Genetic Diversity and Population Structure of Endangered Orchid Cypripedium flavum in Fragmented Habitat Using Fluorescent AFLP Markers
Abstract
:1. Introduction
2. Results
2.1. AFLP Polymorphism
2.2. Genetic Diversity
2.3. Molecular Variance Analysis
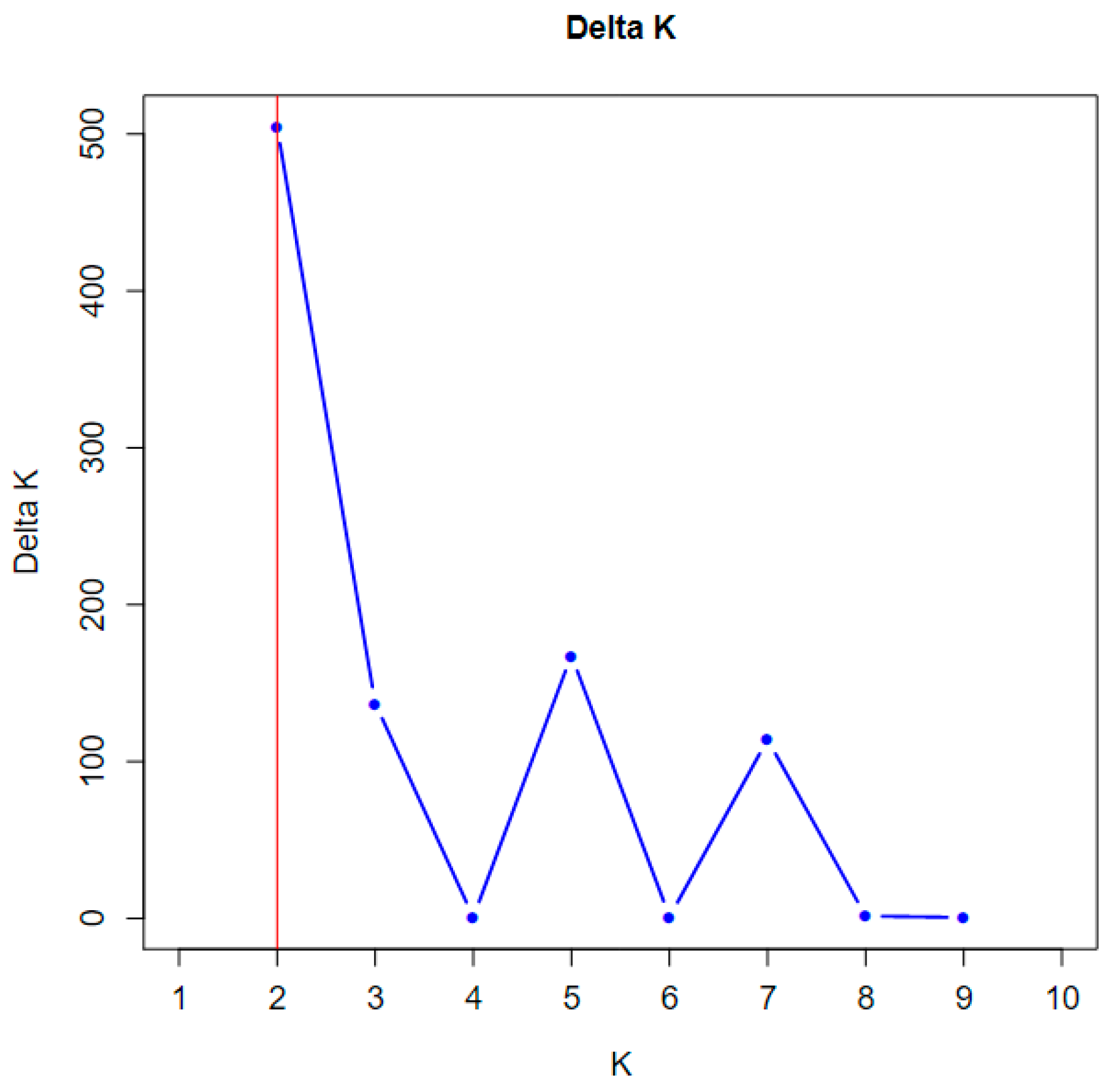
2.4. Genetic Structure of C. Flavum
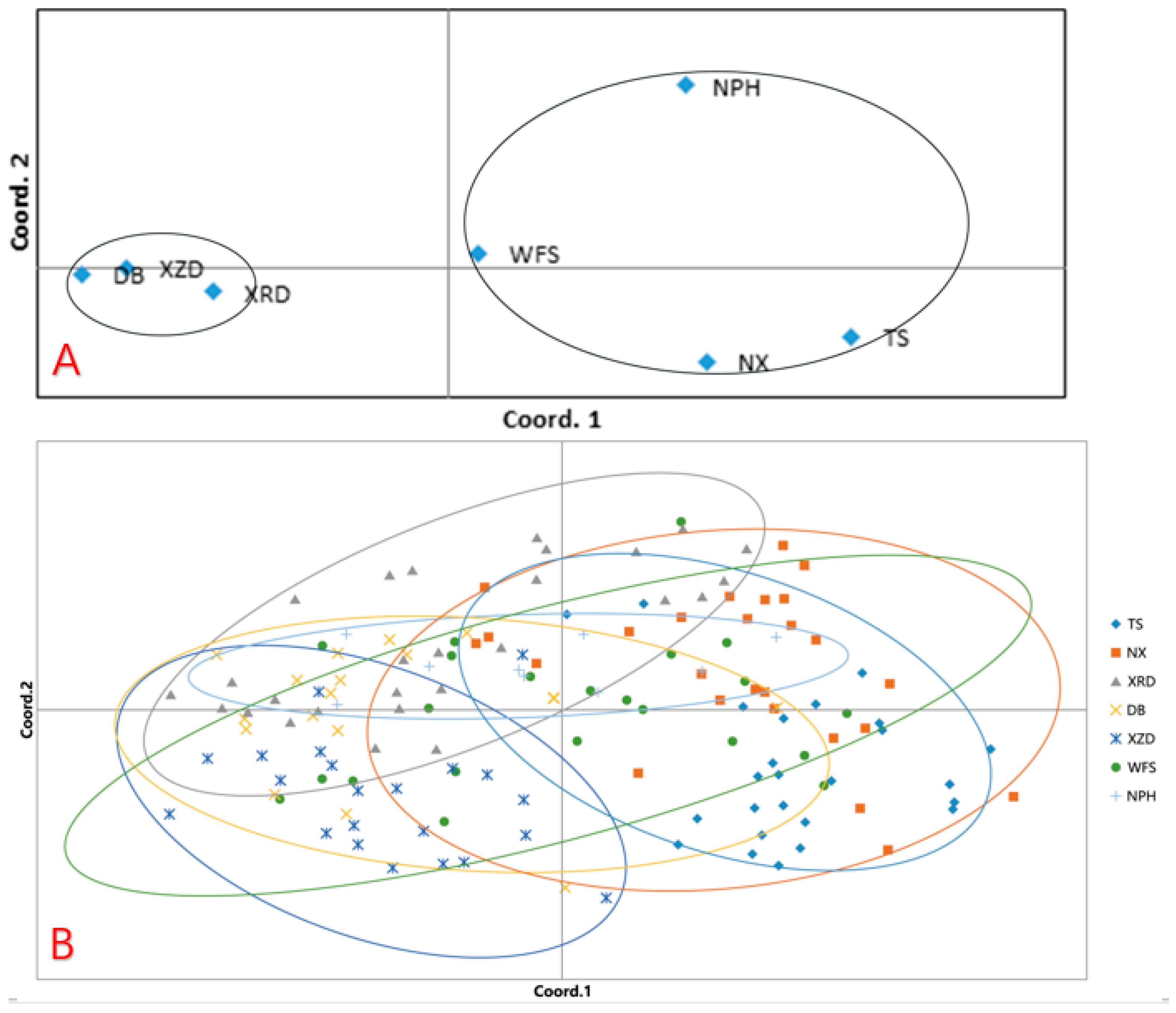
2.5. Genetic Relationship of C. Flavum
3. Discussion
3.1. Genetic Diversity
3.2. Genetic Structure and Population Differentiation
3.3. Effects of Habitat Fragmentation
4. Materials and Methods
4.1. Plant Material and Study Sites
4.2. DNA Extraction and AFLP Analysis
4.3. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, D.Q.; Zhou, N. Genetic diversity and population structure of the endangered conifer Taxus wallichiana var. mairei (Taxaceae) revealed by Simple Sequence Repeat (SSR) markers. Biochem. Syst. Ecol. 2013, 49, 107–114. [Google Scholar] [CrossRef]
- Minh Nguyen, D.; Lan Phan Nguyen, H.; Minh Nguyen, T. Genetic structure of the endemic Dipterocarpus condorensis revealed by microsatellite markers. AoB Plants 2022, 14, plac007. [Google Scholar] [CrossRef] [PubMed]
- Reed, D.H.; Frankham, R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Booy, G.; Hendriks, R.J.J.; Smulders, M.J.M.; van Groenendael, J.M.; Vosman, B. Genetic diversity and the survival of populations. Plant Biol. 2000, 2, 379–395. [Google Scholar] [CrossRef]
- Markert, J.A.; Champlin, D.M.; Gutjahr-Gobell, R.; Grear, J.S.; Kuhn, A.; McGreevy, T.J., Jr.; Roth, A.; Bagley, M.J.; Nacci, D.E. Population genetic diversity and fitness in multiple environments. BMC Evol. Biol. 2010, 10, 205. [Google Scholar] [CrossRef]
- Yamashita, Y.; Satoh, N.; Kurosawa, T.; Kaneko, S. Genetic diversity and structure of the endangered lady’s slipper orchid Cypripedium japonicum Thunb. (Orchidaceae) in Japan. Popul. Ecol. 2023, 65, 54–63. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.L.; Pei, S.Y.; Ning, M.M.; Tang, S.Q. Genetic diversity and population structure of Camellia huana (Theaceae), a limestone species with narrow geographic range, based on chloroplast DNA sequence and microsatellite markers. Plant Divers. 2020, 42, 343–350. [Google Scholar] [CrossRef]
- Aguilar, R.; Quesada, M.; Ashworth, L.; Herrerias-Diego, Y.; Lobo, J. Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol. Ecol. 2008, 17, 5177–5188. [Google Scholar] [CrossRef]
- Mhemmed, G.; Kamel, H.; Chedly, A. Does habitat fragmentation reduce genetic diversity and genetic group connectivity? Ecography 2008, 31, 751–756. [Google Scholar] [CrossRef]
- Minasiewicz, J.; Znaniecka, J.M.; Górniak, M.; Kawiński, A. Spatial genetic structure of an endangered orchid Cypripedium calceolus (Orchidaceae) at a regional scale: Limited gene flow in a fragmented landscape. Conserv. Genet. 2018, 19, 1449–1460. [Google Scholar] [CrossRef]
- Lee, S.R.; Choi, T.Y.; Jung, S.Y. Genetic Diversity on a Rare Terrestrial Orchid, Habenaria linearifolia in South Korea: Implications for Conservation Offered by Genome-Wide Single Nucleotide Polymorphisms. Front. Plant Sci. 2022, 13, 772621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gale, S.W.; Li, J.H.; Fischer, G.A.; Ren, M.X.; Song, X.Q. Pollen-mediated gene flow ensures connectivity among spatially discrete sub-populations of Phalaenopsis pulcherrima, a tropical food-deceptive orchid. BMC Plant Biol. 2019, 19, 597. [Google Scholar] [CrossRef] [PubMed]
- Helsen, K.; Meekers, T.; Vranckx, G.; Roldan-Ruiz, I.; Vandepitte, K.; Honnay, O. A direct assessment of realized seed and pollen flow within and between two isolated populations of the food-deceptive orchid Orchis mascula. Plant Biol. 2016, 18, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Tang, M.; Bi, Y.; Yang, J. Genetic Diversity and Population Structure Analysis of Wild Cymbidium tortisepalum Based on Chloroplast DNA in Yunnan Province of China. J. Am. Soc. Hortic. Sci. 2021, 146, 424–434. [Google Scholar] [CrossRef]
- Xie, H.; Zhang, W.; Luo, H.; Yang, B.; Xiong, D. Study on Population Genetic Structure and Genetic Diversity of Cymbidium kanran from Jiangxi Province. Acta Bot. Boreali.-Occident. Sin. 2020, 40, 785–794. [Google Scholar]
- Torres, E.; Riofrio, M.L.; Iriondo, J.M. Complex fine-scale spatial genetic structure in Epidendrum rhopalostele: An epiphytic orchid. Heredity 2019, 122, 458–467. [Google Scholar] [CrossRef]
- Xie, H.M.; Peng, D.Z.; Chen, Y.R.; Luo, H.L.; Yang, B.Y.; Xiong, D.J. Genetic structure and differentiation of wild populations of Cymbidium goeringii (Rchb. f.) Rchb. f. in the main mountain range of Jiangxi Province, China. Plant Sci. J. 2020, 38, 123–133. [Google Scholar]
- Noh, S.W.; Park, J.-K.; Yu, J.S.; Nam, D.E.; Do, Y.; Chung, K.W. Genetic Diversity and Population Structure of the Spring Orchid Cymbidium goeringii in Korean Distant Islands. Diversity 2020, 12, 486. [Google Scholar] [CrossRef]
- Liang, H.Y.; Wang, X.G.; Chen, W.; Hu, S.N.; Yang, F.; Yang, X.T. Spatial genetic structure of terrestrial orchid Cymbidium faberi in the Qinling Mountains revealed by microsatellite loci. Plant Syst. Evol. 2021, 307, 5. [Google Scholar] [CrossRef]
- Huang, Y.; Li, F.; Chen, K. Analysis of diversity and relationships among Chinese orchid cultivars using EST-SSR markers. Biochem. Syst. Ecol. 2010, 38, 93–102. [Google Scholar] [CrossRef]
- Kurowska, M.; Labocha-Pawlowska, A.; Gnizda, D.; Maluszynski, M.; Szarejko, I. Molecular analysis of point mutations in a barley genome exposed to MNU and gamma rays. Mutat. Res. 2012, 738, 52–70. [Google Scholar] [CrossRef] [PubMed]
- Caballero, A.; Jesus Garcia-Pereira, M.; Quesada, H. Genomic distribution of AFLP markers relative to gene locations for different eukaryotic species. BMC Genom. 2013, 14, 528. [Google Scholar] [CrossRef] [PubMed]
- Vaculna, L.; Majesky, L.; Ali, T.; Seregin, A.P.; Prausova, R.; Kapler, A.; Iakushenko, D.; Thines, M.; Kitner, M. Genetic structure of endangered species Adenophora liliifolia and footprints of postglacial recolonisation in Central Europe. Conserv. Genet. 2021, 22, 1069–1084. [Google Scholar] [CrossRef]
- Nurmansyah; Alghamdi, S.S.; Migdadi, H.M.; Khan, M.A.; Afzal, M. AFLP-Based Analysis of Variation and Population Structure in Mutagenesis Induced Faba Bean. Diversity 2020, 12, 303. [Google Scholar] [CrossRef]
- Lazarevic, M.; Siljak-Yakovlev, S.; Sanino, A.; Niketic, M.; Lamy, F.; Hinsinger, D.D.; Tomovic, G.; Stevanovic, B.; Stevanovic, V.; Robert, T. Genetic Variability in Balkan Paleoendemic Resurrection Plants Ramonda serbica and R. nathaliae across Their Range and in the Zone of Sympatry. Front. Plant Sci. 2022, 13, 873471. [Google Scholar] [CrossRef]
- Shimura, H.; Koda, Y. Enhanced symbiotic seed germination of Cypripedium macranthos var. rebunense following inoculation after cold treatment. Physiol. Plant. 2005, 123, 281–287. [Google Scholar] [CrossRef]
- Chandra, N.; Singh, G.; Rai, I.D.; Mishra, A.P.; Kazmi, M.Y.; Pandey, A.; Jalal, J.S.; Costache, R.; Almohamad, H.; Al-Mutiry, M. Predicting distribution and range dynamics of three threatened Cypripedium species under climate change scenario in Western Himalaya. Forests 2023, 14, 633. [Google Scholar] [CrossRef]
- Jiang, H.; Kong, J.J.; Chen, H.C.; Xiang, Z.Y.; Zhang, W.P.; Han, Z.D.; Liao, P.C.; Lee, Y.I. Cypripedium subtropicum (Orchidaceae) employs aphid colony mimicry to attract hoverfly (Syrphidae) pollinators. New Phytol. 2020, 227, 1213–1221. [Google Scholar] [CrossRef]
- Zheng, G.; Li, P.; Pemberton, R.; Luo, Y. Mixed bumblebee and blowfly pollination of Cypripedium flavum (Orchidaceae) in Sichuan, China. Ecol. Res. 2011, 26, 453–459. [Google Scholar] [CrossRef]
- Luo, Y.B.; Jia, J.S.; Wang, C.L. A general review of the conservation status of Chinese orchids. Biodivers. Sci. 2003, 11, 70–77. [Google Scholar]
- Jacquemyn, H.; Vandepitte, K.; Brys, R.; Honnay, O.; Roldan-Ruiz, I. Fitness variation and genetic diversity in small, remnant populations of the food deceptive orchid Orchis purpurea. Biol. Conserv. 2007, 139, 203–210. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- White, T.L.; Adams, W.T.; Neale, D.B.; White, T.L.; Adams, W.T.; Neale, D.B. Forest Genetics; CABI: Washington, DC, USA, 2007; 682p. [Google Scholar]
- Vinson, C.C.; Mangaravite, E.; Sebbenn, A.M.; Lander, T.A. Using molecular markers to investigate genetic diversity, mating system and gene flow of Neotropical trees. Braz. J. Bot. 2018, 41, 481–496. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, M.; Shi, S.; Kang, Y.; Liu, J. Analysis of Genetic Structure of Magnolia sprengeri Populations Based on ISSR Markers. Sci. Silvae Sin. 2014, 50, 76–81. [Google Scholar]
- Lu, X.; Luan, S.; Kong, J.; Hu, L.; Mao, Y.; Zhong, S. Genome-wide mining, characterization, and development of microsatellite markers in Marsupenaeus japonicus by genome survey sequencing. Chin. J. Oceanol. Limnol. 2017, 35, 203–214. [Google Scholar] [CrossRef]
- Jiang, Y.W.; Sun, X.Q.; Luo, H.L.; Yang, B.Y.; Xiong, D.J. Studies on genetic diversity of Cymbidium kanran populations from the main mountains in Jiangxi Province based on ISSR marker. Acta Hortic. Sin. 2017, 44, 1993–2000. [Google Scholar]
- Pandey, M.; Sharma, J. Characterization of microsatellite loci for Cypripedium kentuckiense (Orchidaceae). Conserv. Genet. Resour. 2013, 5, 1031–1033. [Google Scholar] [CrossRef]
- Hamasha, H.; Schmidt-Lebuhn, A.; Durka, W.; Schleuning, M.; Hensen, I. Bioclimatic regions influence genetic structure of four Jordanian Stipa species. Plant Biol. 2013, 15, 882–891. [Google Scholar] [CrossRef]
- Luo, Y.; Qin, W.; Yan, Y.; Yin, K.; Zang, R.; Du, F.K. Climate change vulnerability and conservation strategies for tertiary relict tree species: Insights from landscape genomics of Taxus cuspidata. Evol. Appl. 2024, 17, e13686. [Google Scholar] [CrossRef]
- Münzbergová, Z.; Cousins, S.A.; Herben, T.; Plačková, I.; Mildén, M.; Ehrlén, J. Historical habitat connectivity affects current genetic structure in a grassland species. Plant Biol. 2013, 15, 195–202. [Google Scholar] [CrossRef]
- Wang, W.; Ren, G.; He, Y.; Zhu, J. Habitat degradation and conservation status assessment of Gallinaceous birds in the Trans-Himalayas, China. J. Wildl. Manag. 2008, 72, 1335–1341. [Google Scholar] [CrossRef]
- Chung, M.Y.; Lopez-Pujol, J.; Moon, M.O.; Maki, M.; Yukawa, T.; Sugiura, N.; Chung, M.G. Population history of the terrestrial orchid Cremastra appendiculata var. variabilis from Korea, inferred from levels and distribution of genetic diversity. Bot. J. Linn. Soc. 2013, 173, 721–732. [Google Scholar] [CrossRef]
- Zettler, J.A.; Adams, K.; Frederick, B.; Gutting, A.; Ingebretsen, N.; Ragsdale, A.; Schrey, A. Genetic structure of Pseudococcus microcirculus (Hemiptera: Pseudococcidae) populations on epiphytic orchids in south Florida. J. Genet. 2017, 96, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tang, M.; Bi, Y. Nuclear genetic diversity and population structure of a vulnerable and endemic orchid (Cymbidium tortisepalum) in Northwestern Yunnan, China. Sci. Hortic. 2017, 219, 22–30. [Google Scholar] [CrossRef]
- Ross-Ibarra, J.; Tenaillon, M.; Gaut, B.S. Historical Divergence and Gene Flow in the Genus Zea. Genetics 2009, 181, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Schlaepfer, D.R.; Braschler, B.; Rusterholz, H.P.; Baur, B. Genetic effects of anthropogenic habitat fragmentation on remnant animal and plant populations: A meta-analysis. Ecosphere 2018, 9, e02488. [Google Scholar] [CrossRef]
- Swarts, N.D.; Sinclair, E.A.; Krauss, S.L.; Dixon, K.W. Genetic diversity in fragmented populations of the critically endangered spider orchid Caladenia huegelii: Implications for conservation. Conserv. Genet. 2009, 10, 1199–1208. [Google Scholar] [CrossRef]
- Le Cadre, S.; Tully, T.; Mazer, S.J.; Ferdy, J.B.; Moret, J.; Machon, N. Allee effects within small populations of Aconitum napellus ssp. lusitanicum, a protected subspecies in northern France. New Phytol. 2008, 179, 1171–1182. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Roldan-Ruiz, I.; Dendauw, J.; Bockstaele, E.V.; Depicker, A.; Loose, M.D. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 2000, 6, 125–134. [Google Scholar] [CrossRef]
- Kosman, E. Nei’s gene diversity and the index of average differences are identical measures of diversity within populations. Plant Pathol. 2003, 52, 533–535. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]


| Primer Code | Total Alleles | Total No. of PCR Products | Average Band per Allele | PPB(%) | PIC | Discrimination Power |
|---|---|---|---|---|---|---|
| EA4MC1 | 74 | 4770 | 65.46 | 100 | 0.490 | 19.68 |
| EA4MC4 | 74 | 4894 | 60.42 | 100 | 0.493 | 19.68 |
| EA4MC6 | 81 | 5310 | 68.96 | 100 | 0.492 | 21.54 |
| EA7MC1 | 77 | 4968 | 70.97 | 100 | 0.490 | 20.48 |
| EA8MC5 | 70 | 4683 | 66.90 | 100 | 0.494 | 18.62 |
| Average | 75.20 | 4925 | 66.54 | 100 | 0.492 | 20.00 |
| Total | 376 | 24652 | - | - | - |
| Population | Sample Size | Na | Ne | I | H | PPB | Mu |
|---|---|---|---|---|---|---|---|
| TS | 25 | 1.926 | 1.575 | 0.498 | 0.334 | 94.15 | 0.348 |
| NX | 26 | 1.949 | 1.613 | 0.525 | 0.354 | 96.01 | 0.368 |
| XRD | 28 | 1.949 | 1.596 | 0.518 | 0.347 | 96.28 | 0.360 |
| DB | 17 | 1.904 | 1.567 | 0.494 | 0.331 | 92.29 | 0.352 |
| XZD | 22 | 1.923 | 1.550 | 0.486 | 0.324 | 94.15 | 0.339 |
| WFS | 22 | 1.976 | 1.610 | 0.529 | 0.355 | 98.14 | 0.372 |
| NPH | 10 | 1.771 | 1.567 | 0.489 | 0.329 | 88.03 | 0.366 |
| Total | 150 | 1.914 | 1.583 | 0.505 | 0.339 | 94.15 | 0.358 |
| POPGEN | AMOVA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | Fis | Fit | Fst | Nm | Source of Variation | df | SSD | MSD | Variance Components | Total Variance |
| 150 | 0.3258 | 0.2966 | 0.0896 | 5.0826 | Among populations | 6 | 1224.904 | 203.651 | 6.443 | 8.75% |
| Within populations | 143 | 9612.236 | 67.218 | 67.218 | 91.25% | |||||
| TS | NX | XRD | DB | XZD | WFS | NPH | |
|---|---|---|---|---|---|---|---|
| TS | 0.000 | ||||||
| NX | 0.052 | 0.000 | |||||
| XRD | 0.106 | 0.068 | 0.000 | ||||
| DB | 0.113 | 0.095 | 0.042 | 0.000 | |||
| XZD | 0.108 | 0.108 | 0.075 | 0.059 | 0.000 | ||
| WFS | 0.067 | 0.060 | 0.056 | 0.068 | 0.056 | 0.000 | |
| NPH | 0.114 | 0.121 | 0.112 | 0.118 | 0.125 | 0.076 | 0.000 |
| Population Code | Altitude(m) | Sample Size | Geographic Coordinates | Habitat Characters |
|---|---|---|---|---|
| TS | 3460 | 25 | 27°47′53″ N, 99° 49′ 58″ E | Northeast slope; Shrubbery |
| NX | 3180 | 26 | 27°58′ 15″ N, 99°34′ 52″ E | Northwest slope; Popular forest |
| XRD | 3460 | 28 | 27° 45′ 57″ N, 99° 39′ 02″ E | Northwest slope; Shrubbery |
| DB | 3170 | 17 | 27° 57′ 56″ N, 99° 35′ 11″ E | Northwest slope; Shrubbery |
| XZD | 3360 | 22 | 27° 28′ 54″ N, 99° 52′ 51″ E | Northwest slope; Fir forest |
| WFS | 3240 | 22 | 27° 47′ 37″ N, 99° 42′ 46″ E | Northwest slope; Shrubbery |
| NPH | 3260 | 10 | 27° 54′ 23″ N, 99° 38′ 13″ E | Valley; Sparse forest |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Wang, M.; Yan, X.; Cheng, X. Genetic Diversity and Population Structure of Endangered Orchid Cypripedium flavum in Fragmented Habitat Using Fluorescent AFLP Markers. Plants 2024, 13, 2851. https://doi.org/10.3390/plants13202851
Hu S, Wang M, Yan X, Cheng X. Genetic Diversity and Population Structure of Endangered Orchid Cypripedium flavum in Fragmented Habitat Using Fluorescent AFLP Markers. Plants. 2024; 13(20):2851. https://doi.org/10.3390/plants13202851
Chicago/Turabian StyleHu, Shijun, Meizhen Wang, Xiaohui Yan, and Xiaomao Cheng. 2024. "Genetic Diversity and Population Structure of Endangered Orchid Cypripedium flavum in Fragmented Habitat Using Fluorescent AFLP Markers" Plants 13, no. 20: 2851. https://doi.org/10.3390/plants13202851
APA StyleHu, S., Wang, M., Yan, X., & Cheng, X. (2024). Genetic Diversity and Population Structure of Endangered Orchid Cypripedium flavum in Fragmented Habitat Using Fluorescent AFLP Markers. Plants, 13(20), 2851. https://doi.org/10.3390/plants13202851







