Genome-Wide Identification of the RALF Gene Family and Expression Pattern Analysis in Zea mays (L.) under Abiotic Stresses
Abstract
1. Introduction
2. Results
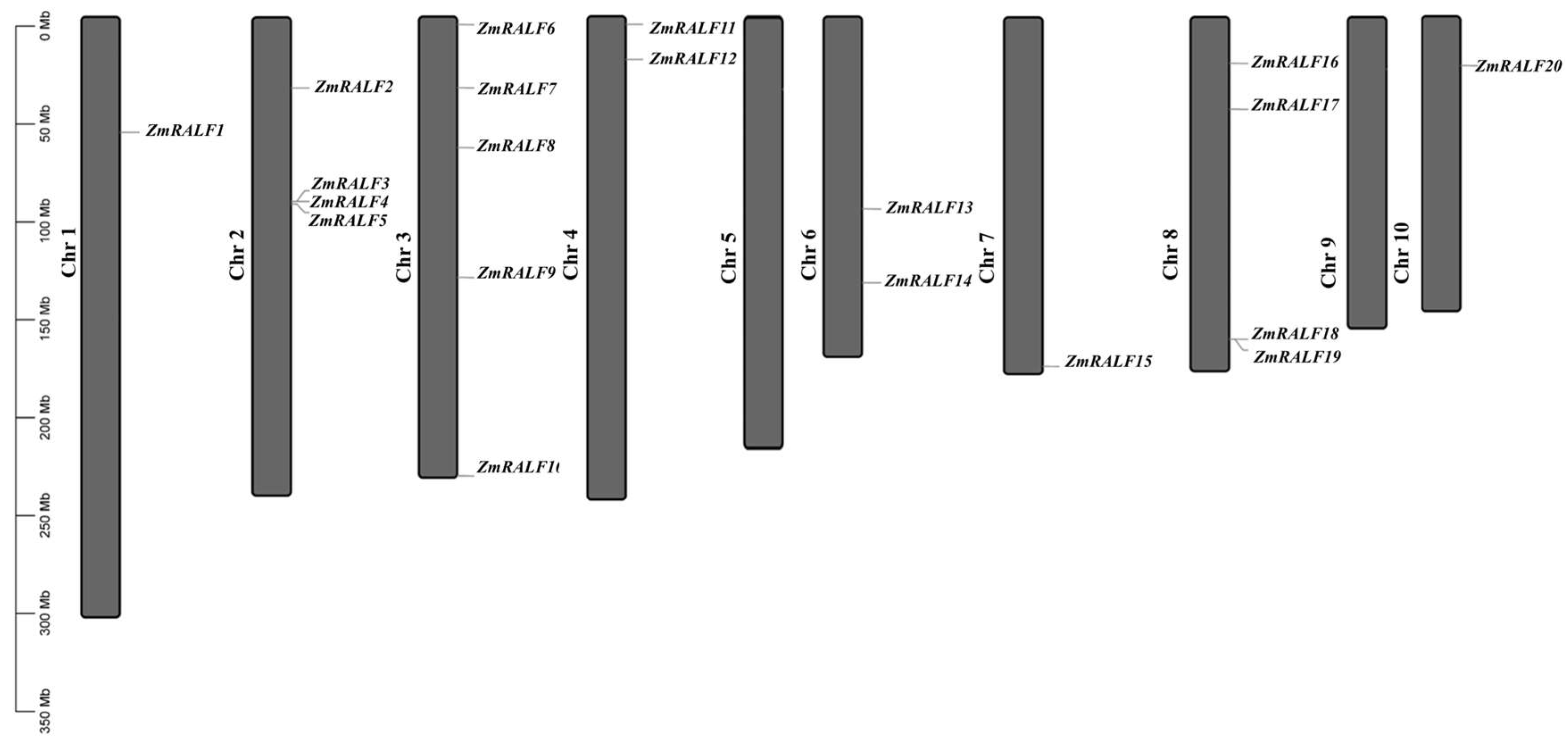
2.1. Whole-Genome Identification of RALF Genes in Maize
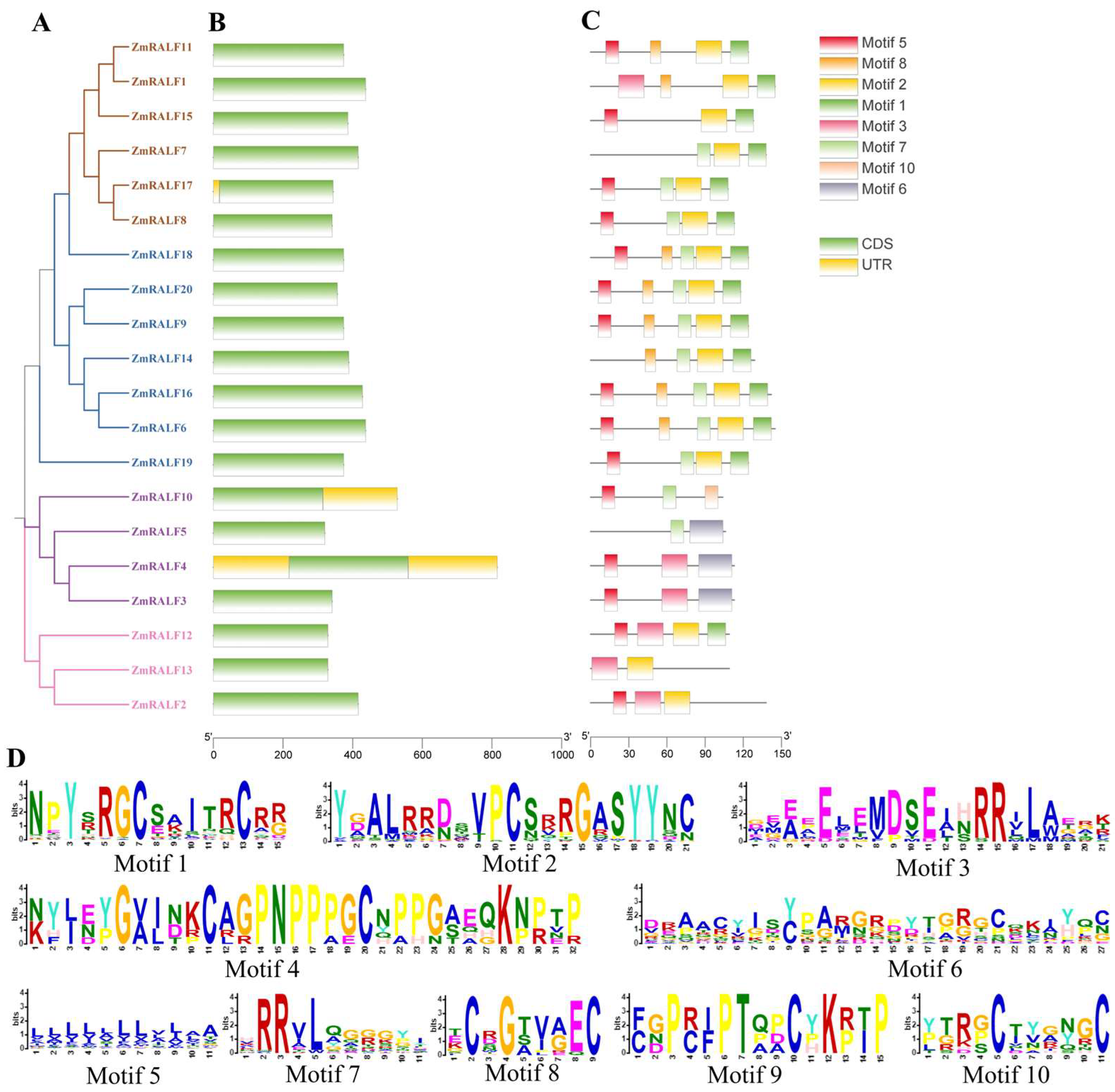
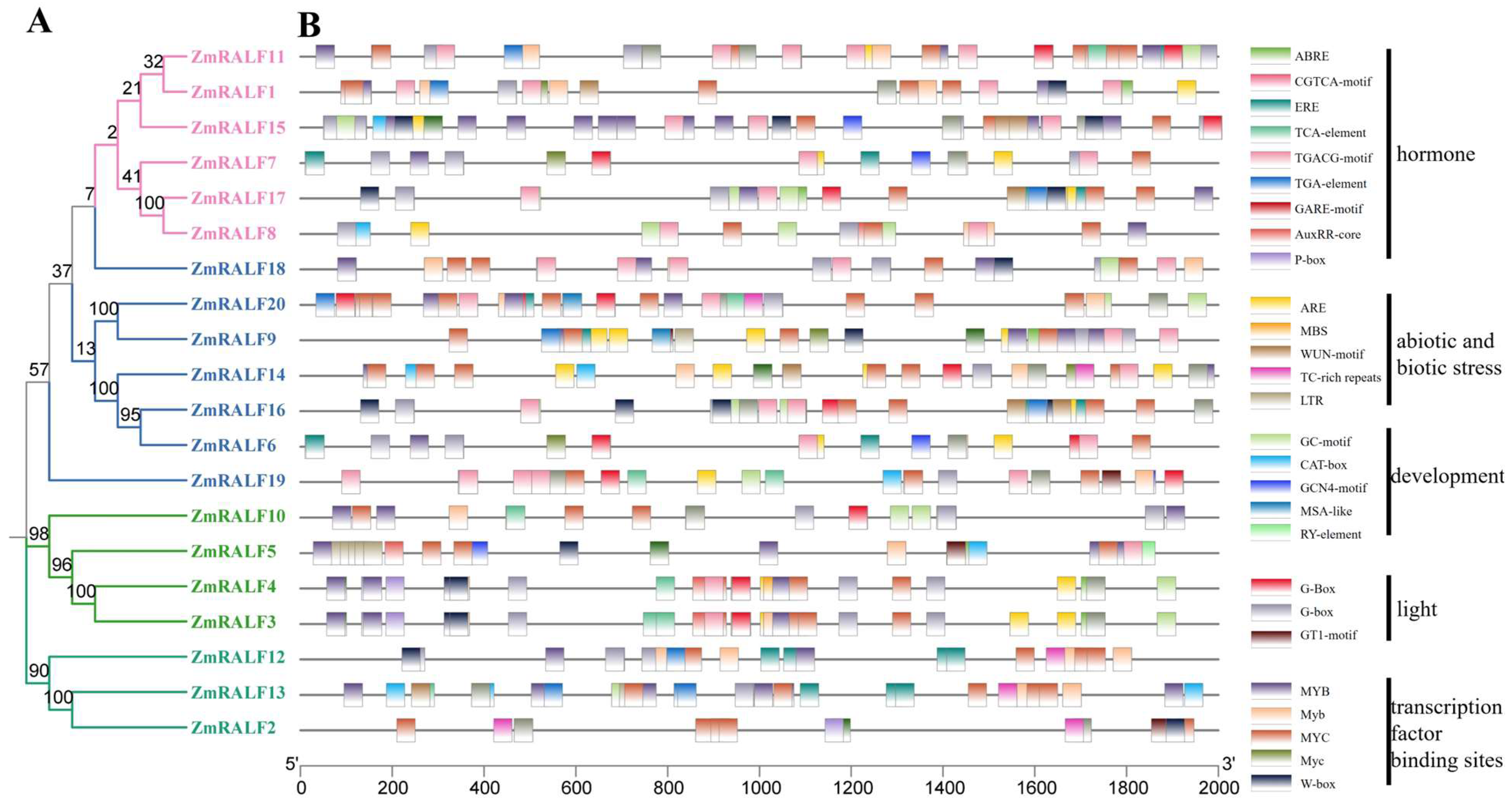
2.2. Sequence Alignment and Phylogenetic Analysis of ZmRALF
2.3. Gene Structure and Protein Conserved Motif of ZmRALF
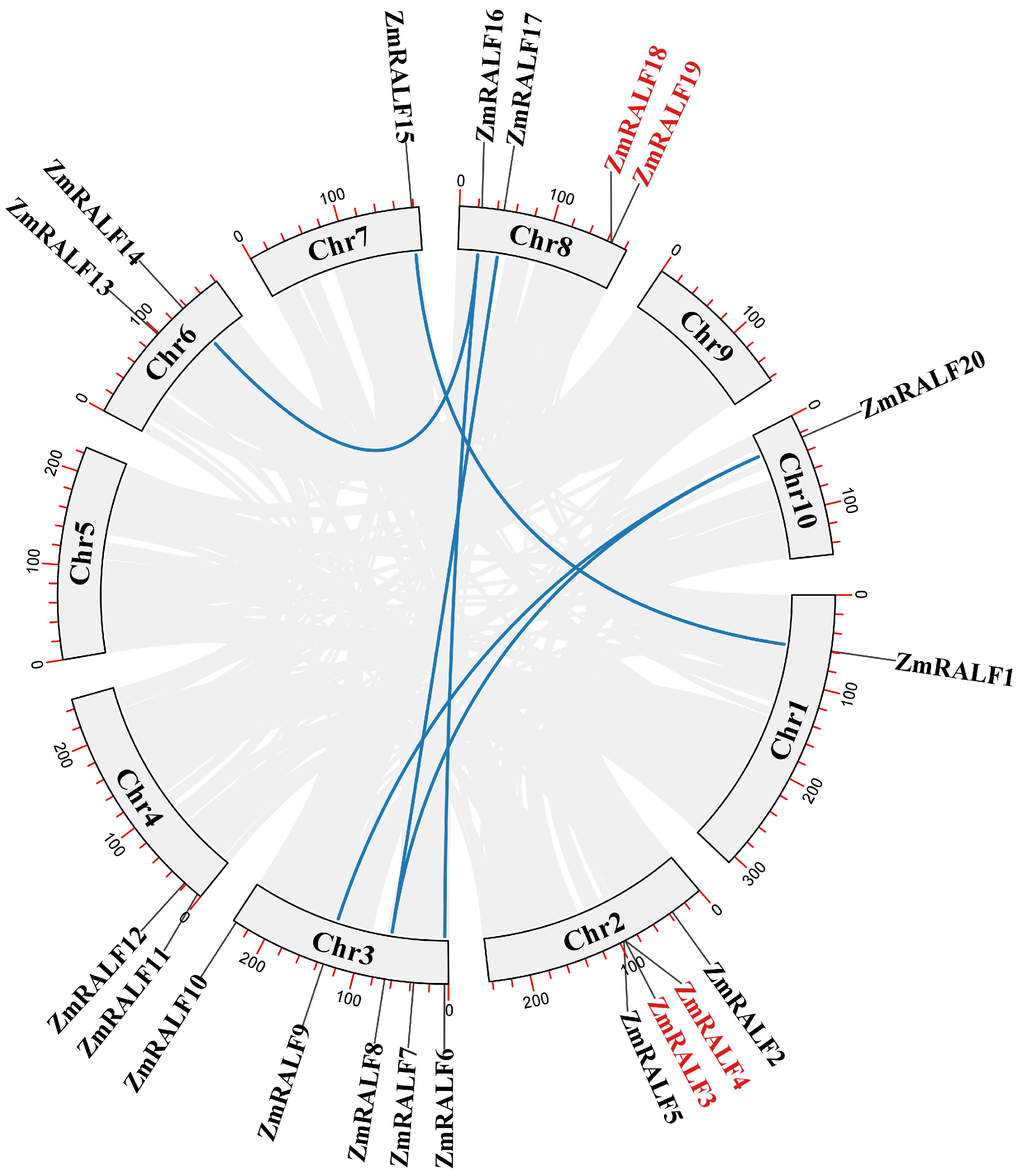
2.4. Homologous Gene Analysis of ZmRALF
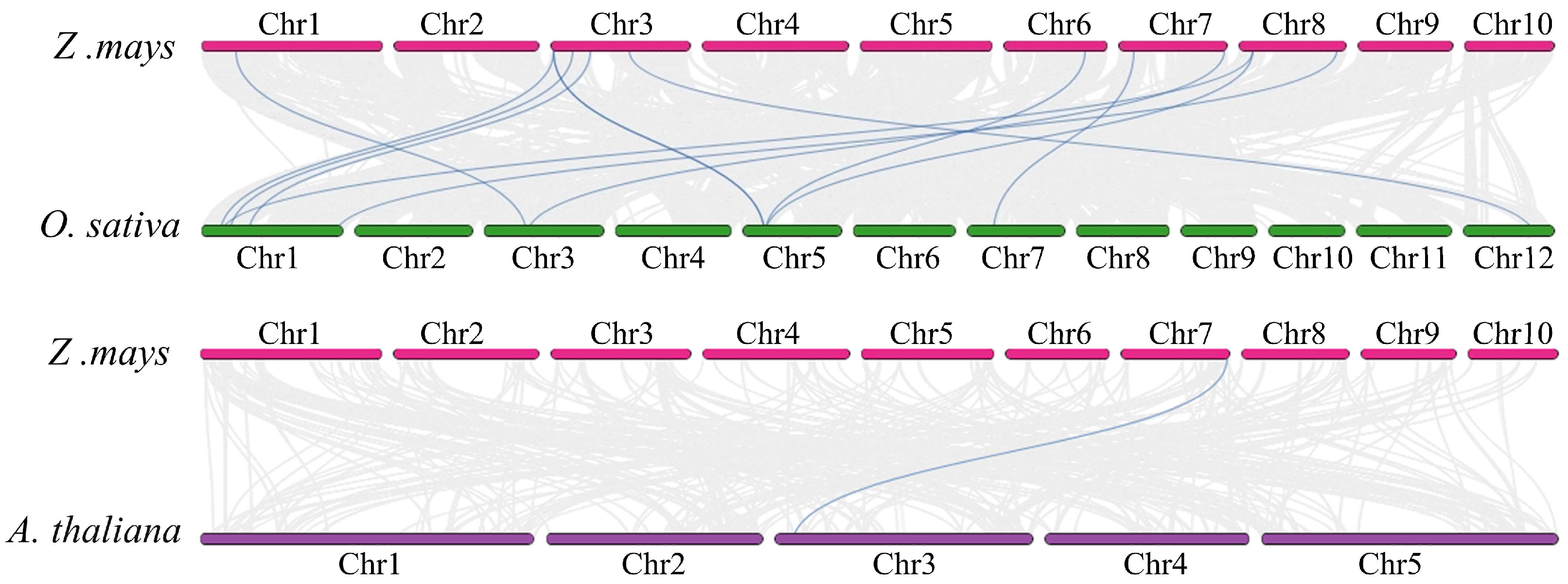
2.5. Collinearity Analysis of ZmRALF Genes with OsRALF and AtRALF
2.6. Analysis of Cis-Elements of ZmRALF Gene
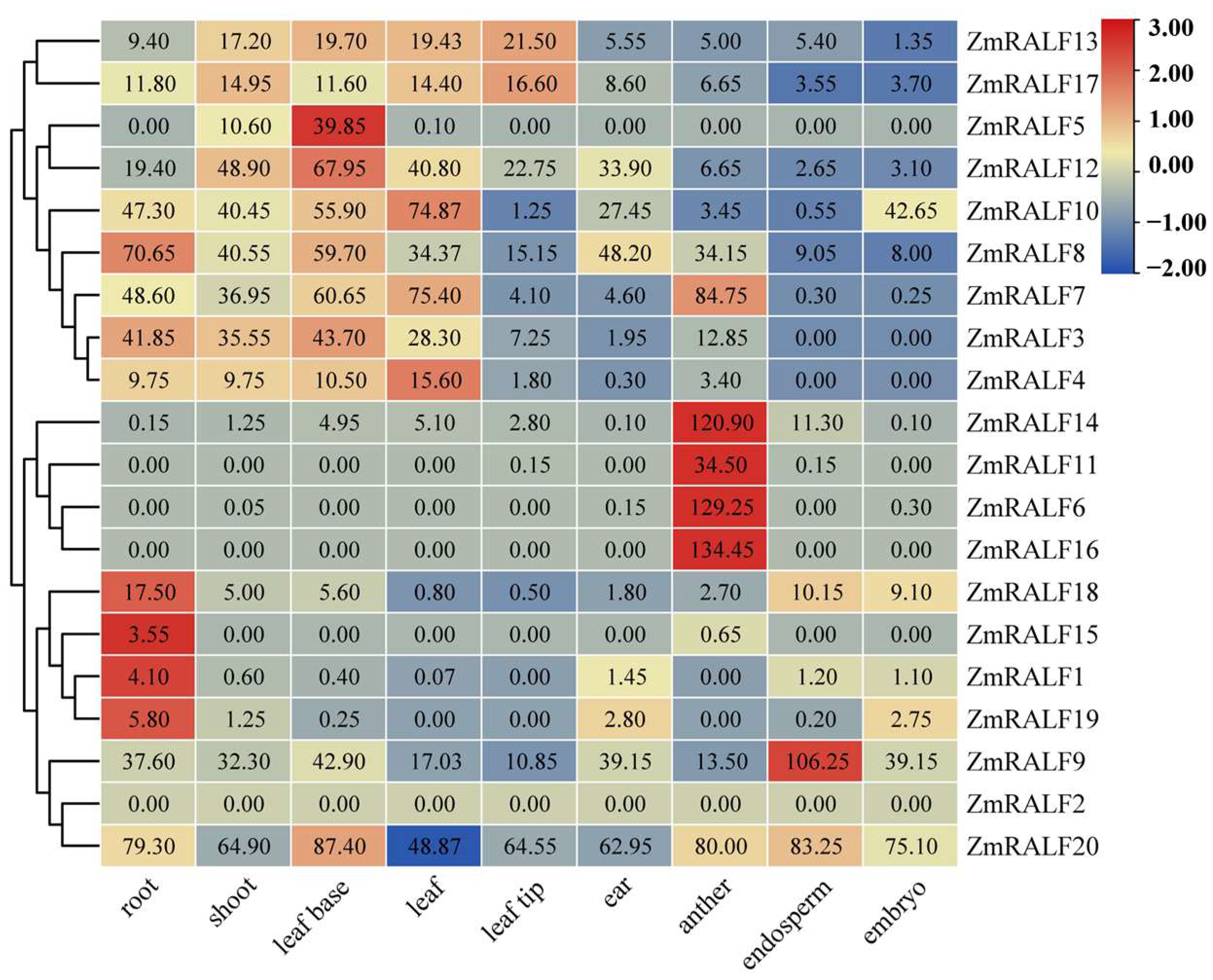
2.7. Tissue-Specific Expression Analysis of ZmRALF
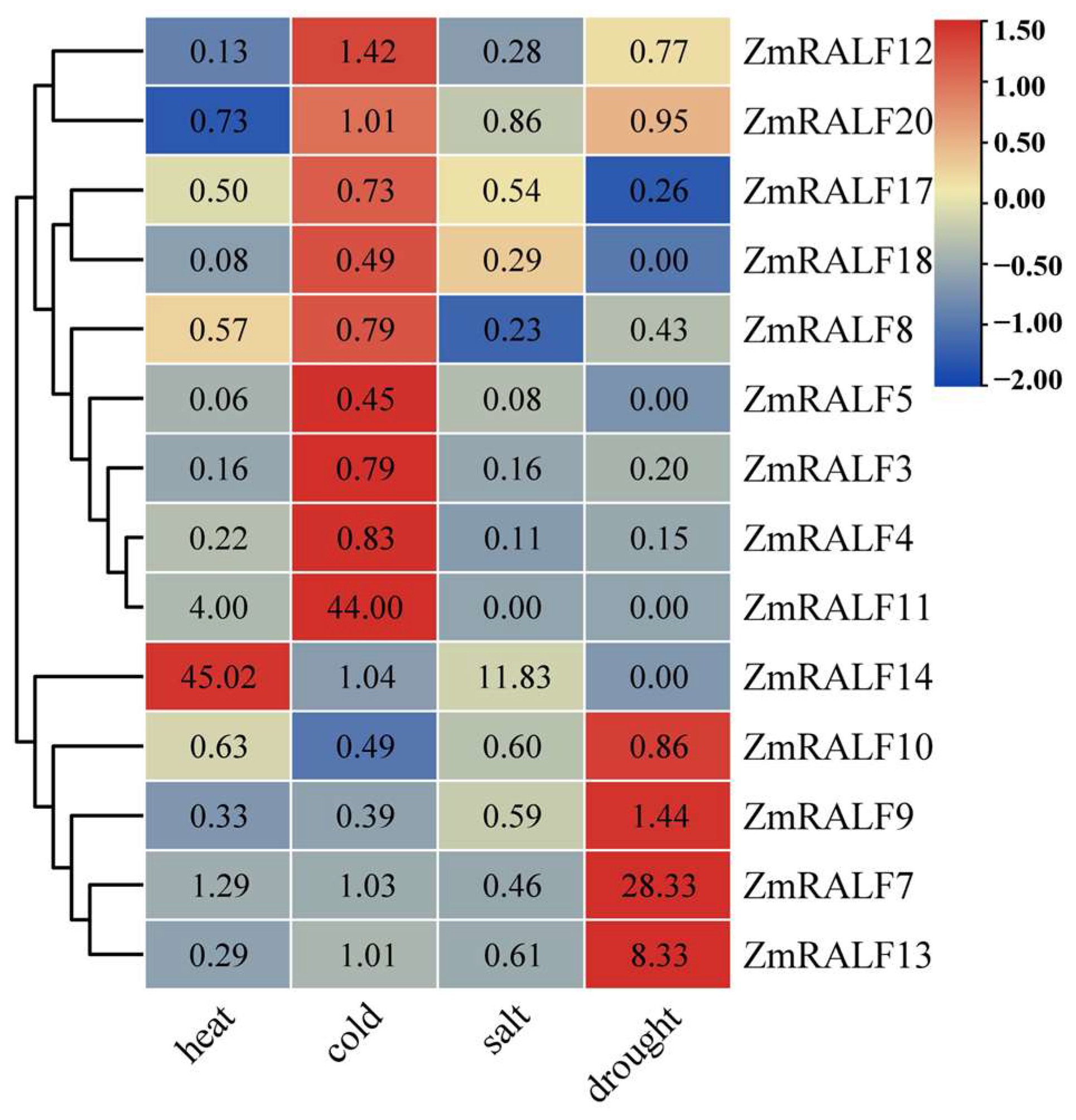
2.8. Expression Patterns of ZmRALF under Abiotic Stresses
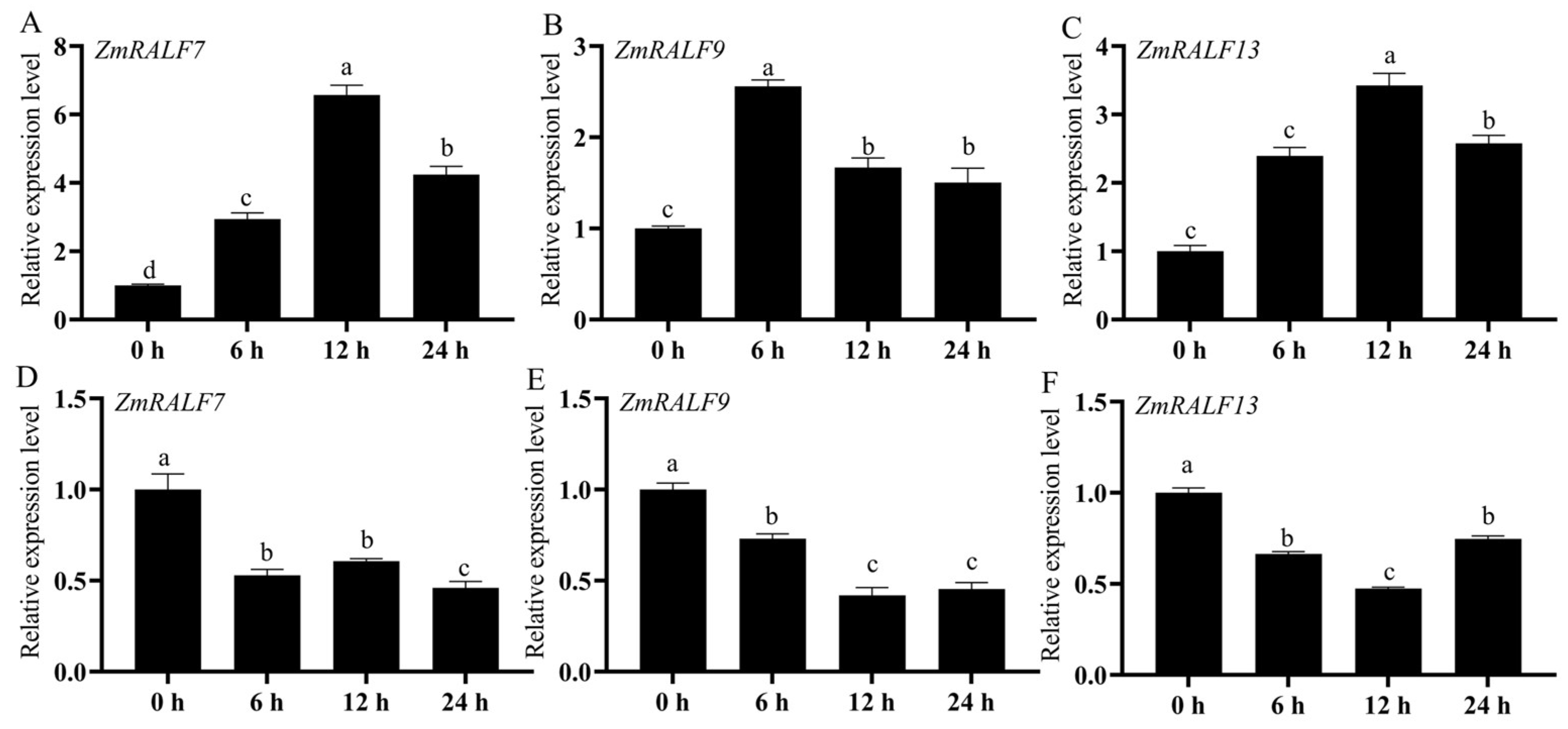
2.9. Expression Analysis of ZmRALF Genes under NaCl and PEG Treatments by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Identification and Phylogenetics of ZmRALF Family Genes in Maize
4.2. Analysis of ZmRALF Gene Structures, Protein Motifs, and Cis-Acting Elements
4.3. Homologous Gene Pair and ka/ks Ratio Analysis of ZmRALF
4.4. ZmRALF Gene Expression Patterns Analysis
4.5. Plant Materials, Growth Conditions, and PEG and NaCl Treatments
4.6. Total RNA Extraction and RT-qPCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P. The Plant Peptidome: An Expanding Repertoire of Structural Features and Biological Functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef] [PubMed]
- Lan, Z.; Song, Z.; Wang, Z.; Li, L.; Liu, Y.; Zhi, S.; Wang, R.; Wang, J.; Li, Q.; Bleckmann, A.; et al. Antagonistic RALF peptides control an intergeneric hybridization barrier on Brassicaceae stigmas. Cell 2023, 186, 4773–4787. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A., Jr. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 2001, 98, 12843–12847. [Google Scholar] [CrossRef] [PubMed]
- Mamaeva, A.; Lyapina, I.; Knyazev, A.; Golub, N.; Mollaev, T.; Chudinova, E.; Elansky, S.; Babenko, V.V.; Veselovsky, V.A.; Klimina, K.M.; et al. RALF peptides modulate immune response in the moss Physcomitrium patens. Front. Plant Sci. 2023, 14, 1077301. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Shi, F. Evolution of the RALF gene family in plants: Gene duplication and selection patterns. Evol. Bioinform. 2012, 8, 271–292. [Google Scholar] [CrossRef]
- He, Y.H.; Zhang, Z.R.; Xu, Y.P.; Chen, S.Y.; Cai, X.Z. Genome-Wide Identification of Rapid Alkalinization Factor Family in Brassica napus and Functional Analysis of BnRALF10 in Immunity to Sclerotinia sclerotiorum. Front. Plant Sci. 2022, 13, 877404. [Google Scholar] [CrossRef]
- Jiang, W.; Li, C.; Li, L.; Li, Y.; Wang, Z.; Yu, F.; Yi, F.; Zhang, J.; Zhu, J.K.; Zhang, H.; et al. Genome-wide analysis of CqCrRLK1L and CqRALF gene families in chenopodium quinoa and their roles in salt stress response. Front. Plant Sci. 2022, 13, 918594. [Google Scholar] [CrossRef]
- Ge, Z.; Bergonci, T.; Zhao, Y. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 2017, 358, 1596–1600. [Google Scholar] [CrossRef]
- Mecchia, M.A.; Santos-Fernandez, G.; Duss, N.N.; Somoza, S.C.; Boisson-Dernier, A.; Gagliardini, V.; Martínez-Bernardini, A.; Fabrice, T.N.; Ringli, C.; Muschietti, J.P.; et al. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017, 358, 1600–1603. [Google Scholar] [CrossRef]
- Xiao, Y.; Stegmann, M.; Han, Z.; DeFalco, T.A.; Parys, K.; Xu, L.; Belkhadir, Y.; Zipfel, C.; Chai, J. Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 2019, 572, 270–274. [Google Scholar] [CrossRef]
- Zhong, S.; Li, L.; Wang, Z.; Ge, Z.; Li, Q.; Bleckmann, A.; Wang, J.; Song, Z.; Shi, Y.; Liu, T.; et al. RALF peptide signaling controls the polytubey block in Arabidopsis. Science 2022, 375, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.K.M.; Walker, C.; Lee, W.S.; Urban, M.; Hammond-Kosack, K.E. Functional evaluation of a homologue of plant rapid alkalinisation factor (RALF) peptides in Fusarium graminearum. Fungal Biol. 2020, 124, 753–765. [Google Scholar] [CrossRef] [PubMed]
- He, Y.H.; Chen, S.Y.; Chen, X.Y.; Xu, Y.P.; Liang, Y.; Cai, X.Z. RALF22 promotes plant immunity and amplifies the Pep3 immune signal. J. Integr. Plant Biol. 2023, 65, 2519–2534. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Assmann, S.M. Inter-relationships between the heterotrimeric Gβ subunit AGB1, the receptor-like kinase FERONIA, and RALF1 in salinity response. Plant Cell Environ. 2018, 41, 2475–2489. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef]
- Xu, Y.; Magwanga, R.O.; Jin, D.; Cai, X.; Hou, Y.; Juyun, Z.; Agong, S.G.; Wang, K.; Liu, F.; Zhou, Z. Comparative transcriptome analysis reveals evolutionary divergence and shared network of cold and salt stress response in diploid D-genome cotton. BMC Plant Biol. 2020, 20, 518. [Google Scholar] [CrossRef]
- Zhang, H.; Jing, X.; Chen, Y.; Liu, Z.; Xin, Y.; Qiao, Y. The Genome-Wide Analysis of RALF-Like Genes in Strawberry (Wild and Cultivated) and Five Other Plant Species (Rosaceae). Genes 2022, 11, 174. [Google Scholar] [CrossRef]
- Dressano, K.; Ceciliato, P.H.O.; Silva, A.L.; Guerrero-Abad, J.C.; Bergonci, T.; Ortiz-Morea, F.A.; Bürger, M.; Silva-Filho, M.C.; Moura, D.S. BAK1 is involved in AtRALF1-induced inhibition of root cell expansion. PLoS Genet. 2017, 13, e1007053. [Google Scholar] [CrossRef]
- Gonneau, M.; Desprez, T.; Martin, M.; Doblas, V.G.; Bacete, L.; Miart, F.; Sormani, R.; Hématy, K.; Renou, J.; Landrein, B.; et al. Receptor Kinase THESEUS1 Is a Rapid Alkalinization Factor 34 Receptor in Arabidopsis. Curr. Biol. 2018, 28, 2452–2458. [Google Scholar] [CrossRef]
- Zhou, L.Z.; Wang, L.; Chen, X.; Ge, Z.; Mergner, J.; Li, X.; Küster, B.; Längst, G.; Qu, L.J.; Dresselhaus, T. The RALF signaling pathway regulates cell wall integrity during pollen tube growth in maize. Plant Cell 2024, 36, 1673–1696. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Li, Y. Genome-Wide Identification and Comparative Analysis of RALF Gene Family in Legume and Non-Legume Species. Int. J. Mol. Sci. 2023, 24, 8842. [Google Scholar] [CrossRef] [PubMed]
- Frederick, R.O.; Haruta, M.; Tonelli, M.; Lee, W.; Cornilescu, G.; Cornilescu, C.C.; Sussman, M.R.; Markley, J.L. Function and solution structure of the Arabidopsis thaliana RALF8 peptide. Protein Sci. 2019, 28, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shi, P.-T.; Zhou, M.; Liu, H.Z.; Xu, X.J.; Liu, W.T.; Chen, K.M. Rapid alkalinization factor: Function, regulation, and potential applications in agriculture. Stress. Biol. 2023, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.Q.; Shi, P.T.; Zhang, R.; Zhou, M.R.; Shalmani, A.; Wang, G.F.; Liu, W.T.; Li, W.Q.; Chen, K.M. Rice kinase OsMRLK63 contributes to drought tolerance by regulating reactive oxygen species production. Plant Physiol. 2024, 194, 2679–2696. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Plant genetics. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Qi, H.; Yu, F.; Deng, J.; Zhang, L.; Yang, P. The high-quality genome of lotus reveals tandem duplicate genes involved in stress response and secondary metabolites biosynthesis. Hortic. Res. 2023, 10, uhad040. [Google Scholar] [CrossRef]
- Shen, C.; Du, H.; Chen, Z.; Lu, H.; Zhu, F.; Chen, H.; Meng, X.; Liu, Q.; Liu, P.; Zheng, L.; et al. The Chromosome-Level Genome Sequence of the Autotetraploid Alfalfa and Resequencing of Core Germplasms Provide Genomic Resources for Alfalfa Research. Mol. Plant 2020, 13, 1250–1261. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Thomas, S.K.; An, H.; Pires, J.C. Mangroves and multiplications: Influence of genome duplications on salt tolerance. Mol. Ecol. 2023, 32, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.; Turner, S.R. A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 2017, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Matos, J.L.; Fiori, C.S.; Filho, M.C.; Moura, D.S. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett. 2008, 582, 3343–3347. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Fu, Q.; Xu, F.; Zheng, H.; Yu, F. New paradigms in cell adaptation: Decades of discoveries on the CrRLK1L receptor kinase signalling network. New Phytol. 2021, 232, 1168–1183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, Z.; Wu, D.; Yu, F. RALF-FERONIA signaling: Linking plant immune response with cell growth. Plant Commun. 2020, 1, 100084. [Google Scholar] [CrossRef] [PubMed]
- Abarca, A.; Franck, C.M.; Zipfel, C. Family-wide evaluation of rapid alkalinization factor peptides. Plant Physiol. 2021, 187, 996–1010. [Google Scholar] [CrossRef]
- Li, L.; Chen, H.; Alotaibi, S.S.; Pěnčík, A.; Adamowski, M.; Novák, O.; Friml, J. RALF1 peptide triggers biphasic root growth inhibition upstream of auxin biosynthesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2121058119. [Google Scholar] [CrossRef]
- Datta, T.; Kumar, R.S.; Sinha, H.; Trivedi, P.K. Small but mighty: Peptides regulating abiotic stress responses in plants. Plant Cell Environ. 2024, 47, 1207–1223. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, X.; Zhang, Y.; Wang, J.; Yang, J.; Ishida, T.; Jiang, W.; Han, X.; Kang, J.; Wang, X.; et al. CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana. Plant Cell Environ. 2019, 42, 1033–1044. [Google Scholar] [CrossRef]
- Roberts, I.; Smith, S.; Stes, E.; De Rybel, B.; Staes, A.; van de Cotte, B.; Njo, M.F.; Dedeyne, L.; Demol, H.; Lavenus, J.; et al. CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. J. Exp. Bot. 2016, 67, 4889–4899. [Google Scholar] [CrossRef]
- Dresselhaus, T.; Sprunck, S.; Wessel, G.M. Fertilization mechanisms in flowering plants. Curr. Biol. 2016, 26, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Li, H.; Abhinandan, K.; Xun, B.; Yao, K.; Shi, J.; Zhao, R.; Li, M.; Wu, Y.; Lan, X. Fatty acid biosynthesis pathways are downregulated during stigma development and are critical during self-incompatible responses in Ornamental kale. Int. J. Mol. Sci. 2022, 23, 13102. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shen, L.; Xiao, Y.; Vyshedsky, D.; Peng, C.; Sun, X.; Liu, Z.; Cheng, L.; Zhang, H.; Han, Z.; et al. Pollen PCP-B peptides unlock a stigma peptide-receptor kinase gating mechanism for pollination. Science 2021, 372, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Garcia-Hernandez, M.; Berardini, T.Z.; Chen, G.; Crist, D.; Doyle, A.; Huala, E.; Knee, E.; Lambrecht, M.; Miller, N.; Mueller, L.A.; et al. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genom. 2002, 2, 239–253. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. Tbtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, H.; Long, Y.; Shu, Y.; Zhai, J. Plant Public RNA-seq Database: A comprehensive online database for expression analysis of 45,000 plant public RNA-Seq libraries. Plant Biotechnol. J. 2022, 20, 806–808. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhou, L.; Li, T.; Ruan, Y.; Zhang, A.; Dong, X.; Zhu, Y.; Li, C.; Fan, J. Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize. Genes 2022, 13, 1682. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, B.; Liang, Z.; Liu, Y.; Li, D.; Liu, C. Genome-Wide Identification of the RALF Gene Family and Expression Pattern Analysis in Zea mays (L.) under Abiotic Stresses. Plants 2024, 13, 2883. https://doi.org/10.3390/plants13202883
Xue B, Liang Z, Liu Y, Li D, Liu C. Genome-Wide Identification of the RALF Gene Family and Expression Pattern Analysis in Zea mays (L.) under Abiotic Stresses. Plants. 2024; 13(20):2883. https://doi.org/10.3390/plants13202883
Chicago/Turabian StyleXue, Baoping, Zicong Liang, Yue Liu, Dongyang Li, and Chang Liu. 2024. "Genome-Wide Identification of the RALF Gene Family and Expression Pattern Analysis in Zea mays (L.) under Abiotic Stresses" Plants 13, no. 20: 2883. https://doi.org/10.3390/plants13202883
APA StyleXue, B., Liang, Z., Liu, Y., Li, D., & Liu, C. (2024). Genome-Wide Identification of the RALF Gene Family and Expression Pattern Analysis in Zea mays (L.) under Abiotic Stresses. Plants, 13(20), 2883. https://doi.org/10.3390/plants13202883




