Genetic Transformation of Triticum dicoccum and Triticum aestivum with Genes of Jasmonate Biosynthesis Pathway Affects Growth and Productivity Characteristics
Abstract
1. Introduction
2. Results
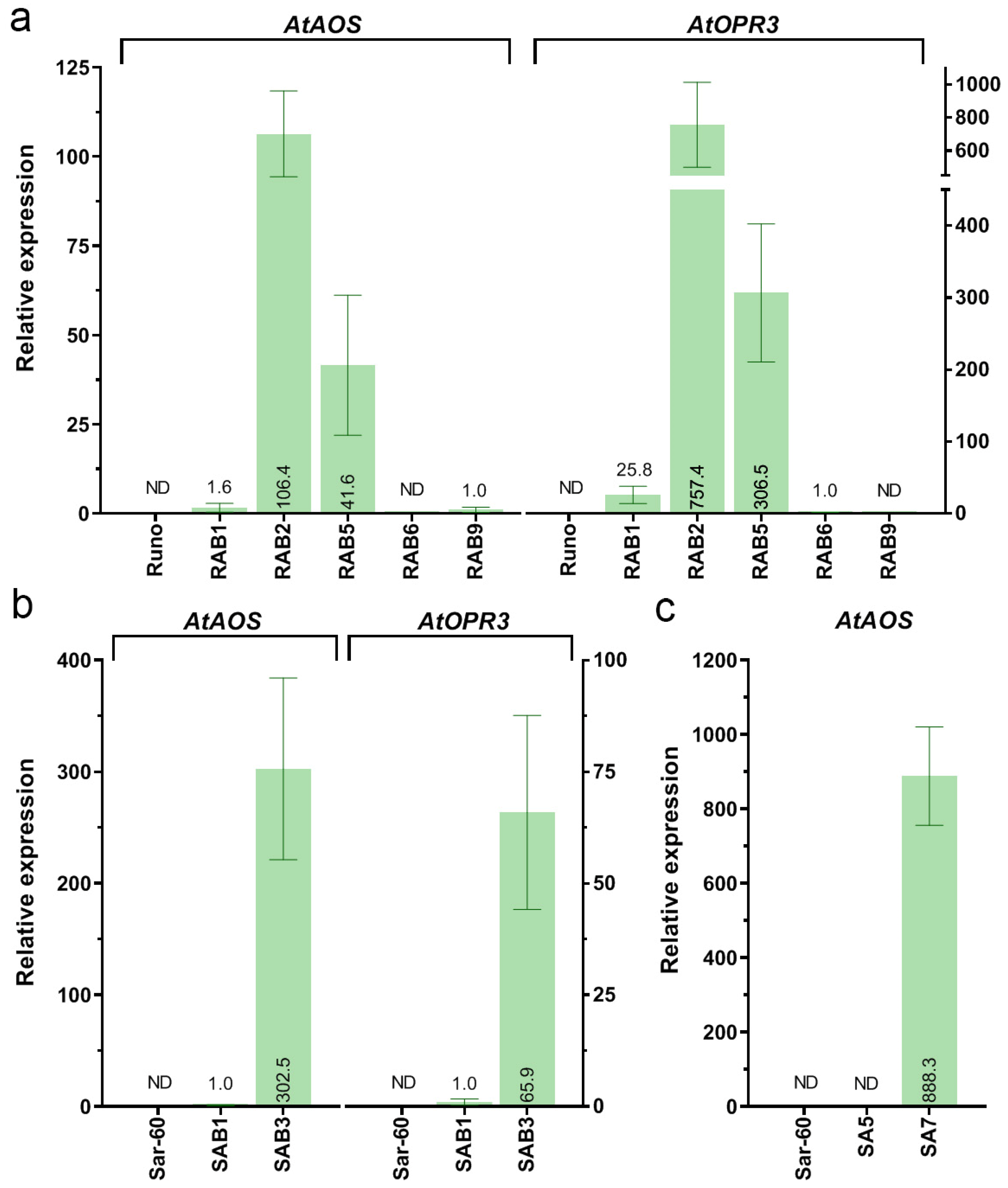
2.1. Generation of Emmer Wheat and Bread Wheat Plants Co-Expressing AtAOS and AtOPR3 Genes
2.1.1. Integration and Overexpression of the AtAOS Gene in Bread Wheat
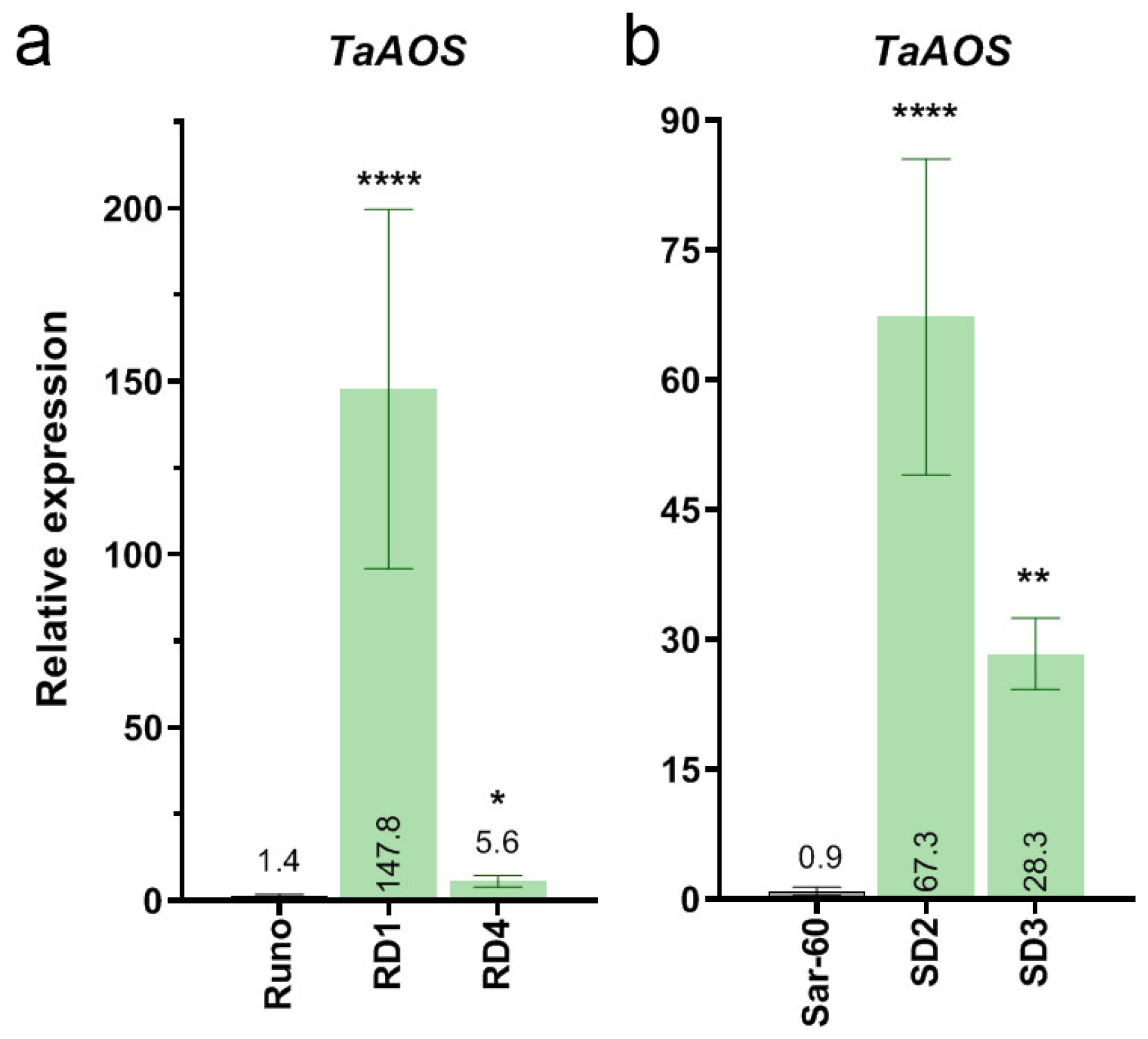
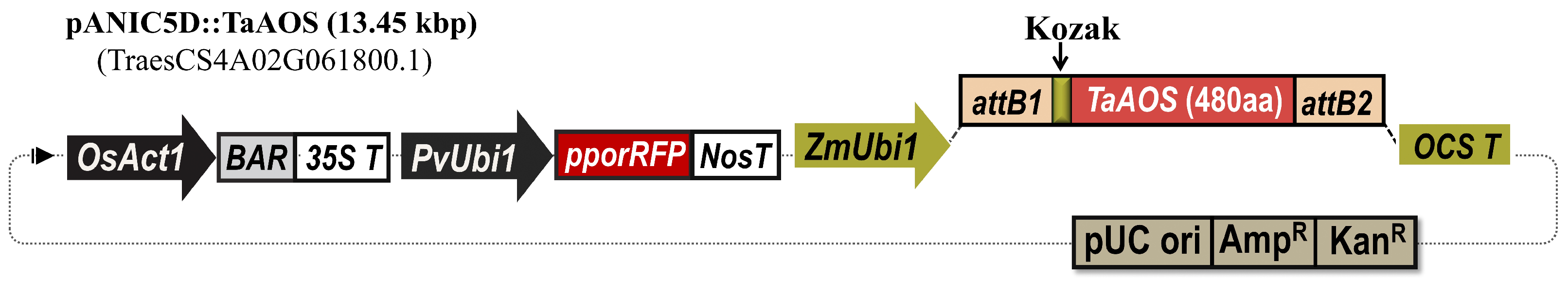
2.1.2. Generation of Transgenic Emmer Wheat and Bread Wheat Plants Overexpressing ALLENE OXIDE SYNTHASE (TaAOS) from Bread Wheat
2.2. Characterization of the Transgenic Wheat Plants
2.2.1. Phytohormone Analysis
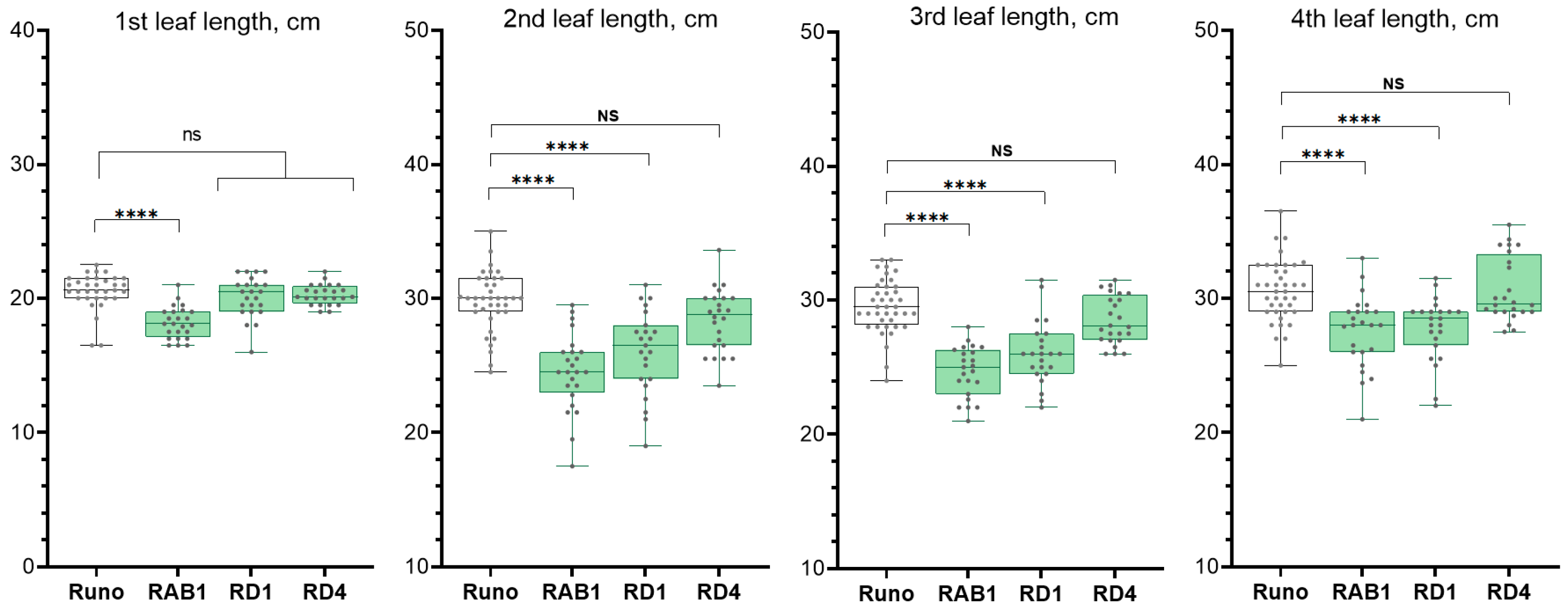
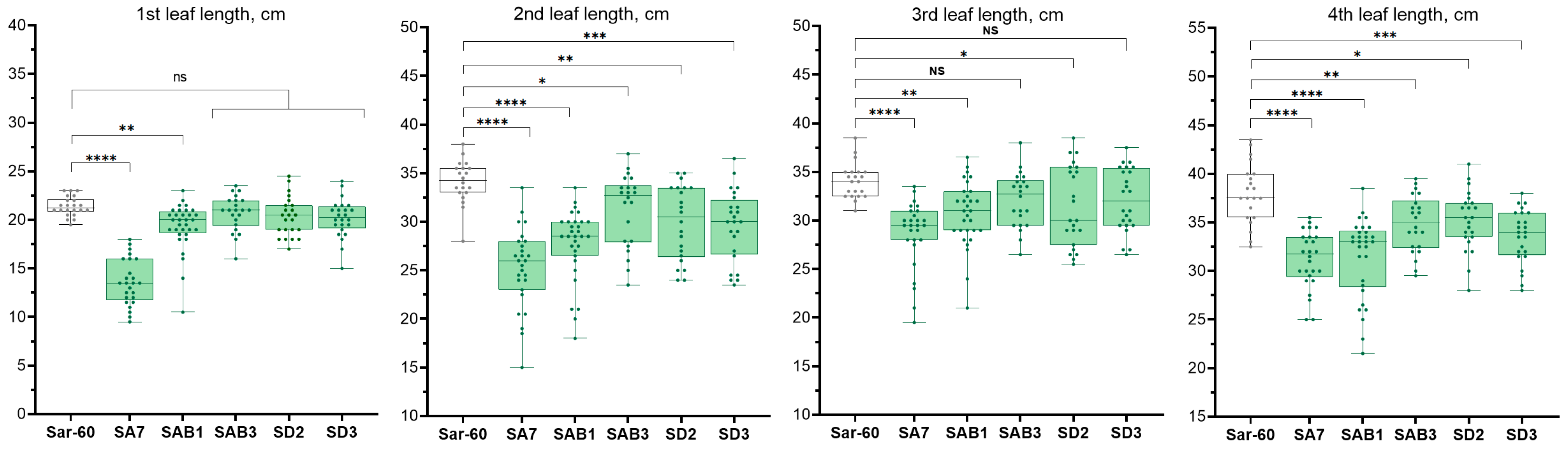
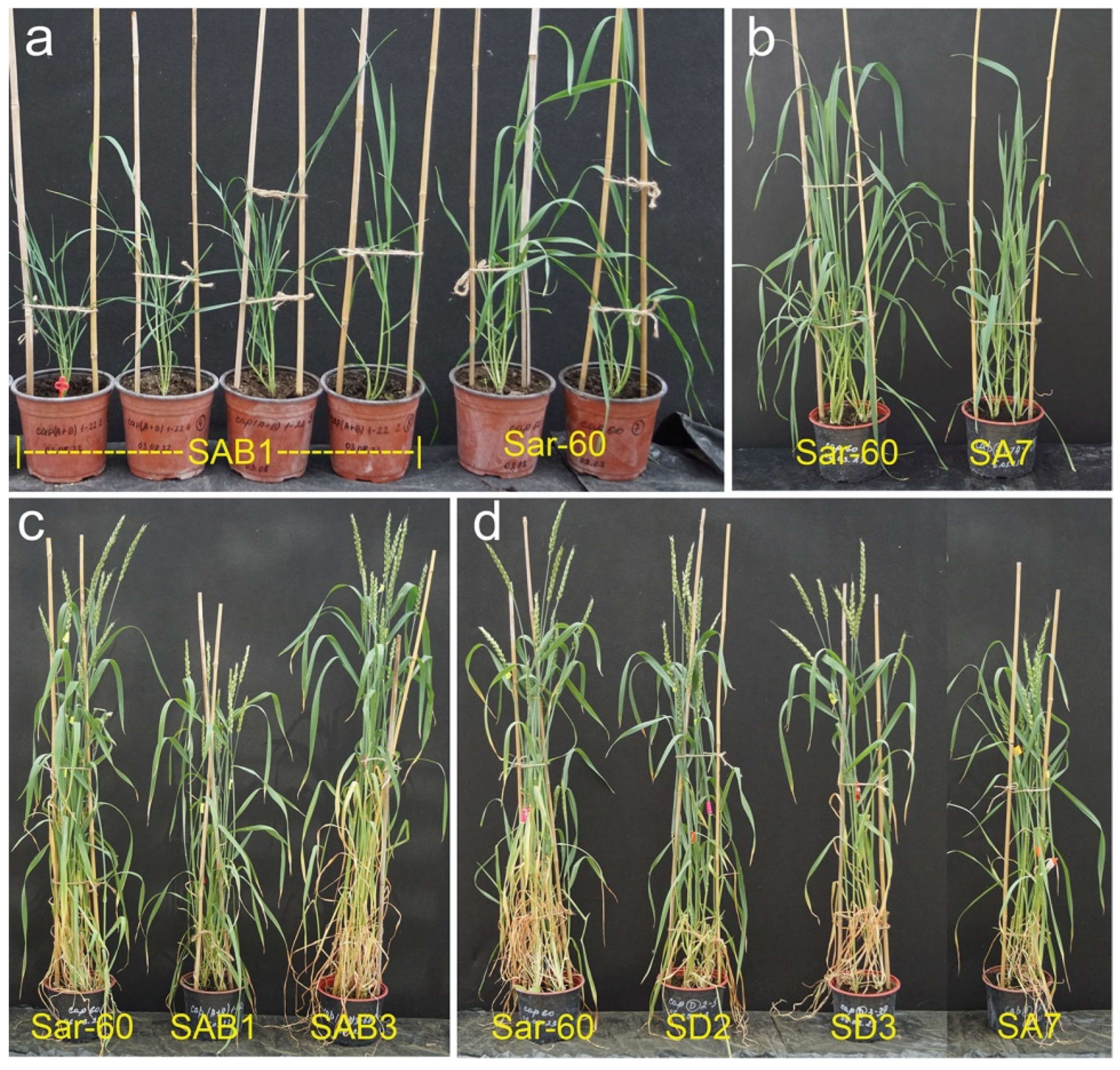
2.2.2. Analysis of the Growth of Transgenic Plants
3. Discussion
4. Materials and Methods
4.1. TaAOS Gene Cloning and Construction of Expression Vectors
4.2. Generation of Transgenic Plants of Emmer Wheat and Bread Wheat
4.3. Analysis of Phytohormones
4.4. Measurements of the Leaf Length
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, S.; Springer, A.; Kang, C.; Wettstein, D.; von Reinbothe, C.; Reinbothe, S.; Pollmann, S. ALLENE OXIDE SYNTHASE and HYDROPEROXIDE LYASE, Two Non-Canonical Cytochrome P450s in Arabidopsis thaliana and Their Different Roles in Plant Defense. Int. J. Mol. Sci. 2019, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Monte, I.; Zamarreño, A.M.; García-Mina, J.M.; Solano, R. Evolution of the jasmonate ligands and their biosynthetic pathways. New Phytol. 2023, 238, 2236–2246. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Y.; Li, S.-S.; Han, G.-Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Schluttenhofer, C. Origin and evolution of jasmonate signaling. Plant Sci. 2020, 298, 110542. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Xin, X.-F. Regulation and integration of plant jasmonate signaling: A comparative view of monocot and dicot. J. Genet. Genomics 2022, 49, 704–714. [Google Scholar] [CrossRef]
- Lyons, R.; Manners, J.M.; Kazan, K. Jasmonate biosynthesis and signaling in monocots: A comparative overview. Plant Cell Rep. 2013, 32, 815–827. [Google Scholar] [CrossRef]
- Okada, K.; Abe, H.; Arimura, G.-I. Jasmonates induce both defense responses and communication in monocotyledonous and dicotyledonous plants. Plant Cell Physiol. 2015, 56, 16–27. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Cheaib, M.; Fournel, M.; Rios, M.; Gantet, P.; Laplaze, L.; Guyomarc’h, S.; Riemann, M.; Heitz, T.; Petitot, A.-S.; et al. Genetic analysis of the rice jasmonate receptors reveals specialized functions for OsCOI2. PLoS ONE 2023, 18, e0291385. [Google Scholar] [CrossRef]
- Okumura, T.; Kitajima, T.; Kaji, T.; Urano, H.; Matsumoto, K.; Inagaki, H.; Miyamoto, K.; Okada, K.; Ueda, M. Difference in the ligand affinity among redundant plant hormone receptors of rice OsCOI1a/1b/2-OsJAZs. Biosci. Biotechnol. Biochem. 2023, 87, 1122–1128. [Google Scholar] [CrossRef]
- Wang, K.-D.; Borrego, E.J.; Kenerley, C.M.; Kolomiets, M.V. Oxylipins Other Than Jasmonic Acid Are Xylem-Resident Signals Regulating Systemic Resistance Induced by Trichoderma virens in Maize. Plant Cell 2020, 32, 166–185. [Google Scholar] [CrossRef] [PubMed]
- Degtyaryov, E.; Pigolev, A.; Miroshnichenko, D.; Frolov, A.; Basnet, A.T.; Gorbach, D.; Leonova, T.; Pushin, A.S.; Alekseeva, V.; Dolgov, S.; et al. 12-Oxophytodienoate Reductase Overexpression Compromises Tolerance to Botrytis cinerea in Hexaploid and Tetraploid Wheat. Plants 2023, 12, 2050. [Google Scholar] [CrossRef] [PubMed]
- Krivina, E.; Degtyaryov, E.; Tebina, E.; Temraleeva, A.; Savchenko, T. Comparative Analysis of the Fatty Acid Profiles of Selected Representatives of Chlorella-Clade to Evaluate Their Biotechnological Potential. Int. J. Plant Biol. 2024, 15, 837–854. [Google Scholar] [CrossRef]
- Park, J.-H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, T.; Huangfu, J.; Li, R.; Lou, Y. Both Allene Oxide Synthases Genes Are Involved in the Biosynthesis of Herbivore-Induced Jasmonic Acid and Herbivore Resistance in Rice. Plants 2021, 10, 442. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Y.; Tang, L.; Tong, X.; Wang, L.; Liu, L.; Huang, S.; Zhang, J. SAPK10-Mediated Phosphorylation on WRKY72 Releases Its Suppression on Jasmonic Acid Biosynthesis and Bacterial Blight Resistance. iScience 2019, 16, 499–510. [Google Scholar] [CrossRef]
- Fan, X.; Tang, H.; Chen, X.; Zeng, F.; Chen, G.; Chen, Z.-H.; Qin, Y.; Deng, F. Allene oxide synthase 1 contributes to limiting grain arsenic accumulation and seedling detoxification in rice. Stress Biol. 2023, 3, 52. [Google Scholar] [CrossRef]
- Maucher, H.; Hause, B.; Feussner, I.; Ziegler, J.; Wasternack, C. Allene oxide synthases of barley (Hordeum vulgare cv. Salome): Tissue specific regulation in seedling development. Plant J. 2000, 21, 199–213. [Google Scholar] [CrossRef]
- Sun, T.; Chen, Y.; Feng, A.; Zou, W.; Wang, D.; Lin, P.; Chen, Y.; You, C.; Que, Y.; Su, Y. The allene oxide synthase gene family in sugarcane and its involvement in disease resistance. Ind. Crops Prod. 2023, 192, 116136. [Google Scholar] [CrossRef]
- Heckmann, A.; Perochon, A.; Doohan, F.M. Genome-wide analysis of salicylic acid and jasmonic acid signalling marker gene families in wheat. Plant Biol. 2024, 26, 691–704. [Google Scholar] [CrossRef]
- Liu, H.-H.; Wang, Y.-G.; Wang, S.-P.; Li, H.-J.; Xin, Q.-G. Improved zinc tolerance of tobacco by transgenic expression of an allene oxide synthase gene from hexaploid wheat. Acta Physiol. Plant. 2014, 36, 2433–2440. [Google Scholar] [CrossRef]
- Grechkin, A.N.; Ogorodnikova, A.V.; Egorova, A.M.; Mukhitova, F.K.; Ilyina, T.M.; Khairutdinov, B.I. Allene Oxide Synthase Pathway in Cereal Roots: Detection of Novel Oxylipin Graminoxins. ChemistryOpen 2018, 7, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Schaller, F.; Biesgen, C.; Müssig, C.; Altmann, T.; Weiler, E.W. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 2000, 210, 979–984. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Pak, H.; Wang, H.; Kim, Y.; Song, U.; Tu, M.; Wu, D.; Jiang, L. Creation of male-sterile lines that can be restored to fertility by exogenous methyl jasmonate for the establishment of a two-line system for the hybrid production of rice (Oryza sativa L.). Plant Biotechnol. J. 2021, 19, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef]
- Mou, Y.; Liu, Y.; Tian, S.; Guo, Q.; Wang, C.; Wen, S. Genome-Wide Identification and Characterization of the OPR Gene Family in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2019, 20, 1914. [Google Scholar] [CrossRef]
- Pigolev, A.V.; Miroshnichenko, D.N.; Dolgov, S.V.; Alekseeva, V.V.; Pushin, A.S.; Degtyaryova, V.I.; Klementyeva, A.; Gorbach, D.; Leonova, T.; Basnet, A.; et al. Endogenously Produced Jasmonates Affect Leaf Growth and Improve Osmotic Stress Tolerance in Emmer Wheat. Biomolecules 2023, 13, 1775. [Google Scholar] [CrossRef]
- Pigolev, A.V.; Miroshnichenko, D.N.; Pushin, A.S.; Terentyev, V.V.; Boutanayev, A.M.; Dolgov, S.V.; Savchenko, T.V. Overexpression of Arabidopsis OPR3 in Hexaploid Wheat (Triticum aestivum L.) Alters Plant Development and Freezing Tolerance. Int. J. Mol. Sci. 2018, 19, 3989. [Google Scholar] [CrossRef]
- Miroshnichenko, D.N.; Pigolev, A.V.; Tikhonov, K.G.; Degtyaryov, E.A.; Leshchenko, E.F.; Alekseeva, V.V.; Pushin, A.S.; Dolgov, S.V.; Basnet, A.; Gorbach, D.P.; et al. Characteristics of the Stress-Tolerant Transgenic Wheat Line Overexpressing the AtOPR3 Gene Encoding the Jasmonate Biosynthesis Enzyme 12-Oxophytodienoate Reductase. Russ. J. Plant Physiol. 2024, 71, 1446. [Google Scholar] [CrossRef]
- Chehab, E.W.; Perea, J.V.; Gopalan, B.; Theg, S.; Dehesh, K. Oxylipin Pathway in Rice and Arabidopsis. J. Integr. Plant Biol. 2007, 49, 43–51. [Google Scholar] [CrossRef]
- Vasil, V.; Castillo, A.M.; Fromm, M.E.; Vasil, I.K. Herbicide Resistant Fertile Transgenic Wheat Plants Obtained by Microprojectile Bombardment of Regenerable Embryogenic Callus. Nat. Biotechnol. 1992, 10, 667–674. [Google Scholar] [CrossRef]
- Shrawat, A.K.; Armstrong, C.L. Development and Application of Genetic Engineering for Wheat Improvement. Crit. Rev. Plant Sci. 2018, 37, 335–421. [Google Scholar] [CrossRef]
- Borisjuk, N.; Kishchenko, O.; Eliby, S.; Schramm, C.; Anderson, P.; Jatayev, S.; Kurishbayev, A.; Shavrukov, Y. Genetic Modification for Wheat Improvement: From Transgenesis to Genome Editing. Biomed Res. Int. 2019, 27, 6216304. [Google Scholar] [CrossRef]
- Miroshnichenko, D.; Klementyeva, A.; Pushin, A.; Dolgov, S. A competence of embryo-derived tissues of tetraploid cultivated wheat species Triticum dicoccum and Triticum timopheevii for efficient and stable transgenesis mediated by particle inflow gun. BMC Plant Biol. 2020, 20, 442. [Google Scholar] [CrossRef]
- Miroshnichenko, D.; Timerbaev, V.; Klementyeva, A.; Pushin, A.; Sidorova, T.; Litvinov, D.; Nazarova, L.; Shulga, O.; Divashuk, M.; Karlov, G.; et al. CRISPR/Cas9-induced modification of the conservative promoter region of VRN-A1 alters the heading time of hexaploid bread wheat. Front. Plant Sci. 2022, 13, 1048695. [Google Scholar] [CrossRef]
- Miroshnichenko, D.; Timerbaev, V.; Divashuk, M.; Pushin, A.; Alekseeva, V.; Kroupin, P.; Bazhenov, M.; Samarina, M.; Ermolaev, A.; Karlov, G.; et al. CRISPR/Cas9-mediated Multiplexed multi-allelic mutagenesis of genes located on A, B and R subgenomes of hexaploid triticale. Plant Cell Rep. 2024, 43, 59. [Google Scholar] [CrossRef]
- Bossio, E.; Díaz Paleo, A.; del Vas, M.; Baroli, I.; Acevedo, A.; Ríos, R.D. Silencing of the glutathione biosynthetic pathway inhibits somatic embryogenesis in wheat. Plant Cell Tissue Organ. Cult. 2013, 112, 239–248. [Google Scholar] [CrossRef]
- Gasparis, S.; Kała, M.; Przyborowski, M.; Orczyk, W.; Nadolska-Orczyk, A. Artificial MicroRNA-Based Specific Gene Silencing of Grain Hardness Genes in Polyploid Cereals Appeared to Be Not Stable Over Transgenic Plant Generations. Front. Plant Sci. 2016, 7, 2017. [Google Scholar] [CrossRef]
- Rana, I.A.; Salomon, S.; Schäfer, W.; Becker, D. Downregulation of Glucan Synthase-Like (TaGSL) genes in wheat leads to inhibition of transgenic plant regeneration. In Vitro Cell. Dev. Biol.-Plant 2014, 50, 696–706. [Google Scholar] [CrossRef]
- Ashikawa, I.; Mori, M.; Nakamura, S.; Abe, F. A transgenic approach to controlling wheat seed dormancy level by using Triticeae DOG1-like genes. Transgenic Res. 2014, 23, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Sparks, C.A.; Jones, H.D. Genetic transformation of wheat via particle bombardment. Methods Mol. Biol. 2014, 1099, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Shi, Q.; Guo, X.; Liu, Y.; Su, H.; Guo, X.; Lv, Z.; Han, F. Site-specific transfer of chromosomal segments and genes in wheat engineered chromosomes. J. Genet. Genomics 2017, 44, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, P.; Ellison, E.; Polley, B.; Bollina, V.; Kulkarni, M.; Ghanbarnia, K.; Song, H.; Gao, C.; Voytas, D.F.; Kagale, S. Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 2018, 8, 6502. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Arndell, T.; Borisjuk, N.; Sharma, N.; Watson-Haigh, N.S.; Tucker, E.J.; Baumann, U.; Langridge, P.; Whitford, R. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 2019, 17, 1905–1913. [Google Scholar] [CrossRef]
- Li, G.; Wu, Y.; Liu, G.; Xiao, X.; Wang, P.; Gao, T.; Xu, M.; Han, Q.; Wang, Y.; Guo, T.; et al. Large-scale Proteomics Combined with Transgenic Experiments Demonstrates An Important Role of Jasmonic Acid in Potassium Deficiency Response in Wheat and Rice. Mol. Cell. Proteomics 2017, 16, 1889–1905. [Google Scholar] [CrossRef]
- Kamińska, M. Role and activity of jasmonates in plants under in vitro conditions. Plant Cell Tissure Organ. Cult. 2021, 146, 425–447. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, W.; Gu, Z.; Wu, S.; Zhou, W.; Lin, J.; Xu, L. Roles of the wound hormone jasmonate in plant regeneration. J. Exp. Bot. 2023, 74, 1198–1206. [Google Scholar] [CrossRef]
- Elhiti, M.; Hebelstrup, K.H.; Wang, A.; Li, C.; Cui, Y.; Hill, R.D.; Stasolla, C. Function of type-2 Arabidopsis hemoglobin in the auxin-mediated formation of embryogenic cells during morphogenesis. Plant J. 2013, 74, 946–958. [Google Scholar] [CrossRef]
- Mira, M.M.; Wally, O.S.D.; Elhiti, M.; El-Shanshory, A.; Reddy, D.S.; Hill, R.D.; Stasolla, C. Jasmonic acid is a downstream component in the modulation of somatic embryogenesis by Arabidopsis Class 2 phytoglobin. J. Exp. Bot. 2016, 67, 2231–2246. [Google Scholar] [CrossRef]
- Ruduś, I.; Weiler, E.W.; Kępczyńska, E. Do stress-related phytohormones, abscisic acid and jasmonic acid play a role in the regulation of Medicago sativa L. somatic embryogenesis? Plant Growth Regul. 2009, 59, 159–169. [Google Scholar] [CrossRef]
- Ahmadi, B.; Shariatpanahi, M.E.; Teixeira da Silva, J.A. Efficient induction of microspore embryogenesis using abscisic acid, jasmonic acid and salicylic acid in Brassica napus L. Plant Cell Tissue Organ. Cult. 2014, 116, 343–351. [Google Scholar] [CrossRef]
- Lukaszewicz, M.; Feuermann1, M.; Jérouville, B.; Stas, A.; Boutry, M. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 2000, 154, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.G.J.; LaFayette, P.R.; Abercrombie, L.L.; King, Z.R.; Mazarei, M.; Halter, M.C.; Poovaiah, C.R.; Baxter, H.; Shen, H.; Dixon, R.A.; et al. Gateway-compatible Vectors for High-throughput Gene Functional Analysis in Switchgrass (Panicum virgatum L.) and Other Monocot Species. Plant Biotechnol. J. 2012, 10, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Tenea, G.N.; Peres Bota, A.; Cordeiro Raposo, F.; Maquet, A. Reference genes for gene expression studies in wheat flag leaves grown under different farming conditions. BMC Res. Notes 2011, 4, 373. [Google Scholar] [CrossRef]
- Balcke, G.U.; Handrick, V.; Bergau, N.; Fichtner, M.; Henning, A.; Stellmach, H.; Tissier, A.; Hause, B.; Frolov, A. An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 2012, 8, 47. [Google Scholar] [CrossRef]
- Leonova, T.; Popova, V.; Tsarev, A.; Henning, C.; Antonova, K.; Rogovskaya, N.; Vikhnina, M.; Baldensperger, T.; Soboleva, A.; Dinastia, E.; et al. Does Protein Glycation Impact on the Drought-Related Changes in Metabolism and Nutritional Properties of Mature Pea (Pisum sativum L.) Seeds? Int. J. Mol. Sci. 2020, 21, 567. [Google Scholar] [CrossRef]



| Line | 12-OPDA | JA | JA-Ile | |||
|---|---|---|---|---|---|---|
| Intact | Wounded | Intact | Wounded | Intact | Wounded | |
| Terraploid emmer wheat | ||||||
| Runo | 13.2 ± 2.2 | 22.2 ± 3.1 | 14.7 ± 2.0 | 38.5 ± 5.5 | 6.9 ± 0.7 | 46.4 ± 2.9 |
| RD1 | 14.4 ± 2.6 | 24.1 ± 4.8 | 5.6 ± 2.1 * | 78.5 ± 14.6 * | 3.8 ± 0.7 * | 91.3 ± 12.6 * |
| RD4 | 14.3 ± 2.9 | 23.7 ± 4.2 | 14.11 ± 4.0 | 67.3 ± 17.6 | 6.2 ± 0.6 | 75.3 ± 4.7 * |
| Hexaploid bread wheat | ||||||
| Sar-60 | 12.3 ± 2.5 | 42.5 ± 20.1 | 7.1 ± 0.7 | 56.9 ± 23.4 | 5.1 ± 0.8 | 68.5 ± 25.8 |
| SD2 | 48.2 ± 20.6 | 159.6 ± 51.8 * | 4.8 ± 1.6 | 72.7 ± 10.9 | 5.0 ± 0.7 | 94.3 ± 7.1 |
| SD3 | 29.3 ± 5.7 * | 238.7 ± 56.9 * | 3.3 ± 1.0 * | 65.6 ± 9.6 | 5.6 ± 1.0 | 153.3 ± 44.0 |
| Cultivar | Gene(s) | Number of Explants | T0 Events | Transformation Efficiency (%) | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Bombarded | Produced Transgenic T0 Plants (GFP/RFP+) | Carrying Insertion of JA Biosynthesis Gene (PCR+) | Overexpression of JA Biosynthesis Gene (RT-PCR+) | Selective Genes | JA Biosynthesis Genes | |||
| Runo (emmer wheat) | AtAOS * | 576 | 10 | 10 | 6 | 1.7 | 1.7 | [12] |
| AtOPR3 * | 236 | 31 | 30 | 26 | 13.1 | 12.7 | [12] | |
| AtAOS + AtOPR3 * | 301 | 10 | 6 | 3 | 3.3 | 2.0 | this study | |
| TaAOS ** | 547 | 3 | 3 | 3 | 0.5 | 0.5 | this study | |
| Empty vector * | 653 | 85 | - | - | 13.0 | - | [35] | |
| Sar-60 (bread wheat) | AtAOS * | 717 | 8 | 7 | 4 | 1.1 | 1.0 | this study |
| AtOPR3 * | 377 | 16 | 15 | 9 | 4.2 | 4.0 | [29] | |
| AtAOS + AtOPR3 * | 358 | 3 | 3 | 3 | 0.8 | 0.8 | this study | |
| TaAOS ** | 1013 | 3 | 3 | 3 | 0.3 | 0.3 | this study | |
| Empty vector * | 420 | 11 | - | - | 2.6 | - | [29] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miroshnichenko, D.N.; Pigolev, A.V.; Pushin, A.S.; Alekseeva, V.V.; Degtyaryova, V.I.; Degtyaryov, E.A.; Pronina, I.V.; Frolov, A.; Dolgov, S.V.; Savchenko, T.V. Genetic Transformation of Triticum dicoccum and Triticum aestivum with Genes of Jasmonate Biosynthesis Pathway Affects Growth and Productivity Characteristics. Plants 2024, 13, 2781. https://doi.org/10.3390/plants13192781
Miroshnichenko DN, Pigolev AV, Pushin AS, Alekseeva VV, Degtyaryova VI, Degtyaryov EA, Pronina IV, Frolov A, Dolgov SV, Savchenko TV. Genetic Transformation of Triticum dicoccum and Triticum aestivum with Genes of Jasmonate Biosynthesis Pathway Affects Growth and Productivity Characteristics. Plants. 2024; 13(19):2781. https://doi.org/10.3390/plants13192781
Chicago/Turabian StyleMiroshnichenko, Dmitry N., Alexey V. Pigolev, Alexander S. Pushin, Valeria V. Alekseeva, Vlada I. Degtyaryova, Evgeny A. Degtyaryov, Irina V. Pronina, Andrej Frolov, Sergey V. Dolgov, and Tatyana V. Savchenko. 2024. "Genetic Transformation of Triticum dicoccum and Triticum aestivum with Genes of Jasmonate Biosynthesis Pathway Affects Growth and Productivity Characteristics" Plants 13, no. 19: 2781. https://doi.org/10.3390/plants13192781
APA StyleMiroshnichenko, D. N., Pigolev, A. V., Pushin, A. S., Alekseeva, V. V., Degtyaryova, V. I., Degtyaryov, E. A., Pronina, I. V., Frolov, A., Dolgov, S. V., & Savchenko, T. V. (2024). Genetic Transformation of Triticum dicoccum and Triticum aestivum with Genes of Jasmonate Biosynthesis Pathway Affects Growth and Productivity Characteristics. Plants, 13(19), 2781. https://doi.org/10.3390/plants13192781






