants 13, no. 19: 2730. https://doi.org/10.3390/plants13192730
APA StyleTang, R., Zhuang, Z., Bian, J., Ren, Z., Ta, W., & Peng, Y. (2024). GWAS and Meta-QTL Analysis of Kernel Quality-Related Traits in Maize. Plants, 13(19), 2730. https://doi.org/10.3390/plants13192730





