Integrative Analysis of Oleosin Genes Provides Insights into Lineage-Specific Family Evolution in Brassicales
Abstract
1. Introduction
2. Results
2.1. Identification of Oleosin Genes in A. trichopoda, Avocado, A. coerulea, and Representative Brassicales Species
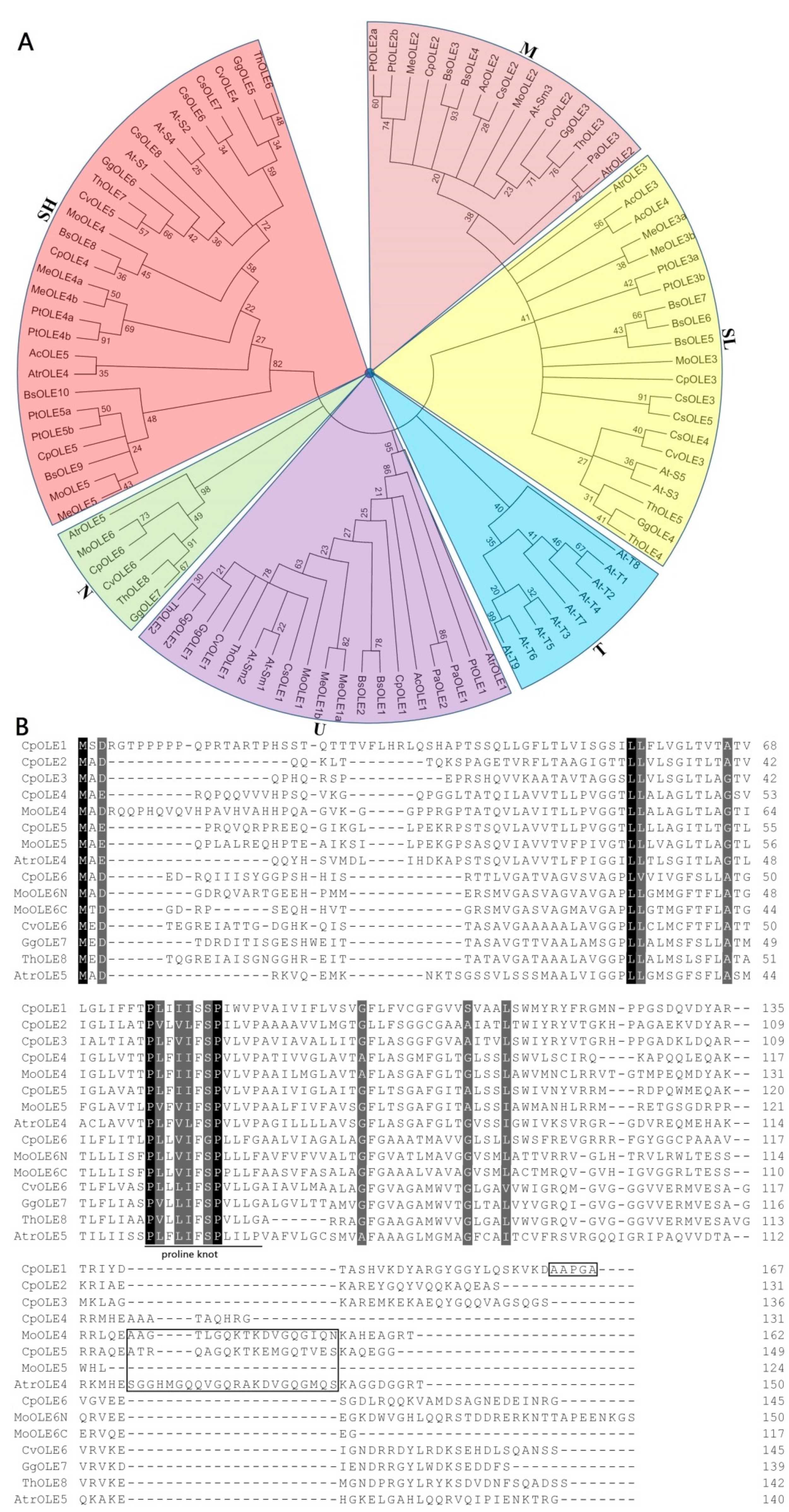
2.2. Evolutionary Analysis and Definition of Orthogroups
2.3. Analysis of Exon–Intron Structure
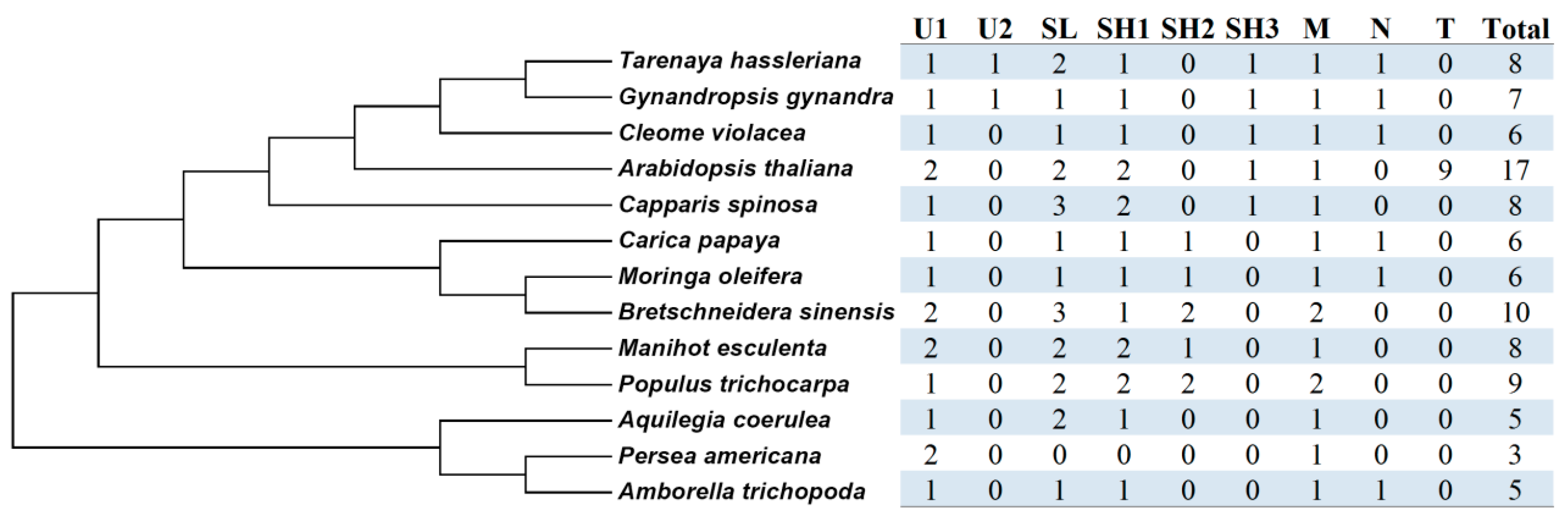
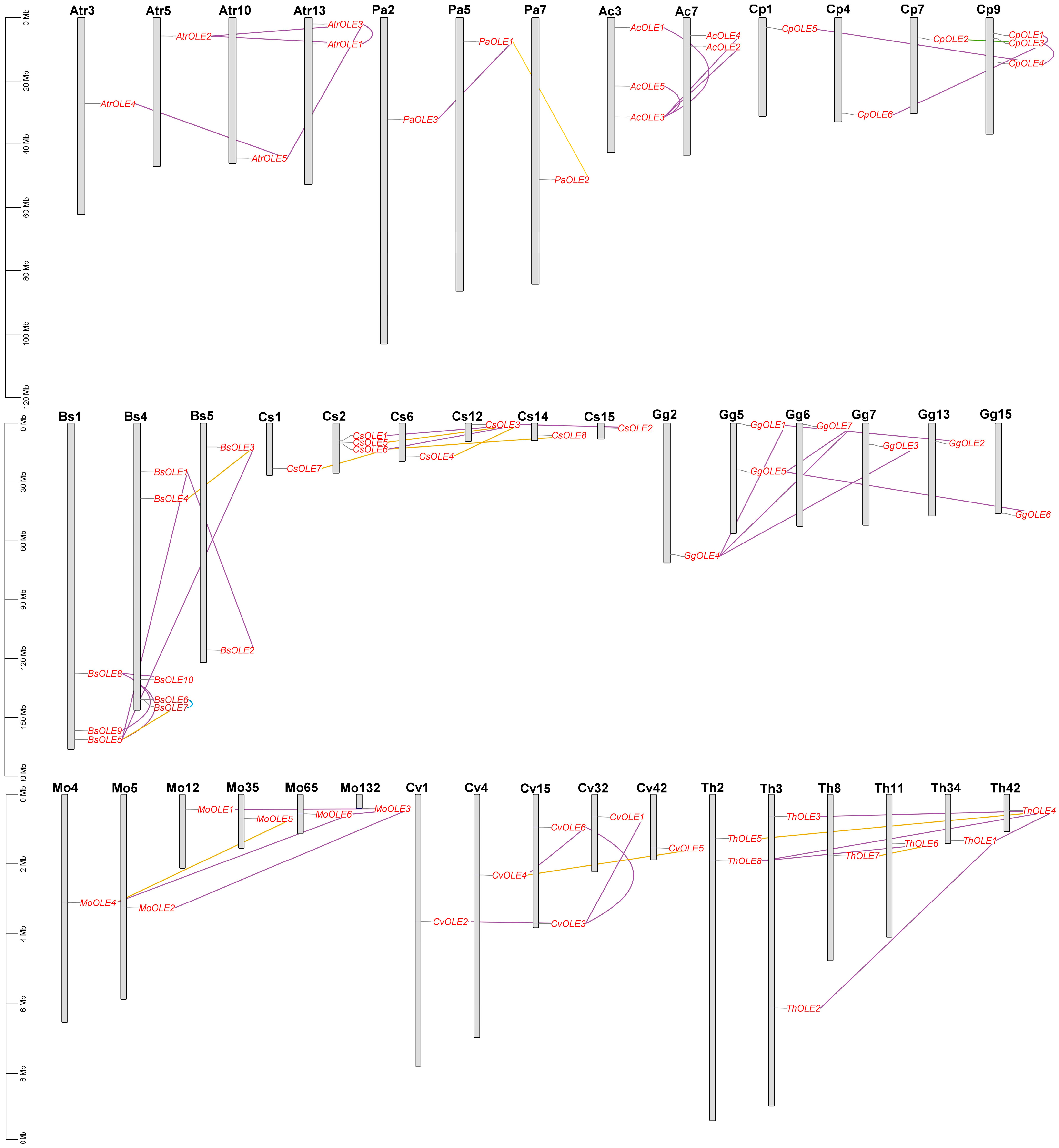
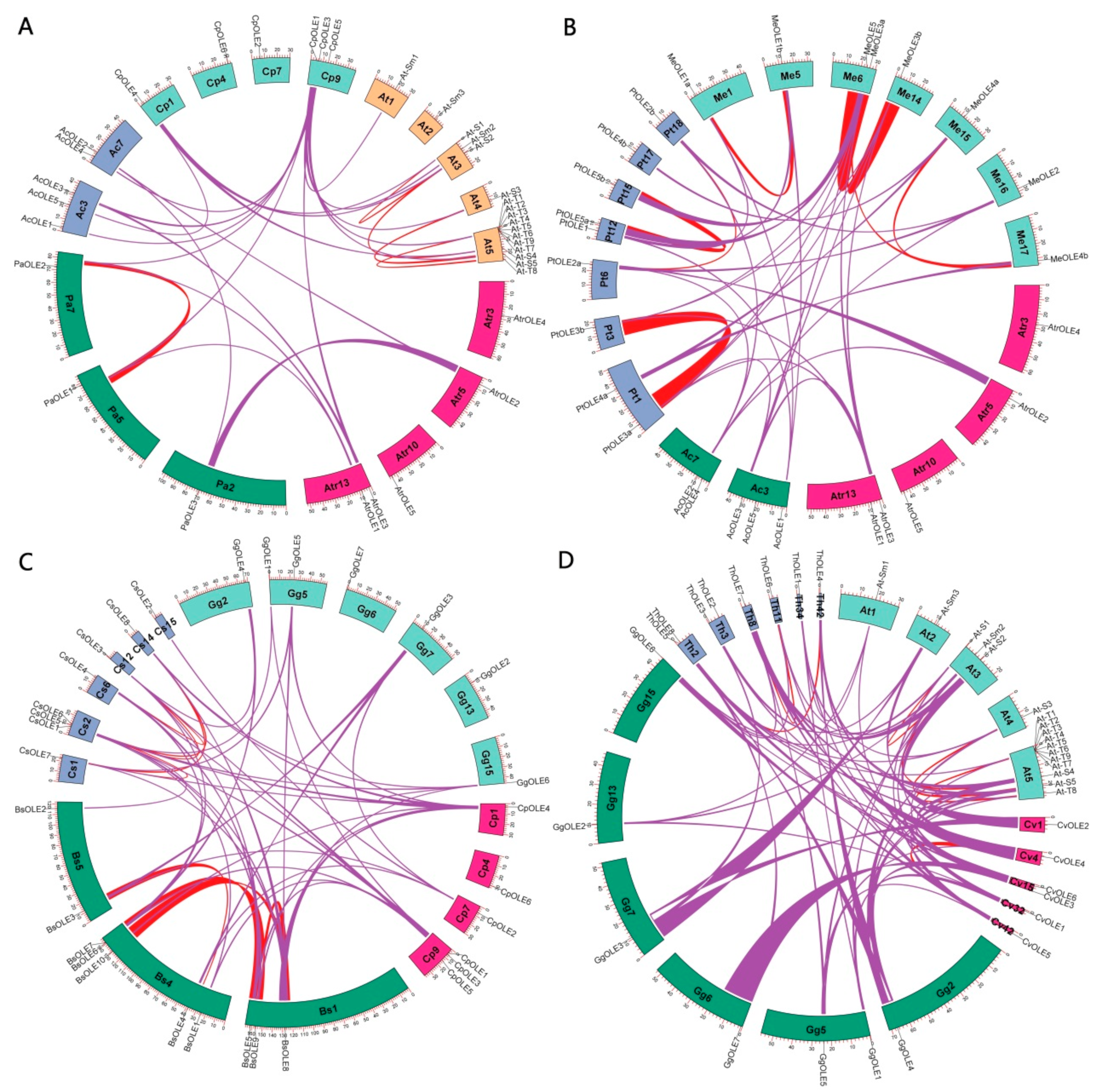
2.4. Gene Localization, Synteny Analysis, and Lineage-Specific Family Evolution in Brassicales
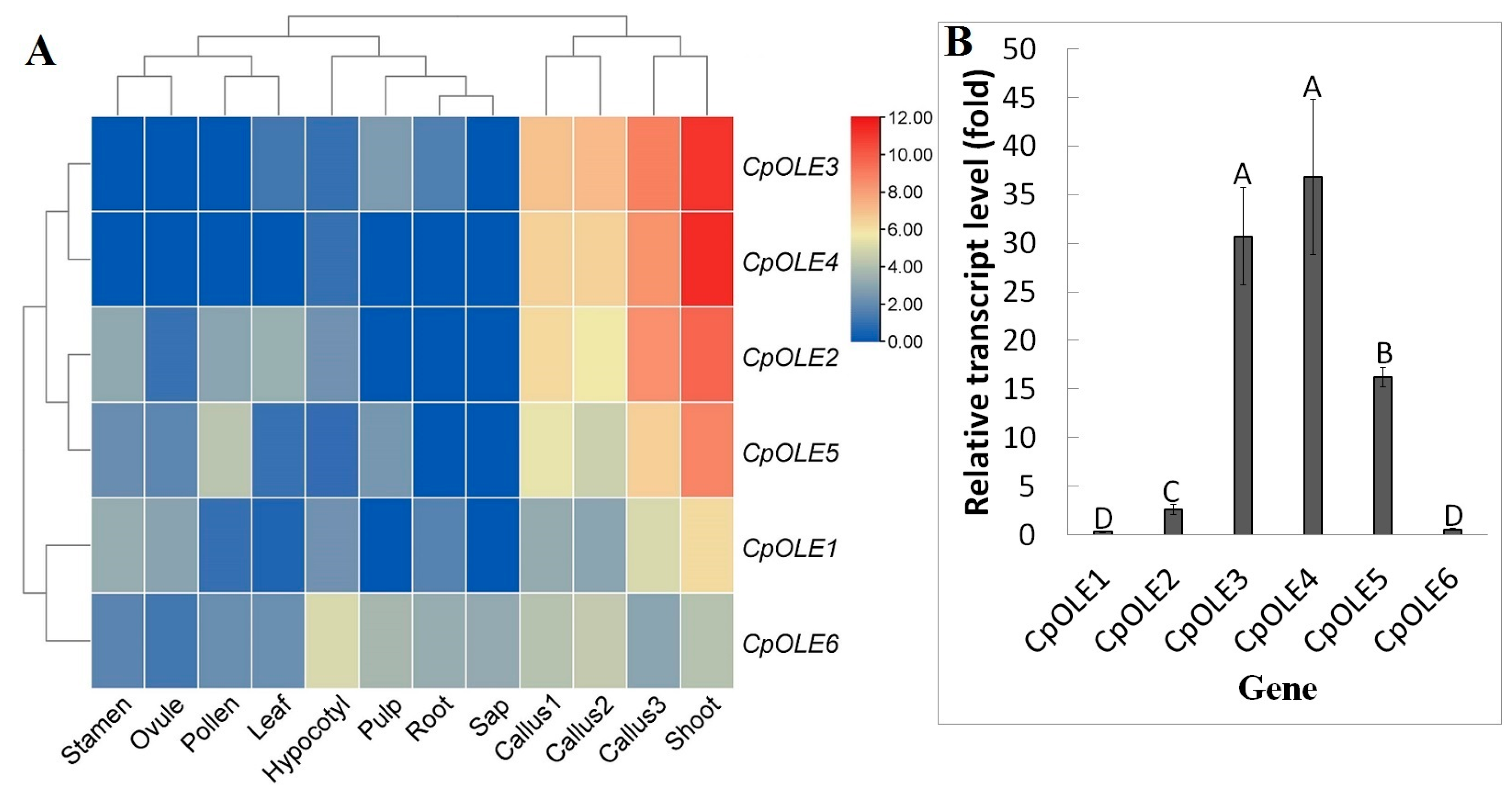
2.5. Expression Divergence of Oleosin Genes
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval and Identification of Oleosin Family Genes
4.2. Sequence Alignment, Evolutionary Analysis, and Definition of Orthogroups
4.3. Gene Localization, Synteny Analysis, and Calculation of Evolutionary Rate
4.4. Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, A.H. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996, 110, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, G.I.; Mundy, J.; Tzen, J.T. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 2001, 112, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Hsieh, K.; Ratnayake, C.; Huang, A.H. A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 2002, 277, 22677–22684. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.D.; Huang, A.H. Subcellular lipid droplets in vanilla leaf epidermis and avocado mesocarp are coated with oleosins of distinct phylogenic lineages. Plant Physiol. 2016, 171, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Zheng, Y.J.; Zhang, Z.T.; Xiao, Y.H.; Xie, Z.N.; Chang, L.L.; Zhang, L.; Zhao, Y.G. Molecular characterization oleosin genes in Cyperus esculentus, a Cyperaceae plant producing oil in underground tubers. Plant Cell Rep. 2023, 42, 1791–1808. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Sun, Y.; Su, W.; Yang, J.; Liu, X.; Wang, Y.; Wang, F.; Li, H.; Li, X. Species-specific size expansion and molecular evolution of the oleosins in angiosperms. Gene 2012, 509, 247–257. [Google Scholar] [CrossRef]
- Fang, Y.; Zhu, R.L.; Mishler, B.D. Evolution of oleosin in land plants. PLoS ONE 2014, 9, e103806. [Google Scholar] [CrossRef][Green Version]
- Huang, M.D.; Huang, A.H. Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol. 2015, 169, 453–470. [Google Scholar] [CrossRef]
- Lu, Y.; Chi, M.; Li, L.; Li, H.; Noman, M.; Yang, Y.; Ji, K.; Lan, X.; Qiang, W.; Du, L.; et al. Genome-wide identification, expression profiling, and functional validation of oleosin gene family in Carthamus tinctorius L. Front. Plant Sci. 2018, 9, 1393. [Google Scholar] [CrossRef]
- Chen, K.; Yin, Y.; Liu, S.; Guo, Z.; Zhang, K.; Liang, Y.; Zhang, L.; Zhao, W.; Chao, H.; Li, M. Genome-wide identification and functional analysis of oleosin genes in Brassica napus L. BMC Plant Biol. 2019, 19, 294. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, X.; Zhang, H.; Liu, C.; Zhang, Y.; Song, X.L.; Gai, S. Genome-wide identification and analysis of oleosin gene family in four cotton species and its involvement in oil accumulation and germination. BMC Plant Biol. 2021, 21, 569. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Zhao, Y.; Zhang, L. Genomic insights into lineage-specific evolution of the oleosin family in Euphorbiaceae. BMC Genom. 2022, 23, 178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xiong, T.; Ye, F.; Chen, J.H.; Chen, Y.R.; Cao, J.J.; Feng, Z.G.; Zhang, Z.B. The lineage-specific evolution of the oleosin family in Theaceae. Gene 2023, 868, 147385. [Google Scholar] [CrossRef] [PubMed]
- Deruyffelaere, C.; Purkrtova, Z.; Bouchez, I.; Collet, B.; Cacas, J.L.; Chardot, T.; Gallois, J.L.; D’Andrea, S. PUX10 is a CDC48A adaptor protein that regulates the extraction of ubiquitinated oleosins from seed lipid droplets in Arabidopsis. Plant Cell 2018, 30, 2116–2136. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.L.; Huang, M.D.; Chen, T.L.; Huang, A.H. Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 2013, 161, 1862–1874. [Google Scholar] [CrossRef] [PubMed]
- Schein, M.; Yang, Z.; Mitchell-Olds, T.; Schmid, K.J. Rapid evolution of a pollen-specific oleosin-like gene family from Arabidopsis thaliana and closely related species. Mol. Biol. Evol. 2004, 21, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Mabry, M.E.; Brose, J.M.; Blischak, P.D.; Sutherland, B.; Dismukes, W.T.; Bottoms, C.A.; Edger, P.P.; Washburn, J.D.; An, H.; Hall, J.C.; et al. Phylogeny and multiple independent whole-genome duplication events in the Brassicales. Am. J. Bot. 2020, 107, 1148–1164. [Google Scholar] [CrossRef]
- Yue, J.; VanBuren, R.; Liu, J.; Fang, J.; Zhang, X.; Liao, Z.; Wai, C.M.; Xu, X.; Chen, S.; Zhang, S.; et al. SunUp and Sunset genomes revealed impact of particle bombardment mediated transformation and domestication history in papaya. Nat. Genet. 2022, 54, 715–724. [Google Scholar] [CrossRef]
- Tian, Y.; Zeng, Y.; Zhang, J.; Yang, C.; Yan, L.; Wang, X.; Shi, C.; Xie, J.; Dai, T.; Peng, L.; et al. High quality reference genome of drumstick tree (Moringa oleifera Lam.), a potential perennial crop. Sci. China Life Sci. 2015, 58, 627–638. [Google Scholar] [CrossRef]
- Zhang, H.; Du, X.; Dong, C.; Zheng, Z.; Mu, W.; Zhu, M.; Yang, Y.; Li, X.; Hu, H.; Shrestha, N.; et al. Genomes and demographic histories of the endangered Bretschneidera sinensis (Akaniaceae). Gigascience 2022, 11, giac050. [Google Scholar] [CrossRef]
- Wang, L.; Fan, L.; Zhao, Z.; Zhang, Z.; Jiang, L.; Chai, M.; Tian, C. The Capparis spinosa var. herbacea genome provides the first genomic instrument for a diversity and evolution study of the Capparaceae family. Gigascience 2022, 11, giac106. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, J.; Sun, X.; Zheng, Q.; Liu, J.; Hua, W.; Liu, J. Integrated global analysis in spider flowers illuminates features underlying the evolution and maintenance of C4 photosynthesis. Hortic. Res. 2023, 10, uhad129. [Google Scholar] [CrossRef]
- Cheng, S.; van den Bergh, E.; Zeng, P.; Zhong, X.; Xu, J.; Liu, X.; Hofberger, J.; de Bruijn, S.; Bhide, A.S.; Kuelahoglu, C.; et al. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 2013, 25, 2813–2830. [Google Scholar] [CrossRef] [PubMed]
- Käfer, J.; Bewick, A.; Andres-Robin, A.; Lapetoule, G.; Harkess, A.; Caïus, J.; Fogliani, B.; Gâteblé, G.; Ralph, P.; de Pamphilis, C.W.; et al. A derived ZW chromosome system in Amborella trichopoda, representing the sister lineage to all other extant flowering plants. New Phytol. 2022, 233, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Nath, O.; Fletcher, S.J.; Hayward, A.; Shaw, L.M.; Masouleh, A.K.; Furtado, A.; Henry, R.J.; Mitter, N. A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes. Hortic. Res. 2022, 9, uhac157. [Google Scholar] [CrossRef]
- Filiault, D.L.; Ballerini, E.S.; Mandáková, T.; Aköz, G.; Derieg, N.J.; Schmutz, J.; Jenkins, J.; Grimwood, J.; Shu, S.; Hayes, R.D.; et al. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife 2018, 7, e36426. [Google Scholar] [CrossRef]
- Sánchez-Albarrán, F.; Suárez-Rodríguez, L.M.; Ruíz-Herrera, L.F.; López-Meza, J.E.; López-Gómez, R. Two oleosins expressed in the mesocarp of native mexican avocado, key genes in the oil content. Plant Foods Hum. Nutr. 2021, 76, 20–25. [Google Scholar] [CrossRef]
- Chang, Y.; Liu, H.; Liu, M.; Liao, X.; Sahu, S.K.; Fu, Y.; Song, B.; Cheng, S.; Kariba, R.; Muthemba, S.; et al. The draft genomes of five agriculturally important African orphan crops. Gigascience 2019, 8, giy152. [Google Scholar] [CrossRef]
- Shyamli, P.S.; Pradhan, S.; Panda, M.; Parida, A. De novo whole-genome assembly of Moringa oleifera helps identify genes regulating drought stress tolerance. Front. Plant Sci. 2021, 12, 766999. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Hsieh, K.; Huang, A.H. Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 2004, 136, 3427–3434. [Google Scholar] [CrossRef] [PubMed]
- Siloto, R.M.P.; Findlay, K.; Lopez, V.A.; Yeung, E.C.; Nykifork, C.L.; Moloney, M.M. The accumulation of oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell 2006, 18, 1961–1974. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.L.; Shimada, T.; Takahashi, H.; Fukao, Y.; Hara-Nishimura, I. A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 2008, 55, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.H. Plant lipid droplets and their associated proteins: Potential for rapid advances. Plant Physiol. 2018, 176, 1894–1918. [Google Scholar] [CrossRef]
- Shao, Q.; Liu, X.; Su, T.; Ma, C.; Wang, P. New insights into the role of seed oil body proteins in metabolism and plant development. Front. Plant Sci. 2019, 10, 1568. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, H.; Hu, Z.; Chu, S.; Yu, K.; Lv, L.; Yang, Y.; Zhang, X.; Chen, X.; Kan, G.; et al. Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet. 2019, 15, e1008267. [Google Scholar] [CrossRef] [PubMed]
- Guzha, A.; Whitehead, P.; Ischebeck, T.; Chapman, K.D. Lipid droplets: Packing hydrophobic molecules within the aqueous cytoplasm. Annu. Rev. Plant Biol. 2023, 74, 195–223. [Google Scholar] [CrossRef]
- Hu, J.; Chen, F.; Zang, J.; Li, Z.; Wang, J.; Wang, Z.; Shi, L.; Xiu, Y.; Lin, S. Native promoter-mediated transcriptional regulation of crucial oleosin protein OLE1 from Prunus sibirica for seed development and high oil accumulation. Int. J. Biol. Macromol. 2023, 253, 126650. [Google Scholar] [CrossRef]
- Jiao, Y.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J.; et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef]
- Vanneste, K.; Baele, G.; Maere, S.; Van de Peer, Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 2014, 24, 1334–1347. [Google Scholar] [CrossRef]
- Hoang, N.V.; Sogbohossou, E.O.D.; Xiong, W.; Simpson, C.J.C.; Singh, P.; Walden, N.; van den Bergh, E.; Becker, F.F.M.; Li, Z.; Zhu, X.G.; et al. The Gynandropsis gynandra genome provides insights into whole-genome duplications and the evolution of C4 photosynthesis in Cleomaceae. Plant Cell 2023, 35, 1334–1359. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa, Torr. & Gray. Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Bredeson, J.V.; Lyons, J.B.; Prochnik, S.E.; Wu, G.A.; Ha, C.M.; Edsinger-Gonzales, E.; Grimwood, J.; Schmutz, J.; Rabbi, I.Y.; Egesi, C.; et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 2016, 34, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Chen, P.Y.; Huang, M.D.; Tsou, C.H.; Jane, W.N.; Huang, A.H. Tandem oleosin genes in a cluster acquired in Brassicaceae created tapetosomes and conferred additive benefit of pollen vigor. Proc. Natl. Acad. Sci. USA 2013, 110, 14480–14485. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Yang, J.H. Genomic analysis of Dof transcription factors in Hevea brasiliensis, a rubber-producing tree. Ind. Crops Prod. 2019, 134, 271–283. [Google Scholar] [CrossRef]
- Zou, Z.; Zheng, Y.J.; Xie, Z.N. Analysis of Carica papaya informs lineage-specific evolution of the aquaporin (AQP) family in Brassicales. Plants 2023, 12, 3847. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, J.H.; Zhang, X.C. Insights into genes encoding respiratory burst oxidase homologs (RBOHs) in rubber tree (Hevea brasiliensis Muell. Arg.). Ind. Crops Prod. 2019, 128, 126–139. [Google Scholar] [CrossRef]
- Zou, Z.; Zhao, Y.G.; Zhang, L.; Xiao, Y.H.; Guo, A.P. Analysis of Cyperus esculentus SMP family genes reveals lineage-specific evolution and seed desiccation-like transcript accumulation during tuber maturation. Ind. Crops Prod. 2022, 187, 115382. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 3, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Zou, Z.; Li, M.Y.; Jia, R.Z.; Zhao, H.; He, P.P.; Zhang, Y.L.; Guo, A.P. Genes encoding light-harvesting chlorophyll a/b-binding proteins in papaya (Carica papaya L.) and insight into lineage-specific evolution in Brassicaceae. Gene 2020, 748, 144685. [Google Scholar] [CrossRef]
- Xu, Y.G.; Zou, Z.; Guo, J.Y.; Kong, H.; Zhu, G.P.; Guo, A.P. Cloning and functional analysis of CpMGT1, a magnesium transporter gene from Carica papaya. Chin. J. Trop. Crop. 2022, 43, 1114–1121. [Google Scholar]
- Zou, Z.; Gong, J.; An, F.; Xie, G.S.; Wang, J.K.; Mo, Y.Y.; Yang, L.F. Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genom. 2015, 16, 1001. [Google Scholar] [CrossRef]
| Gene Name | Locus | Position | Intron No. | AA | MW (kDa) | pI | GRAVY | AI | Duplicate | Mode | Oleosin Location | Clade |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AtrOLE1 | AmTrH2.13G041800 | Chr13:8312320..8313509(−) | 0 | 168 | 17.80 | 10.31 | 0.263 | 105.71 | - | - | 42..154 | U |
| AtrOLE2 | AmTrH2.05G030700 | Chr5:5790540..5791413(+) | 0 | 147 | 15.51 | 9.36 | 0.349 | 105.58 | AtrOLE1 | Dispersed | 22..134 | M |
| AtrOLE3 | AmTrH2.13G011500 | Chr13:2034840..2035253(−) | 0 | 137 | 14.07 | 9.75 | 0.411 | 103.43 | AtrOLE1 | Dispersed | 22..134 | SL |
| AtrOLE4 | AmTrH2.03G086400 | Chr3:27204354..27205476(+) | 0 | 150 | 15.56 | 9.36 | 0.365 | 104.60 | AtrOLE5 | Dispersed | 21..136 | SH |
| AtrOLE5 | AmTrH2.10G130400 | Chr10:44382338..44383409(−) | 0 | 140 | 14.75 | 9.94 | 0.531 | 103.14 | AtrOLE3 | Dispersed | 17..131 | N |
| PaOLE1 | g26506 | Chr5:7463879..7464397(−) | 0 | 172 | 17.99 | 10.01 | 0.294 | 100.41 | - | - | 47..157 | U |
| PaOLE2 | g9736 | Chr7:51190093..51190608(+) | 0 | 171 | 17.75 | 10.00 | 0.322 | 97.02 | PaOLE1 | WGD | 46..157 | U |
| PaOLE3 | g12771 | Chr2:32085665..32086144(+) | 0 | 159 | 17.47 | 9.74 | 0.211 | 98.81 | PaOLE1 | Dispersed | 19..126 | M |
| AcOLE1 | Aqcoe3G048300 | Chr3:3052078..3052669(+) | 0 | 167 | 17.95 | 9.67 | 0.257 | 99.88 | - | - | 41..153 | U |
| AcOLE2 | Aqcoe7G144100 | Chr7:9197082..9198218(−) | 0 | 150 | 16.14 | 9.70 | 0.112 | 94.33 | AcOLE1 | Dispersed | 24..135 | M |
| AcOLE3 | Aqcoe3G267500 | Chr3:31392522..31393370(+) | 0 | 146 | 15.38 | 9.30 | 0.482 | 112.81 | AcOLE1 | Dispersed | 27..137 | SL |
| AcOLE4 | Aqcoe7G093500 | Chr7:5627997..5628401(−) | 0 | 134 | 13.90 | 10.02 | 0.516 | 108.43 | AcOLE3 | Dispersed | 23..119 | SL |
| AcOLE5 | Aqcoe3G202700 | Chr3:21602227..21603086(−) | 0 | 171 | 18.12 | 9.39 | 0.116 | 95.85 | AcOLE3 | Dispersed | 35..158 | SH |
| CpOLE1 | sunset09G0006960 | Chr9:5118166..5118919(−) | 0 | 167 | 18.08 | 9.84 | 0.396 | 99.76 | - | - | 41..152 | U |
| CpOLE2 | sunset07G0007350 | Chr7:6423723..6424251(+) | 0 | 131 | 13.68 | 9.56 | 0.422 | 108.85 | CpOLE2 | Transposed | 17..125 | M |
| CpOLE3 | sunset09G0008730 | Chr9:6575284..6575789(−) | 1 | 136 | 14.14 | 9.89 | 0.347 | 104.78 | CpOLE1 | Dispersed | 15..127 | SL |
| CpOLE4 | sunset01G0003770 | Chr1:3107234..3107629(−) | 0 | 131 | 13.56 | 10.89 | 0.675 | 122.82 | CpOLE4 | Dispersed | 26..129 | SH |
| CpOLE5 | sunset09G0012790 | Chr9:14063375..14064171(+) | 0 | 149 | 15.96 | 10.34 | 0.169 | 106.71 | CpOLE3 | Dispersed | 28..138 | SH |
| CpOLE6 | sunset04G0023010 | Chr4:30227031..30227636(+) | 0 | 145 | 14.83 | 5.56 | 0.720 | 117.72 | CpOLE3 | Dispersed | 27..104 | N |
| MoOLE1 | - | Scf12:425030..425509(+) | 0 | 159 | 17.39 | 10.00 | 0.348 | 94.47 | - | - | 33..145 | U |
| MoOLE2 | GLEAN_10017149 | Scf5:3253661..3254092(+) | 0 | 143 | 15.25 | 9.23 | 0.344 | 100.35 | MoOLE1 | Dispersed | 21..132 | M |
| MoOLE3 | GLEAN_10002091 | Scf132:402521..407622(+) | 1 | 137 | 14.64 | 9.89 | 0.397 | 112.48 | MoOLE1 | Dispersed | 17..127 | SL |
| MoOLE4 | GLEAN_10017990 | Scf4:3104843..3105331(+) | 0 | 162 | 16.81 | 10.28 | 0.355 | 107.22 | MoOLE4 | γ WGD | 37..149 | SH |
| MoOLE5 | GLEAN_10007003 | Scf35:698143..698517(−) | 0 | 124 | 13.15 | 9.95 | 0.784 | 121.13 | MoOLE6 | Dispersed | 29..116 | SH |
| MoOLE6 | GLEAN_10005491 | Scf65:559782..564316(+) | 0 | 267 | 28.01 | 6.07 | 0.471 | 98.58 | MoOLE3 | Dispersed | 25..123 171..265 | N |
| BsOLE1 | BsiG0022789 | Chr4:24719773..24720252(−) | 0 | 159 | 17.43 | 9.84 | 0.394 | 102.45 | - | - | 33..145 | U |
| BsOLE2 | BsiG0031356 | Chr5:115702358..115702837(+) | 0 | 159 | 17.55 | 9.69 | 0.424 | 106.73 | BsOLE1 | Dispersed | 33..145 | U |
| BsOLE3 | BsiG0027711 | Chr5:12196723..12197160(+) | 0 | 145 | 15.60 | 9.52 | 0.239 | 98.97 | BsOLE1 | Dispersed | 21..131 | M |
| BsOLE4 | BsiG0023505 | Chr4:38309092..38309529(−) | 0 | 145 | 15.48 | 9.55 | 0.374 | 106.28 | BsOLE3 | α WGD | 21..132 | M |
| BsOLE5 | BsiG0007300 | Chr1:161375811..161376341(−) | 1 | 139 | 14.61 | 9.77 | 0.397 | 103.17 | BsOLE1 | Dispersed | 18..128 | SL |
| BsOLE6 | BsiG0026540 | Chr4:140933592..140934122(−) | 1 | 135 | 14.12 | 9.52 | 0.400 | 105.48 | BsOLE5 | α WGD | 13..124 | SL |
| BsOLE7 | BsiG0026541 | Chr4:140939897..140940427(−) | 1 | 135 | 14.08 | 9.52 | 0.417 | 107.63 | BsOLE6 | Tandem | 13..124 | SL |
| BsOLE8 | BsiG0004876 | Chr1:127499323..127499826(−) | 0 | 167 | 17.89 | 9.39 | 0.180 | 107.49 | BsOLE5 | Dispersed | 38..147 | SH |
| BsOLE9 | BsiG0006900 | Chr1:156712314..156712805(+) | 0 | 163 | 17.58 | 9.51 | -0.144 | 90.98 | BsOLE8 | γ WGD | 34..138 | SH |
| BsOLE10 | BsiG0025867 | Chr4:130692072..130692554(−) | 0 | 160 | 17.10 | 9.97 | 0.078 | 104.25 | BsOLE9 | α WGD | 23..133 | SH |
| CsOLE1 | Cs02G002030 | Chr2:9103395..9103838(+) | 0 | 147 | 15.81 | 9.35 | 0.563 | 112.18 | - | - | 26..133 | U |
| CsOLE2 | Cs15G003740 | Chr15:2440935..2441444(+) | 0 | 146 | 15.87 | 9.68 | 0.205 | 88.90 | CsOLE1 | Dispersed | 21..132 | M |
| CsOLE3 | Cs12G001310 | Chr12:683951..684453(+) | 1 | 115 | 11.92 | 11.00 | 0.738 | 123.83 | CsOLE1 | Dispersed | 16..114 | SL |
| CsOLE4 | Cs06G005610 | Chr6:16853769..16854634(−) | 1 | 149 | 15.51 | 9.99 | 0.170 | 94.36 | CsOLE3 | β WGD | 21..132 | SL |
| CsOLE5 | Cs02G003290 | Chr2:9784653..9785174(+) | 1 | 134 | 14.18 | 10.20 | 0.352 | 108.51 | CsOLE3 | α WGD | 16..128 | SL |
| CsOLE6 | Cs01G009580 | Chr1:23015766..23016309(−) | 1 | 149 | 15.40 | 9.59 | 0.358 | 110.13 | CsOLE7 | α WGD | 34..147 | SH |
| CsOLE7 | Cs02G004630 | Chr2:10712950..10713929(−) | 1 | 161 | 16.81 | 9.69 | 0.103 | 99.44 | CsOLE3 | Dispersed | 32..148 | SH |
| CsOLE8 | Cs14G006880 | Chr14:6118834..6119582(+) | 1 | 152 | 16.32 | 9.69 | 0.245 | 100.07 | CsOLE7 | β WGD | 29..140 | SH |
| CvOLE1 | Clevi.0032s0439 | Scf32:644486..645807(−) | 1 | 159 | 17.04 | 9.75 | 0.383 | 103.14 | - | - | 38..145 | M |
| CvOLE2 | Clevi.0001s1658 | Scf1:3646632..3647045(−) | 0 | 137 | 14.65 | 9.61 | 0.412 | 104.01 | CvOLE1 | Dispersed | 19..128 | SL |
| CvOLE3 | Clevi.0015s0023 | Scf15:3700181..3701021(−) | 1 | 143 | 14.98 | 10.20 | 0.262 | 96.22 | CvOLE1 | Dispersed | 19..130 | SL |
| CvOLE4 | Clevi.0004s1912 | Scf4:2316861..2318229(+) | 1 | 157 | 16.42 | 9.98 | 0.297 | 104.33 | CvOLE6 | Dispersed | 32..144 | SH |
| CvOLE5 | Clevi.0042s0814 | Scf42:1538196..1539122(−) | 1 | 161 | 16.79 | 9.69 | 0.441 | 106.02 | CvOLE4 | γ WGD | 35..147 | SH |
| CvOLE6 | Clevi.0015s0551 | Scf15:942332..943312(−) | 0 | 145 | 14.89 | 5.25 | 0.560 | 106.34 | CvOLE3 | Dispersed | 27..132 | N |
| GgOLE1 | GG13G018590 | Chr13:9683189 9684092(+) | 1 | 164 | 17.30 | 9.57 | 0.410 | 104.15 | - | - | 43..150 | U |
| GgOLE2 | GG05G000440 | Chr5:273726 274730(−) | 1 | 162 | 17.16 | 9.41 | 0.446 | 107.72 | GgOLE1 | Dispersed | 41..148 | U |
| GgOLE3 | GG07G021290 | Chr7:10989271 10989687(+) | 0 | 138 | 14.79 | 9.72 | 0.442 | 110.22 | GgOLE1 | Dispersed | 19..128 | M |
| GgOLE4 | GG02G144870 | Chr2:66911747 66912474(+) | 1 | 144 | 15.05 | 10.20 | 0.272 | 96.94 | GgOLE1 | Dispersed | 19..130 | SL |
| GgOLE5 | GG05G049880 | Chr5:23864915 23865662(+) | 1 | 159 | 16.78 | 9.89 | 0.302 | 104.91 | GgOLE7 | Dispersed | 34..146 | SH |
| GgOLE6 | GG15G098790 | Chr15:45889738 45890318(−) | 1 | 161 | 16.77 | 9.52 | 0.406 | 109.01 | GgOLE5 | Dispersed | 35..147 | SH |
| GgOLE7 | - | Chr6:796424..801985(−) | 1 | 139 | 14.63 | 4.43 | 0.609 | 110.79 | GgOLE4 | Dispersed | 26..124 | N |
| ThOLE1 | LOC104821850 | Scf34:1318549..1319624(−) | 1 | 155 | 16.50 | 9.63 | 0.665 | 113.81 | - | - | 34..141 | U |
| ThOLE2 | LOC104819676 | Scf3:6116766..6117891(−) | 1 | 156 | 16.49 | 9.39 | 0.485 | 105.64 | ThOLE1 | Dispersed | 35..142 | U |
| ThOLE3 | LOC104818593 | Scf3:633964..634782(−) | 0 | 138 | 14.70 | 9.56 | 0.449 | 106.81 | ThOLE1 | Dispersed | 19..128 | M |
| ThOLE4 | LOC104825056 | Scf42:463230..464045(+) | 1 | 144 | 15.05 | 10.20 | 0.332 | 102.99 | ThOLE1 | Dispersed | 19..130 | SL |
| ThOLE5 | LOC104811538 | Scf2:1261264..1262172(−) | 1 | 144 | 15.22 | 9.90 | 0.273 | 100.28 | ThOLE4 | α WGD | 22..133 | SL |
| ThOLE6 | LOC104805374 | Scf11:1401936..1403042(+) | 1 | 159 | 16.89 | 9.89 | 0.177 | 98.74 | ThOLE8 | Dispersed | 34..146 | SH |
| ThOLE7 | LOC104802395 | Scf8:1757388..1758247(+) | 1 | 161 | 16.80 | 9.69 | 0.455 | 110.81 | ThOLE6 | β WGD | 35..147 | SH |
| ThOLE8 | LOC104811693 | Scf2:1907125..1907884(+) | 0 | 142 | 14.50 | 6.56 | 0.492 | 104.44 | ThOLE4 | Dispersed | 28..99 | N |
| Gene1 | Gene2 | Identity (%) | Ks | Ka/Ks |
|---|---|---|---|---|
| PaOLE1 | PaOLE2 | 77.8 | 0.8501 | 0.1372 |
| MoOLE4 | MoOLE5 | 57.7 | 2.0410 | 0.1831 |
| BsOLE3 | BsOLE4 | 89.3 | 0.2700 | 0.2629 |
| BsOLE5 | BsOLE6 | 89.0 | 0.1962 | 0.2749 |
| BsOLE8 | BsOLE9 | 55.0 | 1.5864 | 0.9415 |
| BsOLE9 | BsOLE10 | 76.6 | 0.2810 | 0.2839 |
| CsOLE3 | CsOLE4 | 54.9 | 1.6273 | 0.1248 |
| CsOLE3 | CsOLE5 | 76.5 | 0.3691 | 0.1054 |
| CsOLE6 | CsOLE7 | 75.8 | 0.5869 | 0.1216 |
| CsOLE7 | CsOLE8 | 60.3 | - | - |
| CvOLE4 | CvOLE5 | 64.0 | 1.9437 | 0.1515 |
| ThOLE4 | ThOLE5 | 86.0 | 0.3409 | 0.1610 |
| ThOLE6 | ThOLE7 | 65.6 | 1.5677 | 0.1797 |
| At-Sm1 | At-Sm2 | 66.9 | 1.3093 | 0.1880 |
| At-S3 | At-S5 | 58.2 | 1.3683 | 0.1592 |
| At-S1 | At-S4 | 53.1 | - | - |
| At-S2 | At-S4 | 61.8 | 1.5782 | 0.1625 |
| PtOLE2a | PtOLE2b | 75.1 | 0.3138 | 0.5186 |
| PtOLE3a | PtOLE3b | 86.2 | 0.1619 | 0.8850 |
| PtOLE4a | PtOLE4b | 90.7 | 0.2091 | 0.3161 |
| PtOLE5a | PtOLE5b | 84.0 | 0.3696 | 0.2675 |
| MeOLE1a | MeOLE1b | 81.1 | 0.7428 | 0.1198 |
| MeOLE3a | MeOLE3b | 76.7 | 0.6126 | 0.2047 |
| MeOLE4a | MeOLE4b | 78.1 | 0.4175 | 0.3827 |
| MeOLE4b | MeOLE5 | 59.6 | 1.7862 | 0.1548 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Z.; Zhang, L.; Zhao, Y. Integrative Analysis of Oleosin Genes Provides Insights into Lineage-Specific Family Evolution in Brassicales. Plants 2024, 13, 280. https://doi.org/10.3390/plants13020280
Zou Z, Zhang L, Zhao Y. Integrative Analysis of Oleosin Genes Provides Insights into Lineage-Specific Family Evolution in Brassicales. Plants. 2024; 13(2):280. https://doi.org/10.3390/plants13020280
Chicago/Turabian StyleZou, Zhi, Li Zhang, and Yongguo Zhao. 2024. "Integrative Analysis of Oleosin Genes Provides Insights into Lineage-Specific Family Evolution in Brassicales" Plants 13, no. 2: 280. https://doi.org/10.3390/plants13020280
APA StyleZou, Z., Zhang, L., & Zhao, Y. (2024). Integrative Analysis of Oleosin Genes Provides Insights into Lineage-Specific Family Evolution in Brassicales. Plants, 13(2), 280. https://doi.org/10.3390/plants13020280







