Biochemical Defence of Plants against Parasitic Nematodes
Abstract
1. Introduction
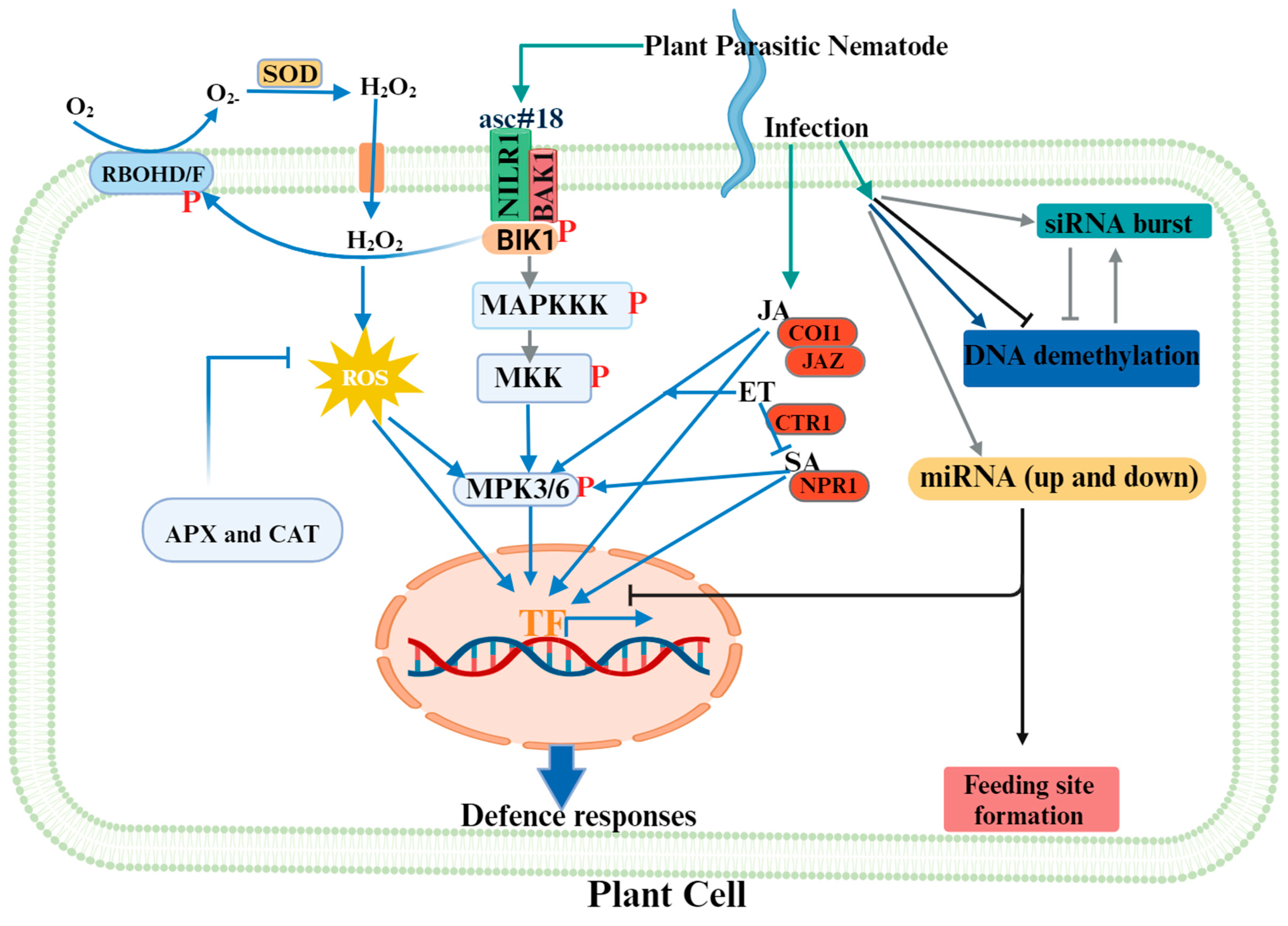
2. Nematode Perception by the Plant
2.1. Perception at the Plasma Membrane
2.2. Intracellular Transmission of the Signal
2.2.1. Receptor-Like Cytoplasmic Kinases (RLCKs)
2.2.2. ROS Production
2.2.3. Calcium Signalling
2.2.4. Mitogen-Activated Protein Kinase Activation
2.2.5. Phytohormones Mediate Plant Defence against Nematodes
2.2.6. Transcription Factors Orchestrating Plant Responses to PPN Infection
3. Epigenetics in the Plant–Nematode Interaction
3.1. DNA Methylation in the Plant–Nematode Interaction
3.2. Histone Modifications in the Plant–Nematode Interaction
3.3. ncRNAs in the Plant–Nematode Interaction
3.4. Intergenerational Acquired Resistance in the Plant–Nematode Interaction
4. Plant Cell Wall Involvement in Defending against PPNs
5. Metabolic Changes and Anti-Nematode Compounds Production
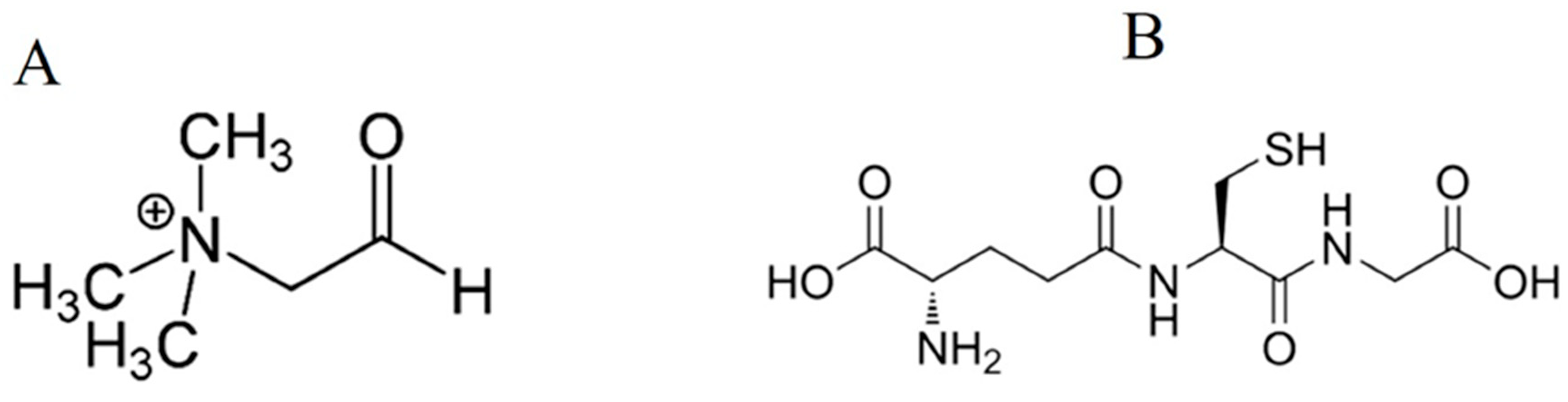
5.1. Glycine Betaine
5.2. Organosulphur Compounds
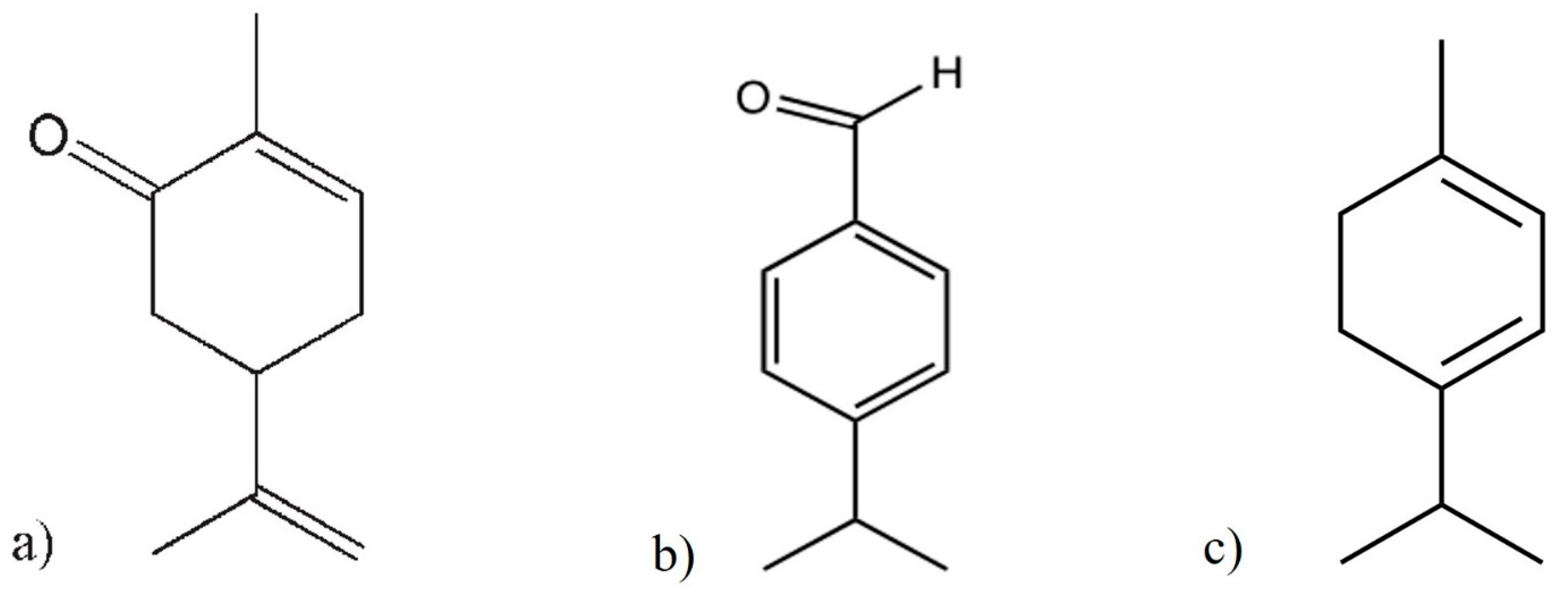
5.3. Terpenoids
5.4. Benzaldehyde
5.5. Benzoxazinoid Compounds
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Decraemer, W.; Hunt, D.J. Structure and classification. In Plant Nematology; Perry, R.N., Moens, M., Eds.; CABI: Wallingford, UK, 2006; pp. 3–32. [Google Scholar] [CrossRef]
- Khan, M.R. Nematode pests of agricultural crops, a global overview. In Novel Biological and Botechnological Applications in Pant Nematode Management; Khan, M.R., Ed.; Springer Nature: Singapore, 2023; p. 3. [Google Scholar]
- Zinoveva, S.V.; Vasyukova, N.I.; Ozeretskovskaya, O.L. Biochemical aspects of plant interactions with phytoparasitic nematodes: A review. Appl. Biochem. Microbiol. 2004, 40, 111–119. [Google Scholar] [CrossRef]
- Desmedt, W.; Mangelinckx, S.; Kyndt, T.; Vanholme, B. A phytochemical perspective on plant defense against nematodes. Front. Plant Sci. 2020, 11, 602079. [Google Scholar] [CrossRef] [PubMed]
- Sikder, M.M.; Vestergård, M. Impacts of root metabolites on soil nematodes. Front. Plant Sci. 2020, 10, 1792. [Google Scholar] [CrossRef] [PubMed]
- Siddique, S.; Coomer, A.; Baum, T.; Williamson, V.M. Recognition and response in plant-nematode interactions. Annu. Rev. Phytopathol. 2022, 60, 143–162. [Google Scholar] [CrossRef]
- Goode, K.; Mitchum, M.G. Pattern-triggered immunity against root-knot nematode infection: A minireview. Physiol. Plant. 2022, 174, e13680. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, X.C.; Cao, J.J.; Yin, L.L.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Heat shock factor HsfA1a is essential for R gene-mediated nematode resistance and triggers H2O2 production. Plant Physiol. 2018, 176, 2456–2471. [Google Scholar] [CrossRef]
- Khan, M.; Khan, A.U. Plant parasitic nematodes effectors and their crosstalk with defense response of host plants: A battle underground. Rhizosphere 2021, 17, 100288. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Al-Khayri, J.M.; Rashmi, R.; Toppo, V.; Chole, P.B.; Banadka, A.; Sudheer, W.N.; Nagella, P.; Shehata, W.F.; Al-Mssallem, M.Q.; Alessa, F.M.; et al. Plant secondary metabolites: The weapons for biotic stress management. Metabolites 2023, 13, 716. [Google Scholar] [CrossRef]
- Klessig, D.F.; Manohar, M.; Baby, S.; Koch, A.; Danquah, W.B.; Luna, E.; Park, H.J.; Kolkman, J.M.; Turgeon, B.G.; Nelson, R.; et al. Nematode ascaroside enhances resistance in a broad spectrum of plant–pathogen systems. J. Phytopathol. 2019, 167, 265–272. [Google Scholar] [CrossRef]
- Manohar, M.; Tenjo-Castano, F.; Chen, S.; Zhang, Y.K.; Kumari, A.; Williamson, V.M.; Wang, X.; Klessig, D.F.; Schroeder, F.C. Plant metabolism of nematode pheromones mediates plant-nematode interactions. Nat. Commun. 2020, 11, 208. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.; Manohar, M.; Von Reuss, S.H.; Chen, S.; Koch, A.; Kaplan, F.; Choe, A.; Micikas, R.J.; Wang, X.; Kogel, K.H.; et al. Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 2015, 6, 7795. [Google Scholar] [CrossRef] [PubMed]
- Mendy, B.; Wangombe, M.W.; Radakovic, Z.S.; Holbein, J.; Ilyas, M.; Chopra, D.; Holton, N.; Zipfel, C.; Grundler, F.M.W.; Siddique, S. Arabidopsis leucine-rich repeat receptor–like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 2017, 13, e1006284. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yuan, Y.; Lewis, C.; Kud, J.; Kuhl, J.C.; Caplan, A.; Dandurand, L.M.; Zasada, I.; Xiao, F. NILR1 perceives a nematode ascaroside triggering immune signaling and resistance. Curr. Biol. 2023, 33, 3992–3997. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Bai, Q.; Li, C.; Wang, L.; Wei, Q.; Ali, K.; Li, W.; Huang, S.; Xu, H.; Li, G.; et al. Pan-brassinosteroid signaling revealed by functional analysis of NILR1 in land plants. New Phytol. 2022, 235, 1455–1469. [Google Scholar] [CrossRef]
- Meresa, B.K.; Ayimut, K.-M.; Weldemichael, M.Y.; Gebremedhin, K.H.; Kassegn, H.H.; Gebremikael, B.A.; Egigu, E.M. Carbohydrate elicitor-induced plant immunity: Advances and prospects. Heliyon 2024, 10, e34871. [Google Scholar] [CrossRef]
- Holbein, J.; Grundler, F.M.W.; Siddique, S. Plant basal resistance to nematodes: An update. J. Exp. Bot. 2016, 67, 2049–2061. [Google Scholar] [CrossRef]
- Neuhaus, B.; Brescianii, J.; Peters, W. Ultrastructure of the pharyngeal cuticle and lectin labelling with wheat germ agglutinin-gold conjugate indicating chitin in the pharyngeal cuticle of Oesophagostornurn dentaturn. Acta Zool. 1997, 78, 205–213. [Google Scholar] [CrossRef]
- Sun, S.; Witte, H.; Sommer, R.J. Chitin contributes to the formation of a feeding structure in a predatory nematode. Curr. Biol. 2023, 33, 15–27. [Google Scholar] [CrossRef]
- Woodruff, G.C. Developmental genetics: The structural basis of malleable teeth. Curr. Biol. 2023, 33, R106–R108. [Google Scholar] [CrossRef]
- Macharia, T.N.; Bellieny-Rabelo, D.; Moleleki, L.N. Transcriptome profiling of potato (Solanum tuberosum L.) responses to root-knot nematode (Meloidogyne javanica) infestation during a compatible interaction. Microorganisms 2020, 8, 1443. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.L.d.S.; de Jesus, J.M.I.; Oliveira, M.I.d.S.; Côrtes, M.V.d.C.B.; de Filippi, M.C.C.; da Rocha, M.R. Biochemical response of resistant and susceptible Capsicum spp. to Meloidogyne enterolobii. J. Phytopathol. 2023, 171, 430–441. [Google Scholar] [CrossRef]
- Channale, S.; Kalavikatte, D.; Thompson, J.P.; Kudapa, H.; Bajaj, P.; Varshney, R.K.; Zwart, R.S.; Thudi, M. Transcriptome analysis reveals key genes associated with root-lesion nematode Pratylenchus thornei resistance in chickpea. Sci. Rep. 2021, 11, 17491. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.B.M.; Jayaswal, P.; Chandra, S.; Jayanthi, M.; Mandal, P.K. Comparative transcriptome profiling of Polianthes tuberosa during a compatible interaction with root-knot nematode Meloidogyne incognita. Mol. Biol. Rep. 2022, 49, 4503–4516. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, Y.; Yang, Y.; Yang, X.; Xue, W.; Wu, M.; Chen, P.; Weng, Y.; Chen, S. Transcriptomic and histological analysis of the response of susceptible and resistant cucumber to Meloidogyne incognita infection revealing complex resistance via multiple signaling pathways. Front. Plant Sci. 2021, 12, 675429. [Google Scholar] [CrossRef]
- Zhang, H.; Kjemtrup-Lovelace, S.; Li, C.; Luo, Y.; Chen, L.P.; Song, B.H. Comparative RNA-seq analysis uncovers a complex regulatory network for soybean cyst nematode resistance in wild soybean (Glycine soja). Sci. Rep. 2017, 7, 9699. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, Q.; Guo, X.; Tan, Y.; Deng, M.; Zhang, L. Mitogen-activated protein kinases MPK3 and MPK6 phosphorylate receptor-like cytoplasmic kinase CDL1 to regulate soybean basal immunity. Plant Cell 2024, 36, 963–986. [Google Scholar] [CrossRef]
- Zhou, D.; Godinez-Vidal, D.; He, J.; Teixeira, M.; Guo, J.; Wei, L.; Van Norman, J.M.; Kaloshian, I. A G-type lectin receptor kinase negatively regulates Arabidopsis immunity against root-knot nematodes. Plant Physiol. 2023, 193, 721–735. [Google Scholar] [CrossRef]
- Kyndt, T.; Zemene, H.Y.; Haeck, A.; Singh, R.; De Vleesschauwer, D.; Denil, S.; De Meyer, T.; Höfte, M.; Demeestere, K.; Gheysen, G. Below-ground attack by the root knot nematode Meloidogyne graminicola predisposes rice to blast disease. Mol. Plant-Microbe Interact. 2017, 30, 255–266. [Google Scholar] [CrossRef]
- Teixeira, M.A.; Wei, L.; Kaloshian, I. Root-knot nematodes induce pattern-triggered immunity in Arabidopsis thaliana roots. New Phytol. 2016, 211, 276–287. [Google Scholar] [CrossRef]
- Peng, H.C.; Kaloshian, I. The tomato leucine-rich repeat receptor-like kinases SlSERK3A and SlSERK3B have overlapping functions in bacterial and nematode innate immunity. PLoS ONE 2014, 9, e93302. [Google Scholar] [CrossRef] [PubMed]
- Klink, V.P.; Darwish, O.; Alkharouf, N.W.; Lawrence, K.S. The impact of PRAP vectors on plant genetic transformation and pathogenesis studies including an analysis of BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1)-mediated resistance. J. Plant Interact. 2021, 16, 270–283. [Google Scholar] [CrossRef]
- Lee, I.H.; Shim, D.; Jeong, J.C.; Sung, Y.W.; Nam, K.J.; Yang, J.W.; Ha, J.; Lee, J.J.; Kim, Y.H. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-resistant and susceptible sweetpotato cultivars. Planta 2019, 249, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.S.; Siddique, S.; Lin, C.J.; Marhavy, P.; Kyndt, T. Redox signalling in plant–nematode interactions: Insights into molecular crosstalk and defense mechanisms. Plant Cell Environ. 2024, 47, 2811–2820. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhou, J. Receptor-like cytoplasmic kinases: Central players in plant receptor kinase–mediated signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Li, L.; Yu, Y.; Zhou, Z.; Zhou, J. Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 2016, 59, 878–888. [Google Scholar] [CrossRef]
- Pant, S.R.; Matsye, P.D.; McNeece, B.T.; Sharma, K.; Krishnavajhala, A.; Lawrence, G.W.; Klink, V.P. Syntaxin 31 functions in Glycine max resistance to the plant parasitic nematode Heterodera glycines. Plant Mol. Biol. 2014, 85, 107–121. [Google Scholar] [CrossRef]
- McNeece, B.T.; Sharma, K.; Lawrence, G.W.; Lawrence, K.S.; Klink, V.P. The mitogen activated protein kinase (MAPK) gene family functions as a cohort during the Glycine max defense response to Heterodera glycines. Plant Physiol. Biochem. 2019, 137, 25–41. [Google Scholar] [CrossRef]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.K.; Sharma, A.; Thakur, J. How do plants defend themselves against pathogens-biochemical mechanisms and genetic interventions? Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef]
- Du, C.; Shen, F.; Li, Y.; Zhao, Z.; Xu, X.; Jiang, J.; Li, J. Effects of salicylic acid, jasmonic acid and reactive oxygen species on the resistance of Solanum peruvianum to Meloidogyne incognita. Sci. Hortic. 2021, 275, 109649. [Google Scholar] [CrossRef]
- Khajuria, A.; Ohri, P. Polyamines induced nematode stress tolerance in Solanum lycopersicum through altered physico-chemical attributes. Physiol. Mol. Plant Pathol. 2020, 112, 101544. [Google Scholar] [CrossRef]
- Noureldeen, A.; Asif, M.; Ansari, T.; Khan, F.; Shariq, M.; Ahmad, F.; Mfarrej, M.F.B.; Khan, A.; Tariq, M.; Siddiqui, M.A.; et al. Effect of individual, simultaneous and sequential inoculation of Pseudomonas fluorescens and Meloidogyne incognita on growth, biochemical, enzymatic and nonenzymatic antioxidants of tomato (Solanum lycopersicum L.). Plants 2021, 10, 1145. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Deepti, S.; Saksham, C.; Kumar, D.; Sharma, R.; Kumar, S. GWAS scans of cereal cyst nematode (Heterodera avenae) resistance in Indian wheat germplasm. Mol. Genet. Genom. 2023, 298, 579–601. [Google Scholar] [CrossRef]
- Zechmann, B. Subcellular roles of glutathione in mediating plant defense during biotic stress. Plants 2020, 9, 1067. [Google Scholar] [CrossRef] [PubMed]
- Al-Zahrani, H.S.; Nahar, K.; Alharby, H.F.; Alsamadany, H.; Hakeem, K.R.; Hasanuzzaman, M. Zinc supplementation enhances glutathione-mediated antioxidant defense and glyoxalase systems to conferring salt tolerance in soybean (Glycine max L.). Agronomy 2022, 12, 1032. [Google Scholar] [CrossRef]
- Chen, X.; Li, S.; Zhao, X.; Zhu, X.; Wang, Y. Modulation of (homo)glutathione metabolism and H2O2 accumulation during soybean cyst nematode infections in susceptible and resistant soybean cultivars. Int. J. Mol. Sci. 2020, 21, 388. [Google Scholar] [CrossRef]
- Sikandar, A.; Wu, F.; He, H.; Ullah, R.M.K.; Wu, H. Growth, physiological, and biochemical variations in tomatoes after infection with different density levels of Meloidogyne enterolobii. Plants 2024, 13, 293. [Google Scholar] [CrossRef]
- Bali, S.; Kaur, P.; Sharma, A.; Ohri, P.; Bhardwaj, R.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Jasmonic acid-induced tolerance to root-knot nematodes in tomato plants through altered photosynthetic and antioxidative defense mechanisms. Protoplasma 2018, 255, 471–484. [Google Scholar] [CrossRef]
- Ali, M.; Ohri, P. Deciphering the synergistic effect of jasmonic acid and spermine in mitigating root-knot nematode stress in tomato plants through enhancing growth and activity of antioxidant enzymes. S. Afr. J. Bot. 2023, 161, 21–35. [Google Scholar] [CrossRef]
- Lee, I.H.; Kim, H.S.; Nam, K.J.; Lee, K.L.; Yang, J.W.; Kwak, S.S.; Lee, J.J.; Shim, D.; Kim, Y.H. The defense response involved in sweetpotato resistance to root-knot nematode Meloidogyne incognita: Comparison of root transcriptomes of resistant and susceptible sweetpotato cultivars with respect to induced and constitutive defense responses. Front. Plant Sci. 2021, 12, 671677. [Google Scholar] [CrossRef]
- Sato, K.; Kadota, Y.; Shirasu, K. Plant immune responses to parasitic nematodes. Front. Plant Sci. 2019, 10, 1165. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S.; Leonetti, P. Inhibition of ROS-scavenging enzyme system is a key event in tomato genetic resistance against root-knot nematodes. Int. J. Mol. Sci. 2023, 24, 7324. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.W.; Park, S.U.; Lee, H.U.; Nam, K.J.; Lee, K.L.; Lee, J.J.; Kim, J.H.; Kwak, S.S.; Kim, H.S.; Kim, Y.H. Differential responses of antioxidant enzymes and lignin metabolism in susceptible and resistant sweetpotato cultivars during root-knot nematode infection. Antioxidants 2023, 12, 1164. [Google Scholar] [CrossRef]
- Hawamda, A.I.M.; Zahoor, A.; Abbas, A.; Ali, M.A.; Bohlmann, H. The Arabidopsis RBOHB encoded by AT1G09090 is important for resistance against nematodes. Int. J. Mol. Sci. 2020, 21, 5556. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Shim, D.; Lee, K.; Nam, K.J.; Yang, J.; Lee, J.J.; Kim, Y. Expression analysis of sweetpotato NADPH oxidase-encoding RBOH genes in response to infection with the root-knot nematode Meloidogyne incognita. Plant Biotechnol. Rep. 2020, 14, 635–642. [Google Scholar] [CrossRef]
- Chopra, D.; Hasan, M.S.; Matera, C.; Chitambo, O.; Mendy, B.; Mahlitz, S.-V.; Naz, A.A.; Szumski, S.; Janakowski, S.; Sobczak, M.; et al. Plant parasitic cyst nematodes redirect host indole metabolism via NADPH oxidase-mediated ROS to promote infection. New Phytol. 2021, 232, 318–331. [Google Scholar] [CrossRef]
- Song, L.-X.; Xu, X.-C.; Wang, F.-N.; Wang, Y.; Xia, X.-J.; Shi, K.; Zhou, Y.-H.; Zhou, J. Brassinosteroids act as a positive regulator for resistance against root-knot nematode involving RESPIRATORY BURST OXIDASEHOMOLOG-dependent activation of MAPKs in tomato. Plant Cell Environ. 2018, 41, 1113–1125. [Google Scholar] [CrossRef]
- Chavan, S.N.; De Kesel, J.; Desmedt, W.; Degroote, E.; Singh, R.R.; Nguyen, G.T.; Demeestere, K.; De Meyer, T.; Kyndt, T. Dehydroascorbate induces plant resistance in rice against root-knot nematode Meloidogyne graminicola. Mol. Plant Pathol. 2022, 23, 1303–1319. [Google Scholar] [CrossRef]
- Han, S.; Smith, J.M.; Du, Y.; Bent, A.F. Soybean transporter AATRhg1 abundance increases along the nematode migration path and impacts vesiculation and ROS. Plant Physiol. 2023, 192, 133–153. [Google Scholar] [CrossRef]
- Cook, D.E.; Lee, T.G.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.M.; Wang, J.; Hughes, T.J.; Willis, D.K.; Clemente, T.E.; et al. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, F.; Bao, A.; You, Q.; Li, Z.; Chen, J.; Cheng, Y.; Zhao, W.; Shen, X.; Zhou, X.; et al. The soybean Rhg1 amino acid transporter gene alters glutamate homeostasis and jasmonic acid-induced resistance to soybean cyst nematode. Mol. Plant Pathol. 2019, 20, 270–286. [Google Scholar] [CrossRef]
- Wang, C.; Luan, S. Calcium homeostasis and signaling in plant immunity. Curr. Opin. Plant Biol. 2024, 77, 102485. [Google Scholar] [CrossRef]
- Davies, L.J.; Brown, C.R.; Elling, A.A. Calcium is involved in the RMc1(Blb)-mediated hypersensitive response against Meloidogyne chitwoodi in potato. Plant Cell Rep. 2015, 34, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Wang, L.; Qin, Y.; Li, P. Activity of chitin/chitosan/chitosan oligosaccharide against plant pathogenic nematodes and potential modes of application in agriculture: A review. Carbohydr. Polym. 2023, 306, 120592. [Google Scholar] [CrossRef] [PubMed]
- Ghaemi, R.; Pourjam, E.; Safaie, N.; Verstraeten, B.; Mahmoudi, S.B.; Mehrabi, R.; De Meyer, T.; Kyndt, T. Molecular insights into the compatible and incompatible interactions between sugar beet and the beet cyst nematode. BMC Plant Biol. 2020, 20, 483. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, J.; Wei, Y.; Su, W.; Li, W.; Wang, B.; Peng, D.; Gheysen, G.; Peng, H.; Dai, L. The nematode effector calreticulin competes with the high mobility group protein OsHMGB1 for binding to the rice calmodulin-like protein OsCML31 to enhance rice susceptibility to Meloidogyne graminicola. Plant Cell Environ. 2024, 47, 732–1746. [Google Scholar] [CrossRef] [PubMed]
- Sidonskaya, E.; Schweighofer, A.; Shubchynskyy, V.; Kammerhofer, N.; Hofmann, J.; Wieczorek, K.; Meskiene, I. Plant resistance against the parasitic nematode Heterodera schachtii is mediated by MPK3 and MPK6 kinases, which are controlled by the MAPK phosphatase AP2C1 in Arabidopsis. J. Exp. Bot. 2016, 67, 107–118. [Google Scholar] [CrossRef]
- Niraula, P.M.; Sharma, K.; McNeece, B.T.; Troell, H.A.; Darwish, O.; Alkharouf, N.W.; Lawrence, K.S.; Klink, V.P. Mitogen activated protein kinase (MAPK) regulated genes with predicted signal peptides function in the Glycine max defense response to the root pathogenic nematode Heterodera glycines. PLoS ONE 2020, 15, e0241678. [Google Scholar] [CrossRef]
- Khatri, R.; Pant, S.R.; Sharma, K.; Niraula, P.M.; Lawaju, B.R.; Lawrence, K.S.; Alkharouf, N.W.; Klink, V.P. Glycine max homologs of DOESN’T MAKE INFECTIONS 1, 2, and 3 function to impair Heterodera glycines parasitism while also regulating mitogen activated protein kinase expression. Front. Plant Sci. 2022, 13, 842597. [Google Scholar] [CrossRef]
- Klink, V.P.; Sharma, K.; Pant, S.R.; McNeece, B.; Niraula, P.; Lawrence, G.W. Components of the SNARE-containing regulon are co-regulated in root cells undergoing defense. Plant Signal. Behav. 2017, 12, e1274481. [Google Scholar] [CrossRef]
- Klink, V.P.; Alkharouf, N.W.; Lawrence, K.S.; Lawaju, B.R.; Sharma, K.; Niraula, P.M.; McNeece, B.T. The heterologous expression of conserved Glycine max (soybean) mitogen activated protein kinase 3 (MAPK3) paralogs suppresses Meloidogyne incognita parasitism in Gossypium hirsutum (upland cotton). Transgenic Res. 2022, 31, 457–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, C.; Li, Q.; Zhang, K.; Lou, Q.; Li, J.; Chen, J. Multi-omics analysis revealed that MAPK signaling and flavonoid metabolic pathway contributed to resistance against Meloidogyne incognita in the introgression line cucumber. J. Proteom. 2020, 220, 103675. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed]
- Khanam, S.; Bauters, L.; Singh, R.R.; Verbeek, R.; Haeck, A.; Sultan, S.M.D.; Demeestere, K.; Kyndt, T.; Gheysen, G. Mechanisms of resistance in the rice cultivar Manikpukha to the rice stem nematode Ditylenchus angustus. Mol. Plant Pathol. 2018, 9, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Yang, F.; Xu, X.; Peng, Y.; Ji, H. Salicylic acid, jasmonate, and ethylene contribute to rice defense against white tip nematodes Aphelenchoides besseyi. Front. Plant Sci. 2022, 12, 755802. [Google Scholar] [CrossRef]
- Asadi-Sardari, A.; Mahdikhani-Moghadam, E.; Zaki-Aghl, M.; Vetukuri, R.R. Constitutive and inducible expression of genes related to salicylic acid and ethylene pathways in a moderately resistant tomato cultivar leads to delayed development of Meloidogyne javanica. Agriculture 2022, 12, 2122. [Google Scholar] [CrossRef]
- Qiao, S.; Ma, J.; Wang, Y.; Chen, J.; Kang, Z.; Bian, Q.; Chen, J.; Yin, Y.; Cao, G.; Zhao, G.; et al. Integrated transcriptome and metabolome analyses reveal details of the molecular regulation of resistance to stem nematode in sweet potato. Plants 2023, 12, 2052. [Google Scholar] [CrossRef]
- Uehara, T.; Sugiyama, S.; Matsuura, H.; Arie, T.; Masuta, C. Resistant and susceptible responses in tomato to cyst nematode are differentially regulated by salicylic acid. Plant Cell Physiol. 2010, 51, 1524–1536. [Google Scholar] [CrossRef]
- Lin, J.; Mazarei, M.; Zhao, N.; Zhu, J.J.; Zhuang, X.; Liu, W.; Pantalone, V.R.; Arelli, P.R.; Stewart, C.N.; Chen, F. Overexpression of a soybean salicylic acid methyltransferase gene confers resistance to soybean cyst nematode. Plant Biotechnol. J. 2013, 11, 1135–1145. [Google Scholar] [CrossRef]
- Sikder, M.M.; Vestergård, M.; Kyndt, T.; Kudjordjie, E.N.; Nicolaisen, M. Phytohormones selectively affect plant parasitic nematodes associated with Arabidopsis roots. New Phytol. 2021, 232, 1272–1285. [Google Scholar] [CrossRef]
- Wubben, M.J.E.; Jin, J.; Baum, T.J. Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol. Plant-Microbe Interact. 2008, 21, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Fu, Y.; Zhang, P.; You, C.; Li, C.; Peng, H. Transcriptome analysis of resistant and susceptible mulberry responses to Meloidogyne enterolobii infection. BMC Plant Biol. 2021, 21, 338. [Google Scholar] [CrossRef]
- Gheysen, G.; Mitchum, M.G. Phytoparasitic nematode control of plant hormone pathways. Plant Physiol. 2019, 179, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- López-Villamor, A.; Nunes Da Silva, M.; Vasconcelos, M.W. Evaluation of plant elicitation with methyl-jasmonate, salicylic acid and benzo (1,2,3)-thiadiazole-7-carbothioic acid-s-methyl ester for the sustainable management of the pine wilt disease. Tree Physiol. 2022, 42, 2596–2613. [Google Scholar] [CrossRef] [PubMed]
- Klink, V.P.; Hosseini, P.; Matsye, P.D.; Alkharouf, N.W.; Matthews, B.F. Syncytium gene expression in Glycine max[PI88788] roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines. Plant Physiol. Biochem. 2010, 48, 176–193. [Google Scholar] [CrossRef]
- Priya, D.B.; Somasekhar, N.; Prasad, J.S.; Kirti, P.B. Transgenic tobacco plants constitutively expressing Arabidopsis NPR1 show enhanced resistance to root-knot nematode, Meloidogyne incognita. BMC Res. Notes 2011, 4, 231. [Google Scholar] [CrossRef]
- Ojeda-Rivera, J.O.; Ulloa, M.; Roberts, P.A.; Kottapalli, P.; Wang, C.; Nájera-González, H.R.; Payton, P.; Lopez-Arredondo, D.; Herrera-Estrella, L. Root-knot nematode resistance in Gossypium hirsutum determined by a constitutive defense-response transcriptional program avoiding a fitness penalty. Front. Plant Sci. 2022, 13, 858313. [Google Scholar] [CrossRef]
- Aerts, N.; Mendes, M.P.; Van Wees, S.C.M. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef]
- Verbeek, R.E.M.; Van Buyten, E.; Alam, M.Z.; De Vleesschauwer, D.; Van Bockhaven, J.; Asano, T.; Kikuchi, S.; Haeck, A.; Demeestere, K.; Gheysen, G.; et al. Jasmonate-induced defense mechanisms in the belowground antagonistic interaction between Pythium arrhenomanes and Meloidogyne graminicola in rice. Front. Plant Sci. 2019, 10, 1515. [Google Scholar] [CrossRef]
- Yang, T.; Jin, W.; Zou, J.; Chen, X.; Zhao, Q.; Yu, J. NBR1a mediates root-knot nematode resistance by modulating antioxidant system, jasmonic acid and selective autophagy in Solanum lycopersicum. Plant Stress 2024, 11, 100390. [Google Scholar] [CrossRef]
- Wiśniewska, A.; Wojszko, K.; Różańska, E.; Lenarczyk, K.; Sobczak, M. Arabidopsis thaliana AtHRS1 gene is involved in the response to Heterodera schachtii infection and its overexpression hampers development of syncytia and involves a jasmonic acid-dependent mechanism. J. Plant Physiol. 2022, 272, 153680. [Google Scholar] [CrossRef] [PubMed]
- Qiao, F.; Kong, L.A.; Peng, H.; Huang, W.K.; Wu, D.Q.; Liu, S.M.; Clarke, J.L.; Qiu, D.W.; Peng, D.L. Transcriptional profiling of wheat (Triticum aestivum L.) during a compatible interaction with the cereal cyst nematode Heterodera avenae. Sci. Rep. 2019, 9, 2184. [Google Scholar] [CrossRef] [PubMed]
- Bali, S.; Kaur, P.; Jamwal, V.L.; Gandhi, S.G.; Sharma, A.; Ohri, P.; Bhardwaj, R.; Ali, M.A.; Ahmad, P. Seed priming with jasmonic acid counteracts root knot nematode infection in tomato by modulating the activity and expression of antioxidative enzymes. Biomolecules 2020, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; You, J.; Li, C.; Williamson, V.M.; Wang, C. Ethylene response pathway modulates attractiveness of plant roots to soybean cyst nematode Heterodera glycines. Sci. Rep. 2017, 7, 41282. [Google Scholar] [CrossRef] [PubMed]
- Piya, S.; Binder, B.M.; Hewezi, T. Canonical and noncanonical ethylene signaling pathways that regulate Arabidopsis susceptibility to the cyst nematode Heterodera schachtii. New Phytol. 2019, 221, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Mantelin, S.; Bhattarai, K.K.; Jhaveri, T.Z.; Kaloshian, I. Mi-1-mediated resistance to Meloidogyne incognita in tomato may not rely on ethylene but hormone perception through ETR3 participates in limiting nematode infection in a susceptible host. PLoS ONE 2013, 8, e63281. [Google Scholar] [CrossRef]
- Fudali, S.L.; Wang, C.; Williamson, V.M. Ethylene signaling pathway modulates attractiveness of host roots to the root-knot nematode Meloidogyne hapla. Mol. Plant-Microbe Interact. 2013, 26, 75–86. [Google Scholar] [CrossRef]
- Singh, R.R.; Verstraeten, B.; Siddique, S.; Tegene, A.M.; Tenhaken, R.; Frei, M.; Haeck, A.; Demeestere, K.; Pokhare, S.; Gheysen, G.; et al. Ascorbate oxidation activates systemic defence against root-knot nematode Meloidogyne graminicola in rice. J. Exp. Bot. 2020, 71, 4271–4284. [Google Scholar] [CrossRef]
- Singh, R.R.; Nobleza, N.; Demeestere, K.; Kyndt, T. Ascorbate oxidase induces systemic resistance in sugar beet against cyst nematode Heterodera schachtii. Front. Plant Sci. 2020, 11, 591715. [Google Scholar] [CrossRef]
- Fujimoto, T.; Abe, H.; Mizukubo, T.; Seo, S. Phytol, a constituent of chlorophyll, induces root-knot nematode resistance in Arabidopsis via the ethylene signaling pathway. Mol. Plant-Microbe Interact. 2021, 34, 279–285. [Google Scholar] [CrossRef]
- De Kesel, J.; Bonneure, E.; Frei, M.; De Meyer, T.; Mangelinckx, S.; Kyndt, T. Diproline-induced resistance to parasitic nematodes in the same and subsequent rice generations: Roles of iron, nitric oxide and ethylene. Front. Plant Sci. 2023, 14, 1112007. [Google Scholar] [CrossRef] [PubMed]
- Arraes, F.B.M.; Vasquez, D.D.N.; Tahir, M.; Pinheiro, D.H.; Faheem, M.; Freitas-Alves, N.S.; Moreira-Pinto, C.E.; Moreira, V.J.V.; Paes-de-Melo, B.; Lisei-de-Sa, M.E.; et al. Integrated omic approaches reveal molecular mechanisms of tolerance during soybean and Meloidogyne incognita interactions. Plants 2022, 11, 2744. [Google Scholar] [CrossRef] [PubMed]
- Petitot, A.S.; Kyndt, T.; Haidar, R.; Dereeper, A.; Collin, M.; De Almeida Engler, J.; Gheysen, G.; Fernandez, D. Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Ann. Bot. 2017, 119, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Yamada, M.; Higaki, T.; Aida, M.; Kubo, M.; Tsai, A.Y.L.; Sawa, S. PUCHI regulates giant cell morphology during root-knot nematode infection in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 755610. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.; Melo, B.P.; Lourenço-Tessutti, I.T.; Ballesteros, H.F.; Ribeiro, K.V.G.; Menuet, K.; Heyman, J.; Hemerly, A.; Sá, M.F.G.; Veylder, L.; et al. The regeneration conferring transcription factor complex ERF115-PAT1 coordinates a wound-induced response in root-knot nematode induced galls. New Phytol. 2024, 241, 878–895. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, S.; Saeki, K.; Toda, K.; Ishida, T.; Sawa, S. The Atypical E2F transcription factor DEL1 modulates growth–defense tradeoffs of host plants during root-knot nematode infection. Sci. Rep. 2020, 10, 8836. [Google Scholar] [CrossRef]
- Kumar, A.; Sichov, N.; Bucki, P.; Miyara, S.B. SlWRKY16 and SlWRKY31 of tomato, negative regulators of plant defense, involved in susceptibility activation following root-knot nematode Meloidogyne javanica infection. Sci. Rep. 2023, 13, 14592. [Google Scholar] [CrossRef]
- Chinnapandi, B.; Bucki, P.; Fitoussi, N.; Kolomiets, M.; Borrego, E.; Braun Miyara, S. Tomato SlWRKY3 acts as a positive regulator for resistance against the root-knot Nematode Meloidogyne javanica by activating lipids and hormone-mediated defense-signaling pathways. Plant Signal. Behav. 2019, 14, 1601951. [Google Scholar] [CrossRef]
- Nie, W.; Liu, L.; Chen, Y.; Luo, M.; Feng, C.; Wang, C.; Yang, Z.; Du, C. Identification of the regulatory role of SlWRKYs in tomato defense against Meloidogyne incognita. Plants 2023, 12, 2416. [Google Scholar] [CrossRef]
- Willig, J.-J.; Guarneri, N.; van Loon, T.; Wahyuni, S.; Astudillo-Estévez, I.E.; Xu, L.; Willemsen, V.; Goverse, A.; Sterken, M.G.; Lozano-Torres, J.L.; et al. Transcription factor WOX11 modulates tolerance to cyst nematodes via adventitious lateral root formation. Plant Physiol. 2024, 195, 799–811. [Google Scholar] [CrossRef]
- Willig, J.; Guarneri, N.; Steenbrugge, J.J.M.; Jong, W.; Chen, J.; Goverse, A.; Torres, L.L.; Sterken, M.G.; Bakker, J.; Smant, G. The Arabidopsis transcription factor TCP9 modulates root architectural plasticity, reactive oxygen species-mediated processes, and tolerance to cyst nematode infections. Plant J. 2022, 112, 1070–1083. [Google Scholar] [CrossRef] [PubMed]
- Pascual, S.; Emiliozzi, M.; Nombela, G. Role of two transcription factors (TGA 1a and TGA 2.1) in the Mi-1-mediated resistance of tomato to the root-knot nematode Meloidogyne javanica. Horticulturae 2024, 10, 134. [Google Scholar] [CrossRef]
- Hamamouch, N.; Winkel, B.S.J.; Li, C.; Davis, E.L. Modulation of Arabidopsis flavonol biosynthesis genes by cyst and root-knot nematodes. Plants 2020, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Abbas, A.; Kreil, D.P.; Bohlmann, H. Overexpression of the transcription factor RAP2.6 leads to enhanced callose deposition in syncytia and enhanced resistance against the beet cyst nematode Heterodera schachtii in Arabidopsis roots. BMC Plant Biol. 2013, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Akagi, A.; Fukushima, S.; Okada, K.; Jiang, C.J.; Yoshida, R.; Nakayama, A.; Shimono, M.; Sugano, S.; Yamane, H.; Takatsuji, H. WRKY45-dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol. Biol. 2014, 86, 171–183. [Google Scholar] [CrossRef]
- Yamamura, C.; Mizutani, E.; Okada, K.; Nakagawa, H.; Fukushima, S.; Tanaka, A.; Maeda, S.; Kamakura, T.; Yamane, H.; Takatsuji, H.; et al. Diterpenoid phytoalexin factor, a BHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 2015, 84, 1100–1113. [Google Scholar] [CrossRef]
- Desmedt, W.; Kudjordjie, E.N.; Chavan, S.N.; Zhang, J.; Li, R.; Yang, B.; Nicolaisen, M.; Mori, M.; Peters, R.J.; Vanholme, B.; et al. Rice diterpenoid phytoalexins are involved in defence against parasitic nematodes and shape rhizosphere nematode communities. New Phytol. 2022, 235, 1231–1245. [Google Scholar] [CrossRef]
- Desmedt, W.; Kudjordjie, E.N.; Chavan, S.N.; Desmet, S.; Nicolaisen, M.; Vanholme, B.; Vestergård, M.; Kyndt, T. Distinct chemical resistance-inducing stimuli result in common transcriptional, metabolic, and nematode community signatures in rice root and rhizosphere. J. Exp. Bot. 2022, 73, 7564–7581. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Escobar, C.; Brown, S.; Mitchum, M.G. Transcriptomic and proteomic analysis of the plant response to nematode infection. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Jones, J., Gheysen, G., Fenoll, C., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 157–174. [Google Scholar]
- Hendrich, B.; Tweedie, S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003, 19, 269–277. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Ashapkin, V.V. DNA methylation in higher plants: Past, present and future. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 360–368. [Google Scholar] [CrossRef]
- Weinhold, A.; Kallenbach, M.; Baldwin, I.T. Progressive 35S promoter methylation increases rapidly during vegetative development in transgenic Nicotiana attenuata plants. BMC Plant Biol. 2013, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- Bewick, A.J.; Schmitz, R.J. Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 2017, 36, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Briffa, A.; Hollwey, E.; Shahzad, Z.; Moore, J.D.; Lyons, D.B.; Howard, M.; Zilberman, D. Millennia-long epigenetic fluctuations generate intragenic DNA methylation variance in Arabidopsis populations. Cell Syst. 2023, 14, 953–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jang, H.; Luo, Z.; Dong, Y.; Xu, Y.; Kantamneni, Y.; Schmitz, R.J. Dynamic evolution of the heterochromatin sensing histone demethylase IBM1. PLoS Genet. 2024, 20, e1011358. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Luger, K.; Mä Der, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Fransz, P.; De Jong, H. From nucleosome to chromosome: A dynamic organization of genetic information. Plant J. 2011, 66, 4–17. [Google Scholar] [CrossRef]
- Nelissen, H.; Boccardi, T.M.; Himanen, K.; Van Lijsebettens, M. Impact of core histone modifications on transcriptional regulation and plant growth. CRC Crit. Rev. Plant Sci. 2007, 26, 243–263. [Google Scholar] [CrossRef]
- Scheid, R.; Chen, J.; Zhong, X. Biological role and mechanism of chromatin readers in plants. Curr. Opin. Plant Biol. 2021, 61, 102008. [Google Scholar] [CrossRef]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.T.; Liu, J.X.; Han, J.J. Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 2021, 63, 438–450. [Google Scholar] [CrossRef]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Kumar, A.; Kaur, H.; Kaur, N. MiRNA: The taskmaster of plant world. Biologia 2021, 76, 1551–1567. [Google Scholar] [CrossRef]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xia, J.; Lii, Y.E.; Barrera-Figueroa, B.E.; Zhou, X.; Gao, S.; Lu, L.; Niu, D.; Chen, Z.; Leung, C.; et al. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012, 13, R20. [Google Scholar] [CrossRef]
- Lisch, D.; Bennetzen, J.L. Transposable element origins of epigenetic gene regulation. Curr. Opin. Plant Biol. 2011, 14, 156–161. [Google Scholar] [CrossRef]
- Datta, R.; Paul, S. Long non-coding RNAs: Fine-tuning the developmental responses in plants. J. Biosci. 2019, 44, 77. [Google Scholar] [CrossRef]
- Pavet, V.; Quintero, C.; Cecchini, N.M.; Rosa, A.L.; Alvarez, M.E. Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol. Plant-Microbe Interact. 2006, 19, 577–587. [Google Scholar] [CrossRef] [PubMed]
- López Sánchez, A.; Stassen, J.H.M.; Furci, L.; Smith, L.M.; Ton, J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, K.; Katakami, H.; Kim, H.J.; Ogawa, E.; Sano, C.M.; Wada, Y.; Sano, H. Epigenetic inheritance in rice plants. Ann. Bot. 2007, 100, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Lane, T.; Piya, S.; Rambani, A.; Rice, J.H.; Staton, M. Cyst nematode parasitism induces dynamic changes in the root epigenome. Plant Physiol. 2017, 174, 405–420. [Google Scholar] [CrossRef]
- Rambani, A.; Rice, J.H.; Liu, J.; Lane, T.; Ranjan, P.; Mazarei, M.; Pantalone, V.; Stewart, C.N.; Staton, M.; Hewezi, T. The methylome of soybean roots during the compatible interaction with the soybean cyst nematode. Plant Physiol. 2015, 168, 1364–1377. [Google Scholar] [CrossRef]
- Atighi, M.R.; Verstraeten, B.; De Meyer, T.; Kyndt, T. Genome-wide DNA hypomethylation shapes nematode pattern-triggered immunity in plants. New Phytol. 2020, 227, 545–558. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Cabrera, J.; Martinez-Argudo, I.; Artaza, H.; Fenoll, C.; Escobar, C. Silenced retrotransposons are major rasiRNAs targets in Arabidopsis galls induced by Meloidogyne javanica. Mol. Plant Pathol. 2018, 19, 2431–2445. [Google Scholar] [CrossRef]
- Leonetti, P.; Molinari, S. Epigenetic and metabolic changes in root-knot nematode-plant interactions. Int. J. Mol. Sci. 2020, 21, 7759. [Google Scholar] [CrossRef]
- Ji, H.; Gheysen, G.; Denil, S.; Lindsey, K.; Topping, J.F.; Nahar, K.; Haegeman, A.; De Vos, W.H.; Trooskens, G.; Van Criekinge, W.; et al. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J. Exp. Bot. 2013, 64, 3885–3898. [Google Scholar] [CrossRef]
- Yan, L.; Fan, G.; Li, X. Genome-wide analysis of three histone marks and gene expression in Paulownia fortunei with phytoplasma infection. BMC Genom. 2019, 20, 234. [Google Scholar] [CrossRef]
- López, A.; Ramírez, V.; García-Andrade, J.; Flors, V.; Vera, P. The RNA silencing enzyme RNA polymerase V is required for plant immunity. PLoS Genet. 2011, 7, e1002434. [Google Scholar] [CrossRef] [PubMed]
- Ayyappan, V.; Kalavacharla, V.; Thimmapuram, J.; Bhide, K.P.; Sripathi, V.R.; Smolinski, T.G.; Manoharan, M.; Thurston, Y.; Todd, A.; Kingham, B.; et al. Genome-wide profiling of histone modifications (H3K9me2 and H4K12ac) and gene expression in rust (Uromyces appendiculatus) inoculated common bean (Phaseolus vulgaris L.). PLoS ONE 2015, 10, e0132176. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Yekondi, S.; Chen, P.W.; Tsai, C.H.; Yu, C.W.; Wu, K.; Zimmerli, L. Environmental history modulates Arabidopsis pattern-triggered immunity in a histone acetyltransferase1-dependent manner. Plant Cell 2014, 26, 2676–2688. [Google Scholar] [CrossRef] [PubMed]
- Atighi, M.R.; Verstraeten, B.; De Meyer, T.; Kyndt, T. Genome-wide shifts in histone modifications at early stage of rice infection with Meloidogyne graminicola. Mol. Plant Pathol. 2021, 22, 440–455. [Google Scholar] [CrossRef]
- Vijayapalani, P.; Hewezi, T.; Pontvianne, F.; Baum, T.J. An effector from the cyst nematode Heterodera schachtii derepresses host RRNA genes by altering histone acetylation. Plant Cell 2018, 30, 2795–2812. [Google Scholar] [CrossRef]
- Bennett, M.; Piya, S.; Baum, T.J.; Hewezi, T. MiR778 mediates gene expression, histone modification, and DNA methylation during cyst nematode parasitism. Plant Physiol. 2022, 189, 2432–2453. [Google Scholar] [CrossRef]
- Piya, S.; Bennett, M.; Rambani, A.; Hewezi, T. Transcriptional activity of transposable elements may contribute to gene expression changes in the syncytium formed by cyst nematode in Arabidopsis roots. Plant Signal. Behav. 2017, 12, e1362521. [Google Scholar] [CrossRef][Green Version]
- Seo, J.S.; Sun, H.X.; Park, B.S.; Huang, C.H.; Yeh, S.D.; Jung, C.; Chua, N.H. ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 2017, 29, 1024–1038. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, Y.-F.; Feng, Y.-Z.; He, H.; Lian, J.-P.; Yang, Y.-W.; Lei, M.-Q.; Zhang, Y.-C.; Chen, Y.-Q. Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol. J. 2020, 18, 679–690. [Google Scholar] [CrossRef]
- Khoei, M.A.; Karimi, M.; Karamian, R.; Amini, S.; Soorni, A. Identification of the complex interplay between nematode-related lncRNAs and their target genes in Glycine max L. Front. Plant Sci. 2021, 12, 779597. [Google Scholar] [CrossRef]
- Li, X.; Xing, X.; Xu, S.; Zhang, M.; Wang, Y.; Wu, H.; Sun, Z.; Huo, Z.; Chen, F.; Yang, T. Genome-wide identification and functional prediction of tobacco lncRNAs responsive to root-knot nematode stress. PLoS ONE 2018, 13, e0204506. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, B.; Atighi, M.R.; Ruiz-Ferrer, V.; Escobar, C.; De Meyer, T.; Kyndt, T. Non-coding RNAs in the interaction between rice and Meloidogyne graminicola. BMC Genom. 2021, 22, 560. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Gu, L.; Song, X.; Cui, X.; Lu, Z.; Zhou, M.; Wang, L.; Hu, F.; Zhai, J.; Meyers, B.C.; et al. Dicer-like 3 produces transposable element-associated 24-NT SiRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 3877–3882. [Google Scholar] [CrossRef] [PubMed]
- Medina, C.; Da Rocha, M.; Magliano, M.; Raptopoulo, A.; Marteu, N.; Lebrigand, K.; Abad, P.; Favery, B.; Jaubert-Possamai, S. Characterization of siRNAs clusters in Arabidopsis thaliana galls induced by the root-knot nematode Meloidogyne incognita. BMC Genom. 2018, 19, 943. [Google Scholar] [CrossRef]
- Cabrera, J.; Barcala, M.; García, A.; Rio-Machín, A.; Medina, C.; Jaubert-Possamai, S.; Favery, B.; Maizel, A.; Ruiz-Ferrer, V.; Fenoll, C.; et al. Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: A functional role for miR390 and its TAS3-derived TASIRNAs. New Phytol. 2016, 209, 1625–1640. [Google Scholar] [CrossRef]
- Koter, M.D.; Święcicka, M.; Matuszkiewicz, M.; Pacak, A.; Derebecka, N.; Filipecki, M. The miRNAome dynamics during developmental and metabolic reprogramming of tomato root infected with potato cyst nematode. Plant Sci. 2018, 268, 18–29. [Google Scholar] [CrossRef]
- Xu, P.; Li, H.; Wang, X.; Zhao, G.; Lu, X.; Dai, S.; Cui, X.; Yuan, M. Integrated analysis of the LncRNA/CircRNA-miRNA-mRNA expression profiles reveals novel insights into potential mechanisms in response to root-knot nematodes in peanut. BMC Genom. 2022, 23, 239. [Google Scholar] [CrossRef]
- Tian, B.; Wang, S.; Todd, T.C.; Johnson, C.D.; Tang, G.; Trick, H.N. Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genom. 2017, 18, 572. [Google Scholar] [CrossRef]
- Zhao, W.; Li, Z.; Fan, J.; Hu, C.; Yang, R.; Qi, X.; Chen, H.; Zhao, F.; Wang, S. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J. Exp. Bot. 2015, 66, 4653–4667. [Google Scholar] [CrossRef]
- Nahar, K.; Kyndt, T.; de Vleesschauwer, D.; Höfte, M.; Gheysen, G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011, 157, 305–316. [Google Scholar] [CrossRef]
- Cooper, W.R.; Jia, L.; Goggin, L. Effects of jasmonate-induced defenses on root-knot nematode infection of resistant and susceptible tomato cultivars. J. Chem. Ecol. 2005, 31, 1953–1967. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Baum, T.J. Complex feedback regulations govern the expression of miRNA396 and its GRF target genes. Plant Signal. Behav. 2012, 7, 749–751. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Piya, S.; Qi, M.; Balasubramaniam, M.; Rice, J.H.; Baum, T.J. Arabidopsis miR827 mediates post-transcriptional gene silencing of its ubiquitin E3 ligase target gene in the syncytium of the cyst nematode Heterodera schachtii to enhance susceptibility. Plant J. 2016, 88, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Maier, T.R.; Nettleton, D.; Baum, T.J. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012, 159, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Manzano, F.E.; Cabrera, J.; Ripoll, J.-J.; del Olmo, I.; Fe Andres, M.; Silva, A.C.; Barcala, M.; Sanchez, M.; Ruiz-Ferrer, V.; de Almeida-Engler, J.; et al. A role for the gene regulatory module microRNA172 TARGET OF EARLY ACTIVATION TAGGED 1/FLOWERING LOCUS T (miRNA172/TOE1/FT) in the feeding sites induced by Meloidogyne javanica in Arabidopsis thaliana. New Phytol. 2018, 217, 813–827. [Google Scholar] [CrossRef]
- Noureddine, Y.; Mejias, J.; da Rocha, M.; Thomine, S.; Quentin, M.; Abad, P.; Favery, B.; Jaubert-Possamai, S. Copper microRNAs modulate the formation of giant feeding cells induced by the root knot nematode Meloidogyne incognita in Arabidopsis thaliana. New Phytol. 2022, 236, 283–295. [Google Scholar] [CrossRef]
- Grunewald, W.; Cannoot, B.; Friml, J.; Gheysen, G. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 2009, 5, e1000266. [Google Scholar] [CrossRef]
- Pan, X.; Nichols, R.L.; Li, C.; Zhang, B. microRNA-target gene responses to root knot nematode (Meloidogyne incognita) infection in cotton (Gossypium hirsutum L.). Genomics 2019, 111, 383–390. [Google Scholar] [CrossRef]
- Noureddine, Y.; Da Rocha, M.; An, J.; Médina, C.; Mejias, J.; Mulet, K.; Quentin, M.; Abad, P.; Zouine, M.; Favery, B.; et al. AUXIN RESPONSIVE FACTOR8 regulates development of the feeding site induced by root-knot nematodes in tomato. J. Exp. Bot. 2023, 74, 5752–5766. [Google Scholar] [CrossRef]
- Kyndt, T.; Goverse, A.; Haegeman, A.; Warmerdam, S.; Wanjau, C.; Jahani, M.; Engler, G.; De Almeida Engler, J.; Gheysen, G. Redirection of auxin flow in Arabidopsis thaliana roots after infection by root-knot nematodes. J. Exp. Bot. 2016, 67, 4559–4570. [Google Scholar] [CrossRef]
- Abril-Urias, P.; Ruiz-Ferrer, V.; Cabrera, J.; Olmo, R.; Silva, A.C.; Díaz-Manzano, F.E.; Domínguez-Figueroa, J.; Martínez-Gómez, Á.; Gómez-Rojas, A.; Moreno-Risueno, M.Á.; et al. Divergent regulation of auxin responsive genes in root-knot and cyst nematodes feeding sites formed in Arabidopsis. Front. Plant Sci. 2023, 14, 1024815. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Qi, N.; Yan, J.; Zhu, X.; Liu, X.; Xuan, Y.; Fan, H.; Chen, L.; Duan, Y.; Wang, Y. Genome-wide identification of small interfering RNAs from SRNA libraries constructed from soybean cyst nematode resistant and susceptible cultivars. Gene 2022, 832, 146557. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Park, C.M. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 2007, 225, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Replogle, A.; Wang, J.; Paolillo, V.; Smeda, J.; Kinoshita, A.; Durbak, A.; Tax, F.E.; Wang, X.; Sawa, S.; Mitchum, M.G. Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis. Mol. Plant-Microbe Interact. 2013, 26, 87–96. [Google Scholar] [CrossRef]
- Replogle, A.; Wang, J.; Bleckmann, A.; Hussey, R.S.; Baum, T.J.; Sawa, S.; Davis, E.L.; Wang, X.; Simon, R.; Mitchum, M.G. Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 2011, 65, 430–440. [Google Scholar] [CrossRef]
- Bräutigam, K.; Vining, K.J.; Lafon-Placette, C.; Fossdal, C.G.; Mirouze, M.; Marcos, J.G.; Fluch, S.; Fraga, M.F.; Guevara, M.Á.; Abarca, D.; et al. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol. Evol. 2013, 3, 399–415. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef]
- Lankau, R.A.; Strauss, S.Y. Community complexity drives patterns of natural selection on a chemical defense of Brassica nigra. Am. Nat. 2008, 171, 150–161. [Google Scholar] [CrossRef]
- Callahan, H.S.; Dhanoolal, N.; Ungerer, M.C. Plasticity genes and plasticity costs: A new approach using an Arabidopsis recombinant inbred population. New Phytol. 2005, 166, 129–140. [Google Scholar] [CrossRef]
- Alpert, P.; Simms, E.L. The relative advantages of plasticity and fixity in different environments: When is it good for a plant to adjust? Evol. Ecol. 2002, 16, 285–297. [Google Scholar] [CrossRef]
- Meijer, A.; Atighi, M.R.; Demeestere, K.; De Meyer, T.; Vandepoele, K.; Kyndt, T. Dicer-like 3a mediates intergenerational resistance against root-knot nematodes in rice via hormone responses. Plant Physiol. 2023, 193, 2071–2085. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.J.; Anjam, M.S.; Mendy, B.; Anwer, M.A.; Habash, S.S.; Lozano-Torres, J.L.; Grundler, F.M.W.; Siddique, S. Damage-associated responses of the host contribute to defence against cyst nematodes but not root-knot nematodes. J. Exp. Bot. 2017, 68, 5949–5960. [Google Scholar] [CrossRef]
- Acharya, S.; Troell, H.A.; Billingsley, R.L.; Lawrence, K.S.; Mckirgan, D.S.; Alkharouf, N.W.; Klink, V.P. Data analysis of polygalacturonase inhibiting proteins (PGIPs) from agriculturally important proteomes. Data Brief 2024, 52, 109831. [Google Scholar] [CrossRef]
- Acharya, S.; Troell, H.A.; Billingsley, R.L.; McKirgan, D.S.; Lawrence, K.S.; Alkharouf, N.W.; Klink, V. Glycine max polygalacturonase inhibiting protein (PGIP) functions in the root to suppress Heterodera glycines parasitism. Plant Physiol. Biochem. 2024, 7, 108755. [Google Scholar] [CrossRef]
- Kyndt, T.; Vieira, P.; Gheysen, G.; de Almeida-Engler, J. Nematode feeding sites: Unique organs in plant roots. Planta 2013, 238, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Morales-quintana, L.; Beltrán, D.; Mendez-Yañez, Á.; Valenzuela-Riff, F.; Herrera, R.; Moya-León, M.A. Characterization of FcXTH2, a novel xyloglucan endotransglycosylase/hydrolase enzyme of Chilean strawberry with hydrolase activity. Int. J. Mol. Sci. 2020, 21, 3380. [Google Scholar] [CrossRef] [PubMed]
- Matsye, P.D.; Kumar, R.; Hosseini, P.; Jones, C.M.; Tremblay, A.; Alkharouf, N.W.; Matthews, B.F.; Klink, V.P. Mapping cell fate decisions that occur during soybean defense responses. Plant Mol. Biol. 2011, 77, 513–528. [Google Scholar] [CrossRef]
- Sato, K.; Uehara, T.; Holbein, J.; Sasaki-Sekimoto, Y.; Gan, P.; Bino, T.; Yamaguchi, K.; Ichihashi, Y.; Maki, N.; Shigenobu, S.; et al. Transcriptomic analysis of resistant and susceptible responses in a new model root-knot nematode infection system using Solanum torvum and Meloidogyne arenaria. Front. Plant Sci. 2021, 12, 680151. [Google Scholar] [CrossRef]
- Niraula, P.M.; Lawrence, K.S.; Klink, V.P. The heterologous expression of a soybean (Glycine max) xyloglucan endotransglycosylase/hydrolase (XTH) in cotton (Gossypium hirsutum) suppresses parasitism by the root knot nematode Meloidogyne incognita. PLoS ONE 2020, 15, e0235344. [Google Scholar] [CrossRef]
- Niraula, P.M.; Zhang, X.; Jeremic, D.; Lawrence, K.S.; Klink, V.P. Xyloglucan endotransglycosylase/hydrolase increases tightly-bound xyloglucan and chain number but decreases chain length contributing to the defense response that Glycine max has to Heterodera glycines. PLoS ONE 2021, 16, e0244305. [Google Scholar] [CrossRef]
- Wan, J.; He, M.; Hou, Q.; Zou, L.; Yang, Y.; Wei, Y.; Chen, X. Cell wall associated immunity in plants. Stress Biol. 2021, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvo, P.; López, G.; Martín-Dacal, M.; Aitouguinane, M.; Carrasco-López, C.; González-Bodí, S.; Bacete, L.; Mélida, H.; Sánchez-Vallet, A.; Molina, A. Leucine rich repeat-malectin receptor kinases IGP1/CORK1, IGP3 and IGP4 are required for Arabidopsis immune responses triggered by β-1,4-D-xylo-oligosaccharides from plant cell walls. Cell Surf. 2024, 11, 100124. [Google Scholar] [CrossRef] [PubMed]
- Holbein, J.; Franke, R.B.; Marhavy, P.; Fujita, S.; Gorecka, M.; Sobczak, M.; Geldner, N.; Schreiber, L.; Grundler, F.M.W.; Siddique, S. Root endodermal barrier system contributes to defence against plant-parasitic cyst and root-knot nematodes. Plant J. 2019, 100, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Cabasan, M.T.N.; Kumar, A.; Bellafiore, S.; De Waele, D. Histopathology of the rice root-knot nematode, Meloidogyne graminicola, on Oryza sativa and O. glaberrima. Nematology 2014, 16, 73–81. [Google Scholar] [CrossRef]
- Galeng-Lawilao, J.; Kumar, A.; Cabasan, M.T.N.; De Waele, D. Comparison of the penetration, development and reproduction of Meloidogyne graminicola, and analysis of lignin and total phenolic content in partially resistant and resistant recombinant inbred lines of Oryza sativa. Trop. Plant Pathol. 2019, 44, 171–182. [Google Scholar] [CrossRef]
- Singh, D.; Dutta, T.K.; Shivakumara, T.N.; Dash, M.; Bollinedi, H.; Rao, U. Suberin biopolymer in rice root exodermis reinforces preformed barrier against Meloidogyne graminicola infection. Rice Sci. 2021, 28, 301–312. [Google Scholar] [CrossRef]
- Veronico, P.; Paciolla, C.; Pomar, F.; De Leonardis, S.; García-Ulloa, A.; Melillo, M.T. Changes in lignin biosynthesis and monomer composition in response to benzothiadiazole and root-knot nematode Meloidogyne incognita infection in tomato. J. Plant Physiol. 2018, 230, 40–50. [Google Scholar] [CrossRef]
- Bali, S.; Vining, K.; Gleason, C.; Majtahedi, H.; Brown, C.R.; Sathuvalli, V. Transcriptome profiling of resistance response to Meloidogyne chitwoodi introgressed from wild species Solanum bulbocastanum into cultivated potato. BMC Genom. 2019, 20, 907. [Google Scholar] [CrossRef]
- Lee, K.H.; Lee, K.L.; Nam, K.J.; Yang, J.W.; Lee, J.J.; Shim, D.; Kim, Y.H. Expression analysis of sweetpotato cinnamyl alcohol dehydrogenase genes in response to infection with the root-knot nematode Meloidogyne incognita. Plant Biotechnol. Rep. 2022, 16, 487–492. [Google Scholar] [CrossRef]
- Hatzade, B.; Singh, D.; Phani, V.; Kumbhar, S.; Rao, U. Profiling of defense responsive pathway regulatory genes in Asian rice (Oryza sativa) against infection of Meloidogyne graminicola (Nematoda: Meloidogynidae). 3 Biotech 2020, 10, 1–16. [Google Scholar] [CrossRef]
- Modesto, I.; Mendes, A.; Carrasquinho, I.; Miguel, C.M. Molecular defense response of pine trees (Pinus spp.) to the parasitic nematode Bursaphelenchus xylophilus. Cells 2022, 11, 3208. [Google Scholar] [CrossRef] [PubMed]
- Klink, V.P.; Hosseini, P.; Matsye, P.; Alkharouf, N.W.; Matthews, B.F. A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol. Biol. 2009, 71, 525–567. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, F.; Ashraf, M.; Siddique, K.H.M. Role of glycine betaine in the thermotolerance of plants. Agronomy 2022, 12, 276. [Google Scholar] [CrossRef]
- Shafiq, S.; Akram, N.A.; Ashraf, M.; García-Caparrós, P.; Ali, O.M.; Abdel-Latef, A.A.-H. Influence of glycine betaine (natural and synthetic) on growth, metabolism and yield production of drought-stressed maize (Zea mays L.) plants. Plants 2021, 10, 2540. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.; Jamwal, V.L.; Sharma, A.; Gandhi, S.G.; Ohri, P.; Bhardwaj, R.; Al-Huqail, A.A.; Siddiqui, M.H.; Marraiki, N.; Ahmad, P. Evaluation of the role of rhizobacteria in controlling root-knot nematode infection in Lycopersicon esculentum plants by modulation in the secondary metabolite profiles. AoB Plants 2019, 11, plz069. [Google Scholar] [CrossRef]
- Khanna, K.; Sharma, A.; Ohri, P.; Bhardwaj, R.; Abd-Allah, E.F.; Hashem, A.; Ahmad, P. Impact of plant growth promoting rhizobacteria in the orchestration of Lycopersicon esculentum Mill. resistance to plant parasitic nematodes: A metabolomic approach to evaluate defense responses under field conditions. Biomolecules 2019, 9, 676. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Ling, J.; Zhao, J.; Li, Y.; Mao, Z.; Cheng, X.; Xie, B. NRPS-like ATRR in plant-parasitic nematodes involved in glycine betaine metabolism to promote parasitism. Int. J. Mol. Sci. 2024, 25, 4275. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Dubreuil-Maurizi, C.; Poinssot, B. Role of glutathione in plant signaling under biotic stress. Plant Signal. Behav. 2012, 7, 210–212. [Google Scholar] [CrossRef]
- Dorion, S.; Ouellet, J.C.; Rivoal, J. Glutathione metabolism in plants under stress: Beyond reactive oxygen species detoxification. Metabolites 2021, 11, 641. [Google Scholar] [CrossRef]
- Rahaman, M.M.; Zwart, R.S.; Rupasinghe, T.W.T.; Hayden, H.L.; Thompson, J.P. Metabolomic profiling of wheat genotypes resistant and susceptible to root-lesion nematode Pratylenchus thornei. Plant Mol. Biol. 2021, 106, 381–406. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.W.; Kim, J.; Yang, J.W.; Shim, D.; Kim, Y.H. Transcriptome-based comparative expression profiling of sweet potato during a compatible response with root-knot nematode Meloidogyne incognita infection. Genes 2023, 14, 2074. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.S.; Chopra, D.; Damm, A.; Koprivova, A.; Kopriva, S.; Meyer, A.J.; Müller-Schüssele, S.; Grundler, F.M.W.; Siddique, S. Glutathione contributes to plant defence against parasitic cyst nematodes. Mol. Plant Pathol. 2022, 23, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, V.; Tsednee, M.; Yeh, K.C. ZINC TOLERANCE INDUCED BY IRON 1 reveals the importance of glutathione in the cross-homeostasis between zinc and iron in Arabidopsis thaliana. Plant J. 2012, 69, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Deckers, J.; Hendrix, S.; Prinsen, E.; Vangronsveld, J.; Cuypers, A. Glutathione is required for the early alert response and subsequent acclimation in cadmium-exposed Arabidopsis thaliana plants. Antioxidants 2022, 11, 6. [Google Scholar] [CrossRef]
- Datta, R.; Chattopadhyay, S. Glutathione as a crucial modulator of phytohormone signalling during pathogen defence in plants. Proc. Indian Natl. Sci. Acad. 2018, 84, 581–597. [Google Scholar] [CrossRef]
- Velasco-Azorsa, R.; Cruz-Santiago, H.; Del Prado-Vera, I.C.; Ramirez-Mares, M.V.; Gutiérrez-Ortiz, M.D.R.; Santos-Sánchez, N.F.; Salas-Coronado, R.; Villanueva-Cañongo, C.; León, K.I.L.; Hernández-Carlos, B. Chemical characterization of plant extracts and evaluation of their nematicidal and phytotoxic potential. Molecules 2021, 26, 2216. [Google Scholar] [CrossRef]
- Elsharkawy, M.M.; Al-Askar, A.A.; Behiry, S.I.; Abdelkhalek, A.; Saleem, M.H.; Kamran, M.; Derbalah, A. Resistance induction and nematicidal activity of certain monoterpenes against tomato root-knot caused by Meloidogyne incognita. Front. Plant Sci. 2022, 13, 982414. [Google Scholar] [CrossRef]
- El-Habashy, D.E.; Abdel Rasoul, M.A.; Abdelgaleil, S.A.M. Nematicidal activity of phytochemicals and their potential use for the control of Meloidogyne javanica infected eggplant in the greenhouse. Eur. J. Plant Pathol. 2020, 158, 381–390. [Google Scholar] [CrossRef]
- Mwamba, S.; Kihika-Opanda, R.; Murungi, L.K.; Losenge, T.; Beck, J.J.; Torto, B. Identification of repellents from four non-host Asteraceae plants for the root knot nematode, Meloidogyne incognita. J. Agric. Food Chem. 2021, 69, 15145–15156. [Google Scholar] [CrossRef]
- Barbosa, P.; Faria, J.M.S.; Cavaco, T.; Figueiredo, A.C.; Mota, M.; Vicente, S.L. Nematicidal activity of phytochemicals against the root-lesion nematode Pratylenchus penetrans. Plants 2024, 13, 726. [Google Scholar] [CrossRef] [PubMed]
- Dash, M.; Singh, V.; Roli, S.; Jeffrey, B.; Rohit, G.; Uma, N.S. A rice root-knot nematode Meloidogyne graminicola-resistant mutant rice line shows early expression of plant-defence genes. Planta 2021, 253, 108. [Google Scholar] [CrossRef] [PubMed]
- Derbalah, A.; Shebl, A.M.; Elgobashy, S.F.; Ahmad, A.A.; Ramadan, N.E.; Behiry, S.I.; Abdelkhalek, A.; Saleem, M.H.; Al-Askar, A.A.; Kamran, M.; et al. Resistance induction and direct antifungal activity of some monoterpenes against Rhizoctonia solani, the causal of root rot in common bean. Life 2022, 12, 1040. [Google Scholar] [CrossRef] [PubMed]
- Marei, G.K.; Abdel Rasoul, M.A.; Abdelgaleil, S.A.M. Comparative antifungal activities and biochemical effects of monoterpenes on plant pathogenic fungi. Pestic. Biochem. Physiol. 2012, 103, 56–61. [Google Scholar] [CrossRef]
- Desmedt, W.; Jonckheere, W.; Nguyen, V.H.; Ameye, M.; Zutter, N.; Kock, K.; Debode, J.; Leeuwen, T.; Audenaert, K.; Vanholme, B.; et al. The phenylpropanoid pathway inhibitor piperonylic acid induces broad-spectrum pest and disease resistance in plants. Plant Cell Environ. 2021, 44, 3122–3139. [Google Scholar] [CrossRef]
- Yamane, H. Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci. Biotechnol. Biochem. 2013, 77, 1141–1148. [Google Scholar] [CrossRef]
- Van Moerkercke, A.; Schauvinhold, I.; Pichersky, E.; Haring, M.A.; Schuurink, R.C. A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 2009, 60, 292–302. [Google Scholar] [CrossRef]
- Jardim, I.N.; Oliveira, D.F.; Silva, G.H.; Campos, V.P.; de Souza, P.E. (E)-cinnamaldehyde from the essential oil of Cinnamomum cassia controls Meloidogyne incognita in soybean plants. J. Pest Sci. 2018, 91, 479–487. [Google Scholar] [CrossRef]
- Barros, A.F.; Campos, V.P.; De Oliveira, D.F.; De Jesus Silva, F.; Jardim, I.N.; Costa, V.A.; Matrangolo, C.A.R.; Ribeiro, R.C.F.; Silva, G.H. Activities of essential oils from three Brazilian plants and benzaldehyde analogues against Meloidogyne incognita. Nematology 2019, 21, 1081–1089. [Google Scholar] [CrossRef]
- Caboni, P.; Aissani, N.; Cabras, T.; Falqui, A.; Marotta, R.; Liori, B.; Ntalli, N.; Sarais, G.; Sasanelli, N.; Tocco, G. Potent nematicidal activity of phthalaldehyde, salicylaldehyde, and cinnamic aldehyde against Meloidogyne incognita. J. Agric. Food Chem. 2013, 61, 1794–1803. [Google Scholar] [CrossRef]
- Aissani, N.; Aissani, R.; Zouidi, F.; Sebai, H. Nematicidal activity of o-hydroxybenzaldehyde from common buckwheat methanol extract on Meloidogyne incognita. J. Helminthol. 2023, 97, e60. [Google Scholar] [CrossRef] [PubMed]
- Sircar, D.; Mukherjee, C. Characterization of p-hydroxybenzaldehyde dehydrogenase, the final enzyme of p-hydroxybenzoic acid biosynthesis in hairy roots of Daucus carota. Acta Physiol. Plant. 2011, 33, 2019–2024. [Google Scholar] [CrossRef]
- Nguyen, D.; Seo, D.; Kim, K.; Park, R.; Kim, D.; Han, Y.; Kim, T.; Jung, W. Nematicidal activity of 3, 4-dihydroxybenzoic acid purified from Terminalia nigrovenulosa Bark against Meloidogyne incognita. Microb. Pathog. 2013, 59–60, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Akhter, M.; Khatoon, Z. Nematicidal natural products from the aerial parts of Rubus niveus. Nat. Prod. Res. 2010, 24, 407–415. [Google Scholar] [CrossRef]
- Yates, P.; Janiol, J.; Li, C.; Song, B. Nematocidal potential of phenolic acids: A phytochemical seed-coating approach to soybean cyst nematode management. Plants 2024, 13, 319. [Google Scholar] [CrossRef]
- Sikder, M.; Vestergård, M.; Kyndt, T.; Fomsgaard, I.S.; Kudjordjie, E.N. Benzoxazinoids selectively affect maize root-associated nematode taxa. J. Exp. Bot. 2021, 72, 3835–3845. [Google Scholar] [CrossRef]
- Hussain, M.I.; Araniti, F.; Schulz, M.; Baerson, S.; Vieites-Álvarez, Y.; Rempelos, L.; Bilsborrow, P.; Chinchilla, N.; Macías, F.A.; Weston, L.A.; et al. Benzoxazinoids in wheat allelopathy-from discovery to application for sustainable weed management. Environ. Exp. Bot. 2022, 202, 104997. [Google Scholar] [CrossRef]
- Hama, J.R.; Hooshmand, K.; Laursen, B.B.; Vestergård, M.; Fomsgaard, I.S. Clover root uptake of cereal benzoxazinoids (BXs) caused accumulation of BXs and BX transformation products concurrently with substantial increments in clover flavonoids and abscisic acid. J. Agric. Food Chem. 2022, 70, 14633–14640. [Google Scholar] [CrossRef]
- Hama, J.R.; Fomsgaard, I.S.; Topalovic, O.; Vestergård, M. Root uptake of cereal benzoxazinoids grants resistance to root-knot nematode invasion in white clover. Plant Physiol. Biochem. 2024, 210, 108636. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meresa, B.K.; Matthys, J.; Kyndt, T. Biochemical Defence of Plants against Parasitic Nematodes. Plants 2024, 13, 2813. https://doi.org/10.3390/plants13192813
Meresa BK, Matthys J, Kyndt T. Biochemical Defence of Plants against Parasitic Nematodes. Plants. 2024; 13(19):2813. https://doi.org/10.3390/plants13192813
Chicago/Turabian StyleMeresa, Birhanu Kahsay, Jasper Matthys, and Tina Kyndt. 2024. "Biochemical Defence of Plants against Parasitic Nematodes" Plants 13, no. 19: 2813. https://doi.org/10.3390/plants13192813
APA StyleMeresa, B. K., Matthys, J., & Kyndt, T. (2024). Biochemical Defence of Plants against Parasitic Nematodes. Plants, 13(19), 2813. https://doi.org/10.3390/plants13192813







