Integration of mRNA-miRNA Reveals the Possible Role of PyCYCD3 in Increasing Branches Through Bud-Notching in Pear (Pyrus bretschneideri Rehd.)
Abstract
1. Introduction
2. Results
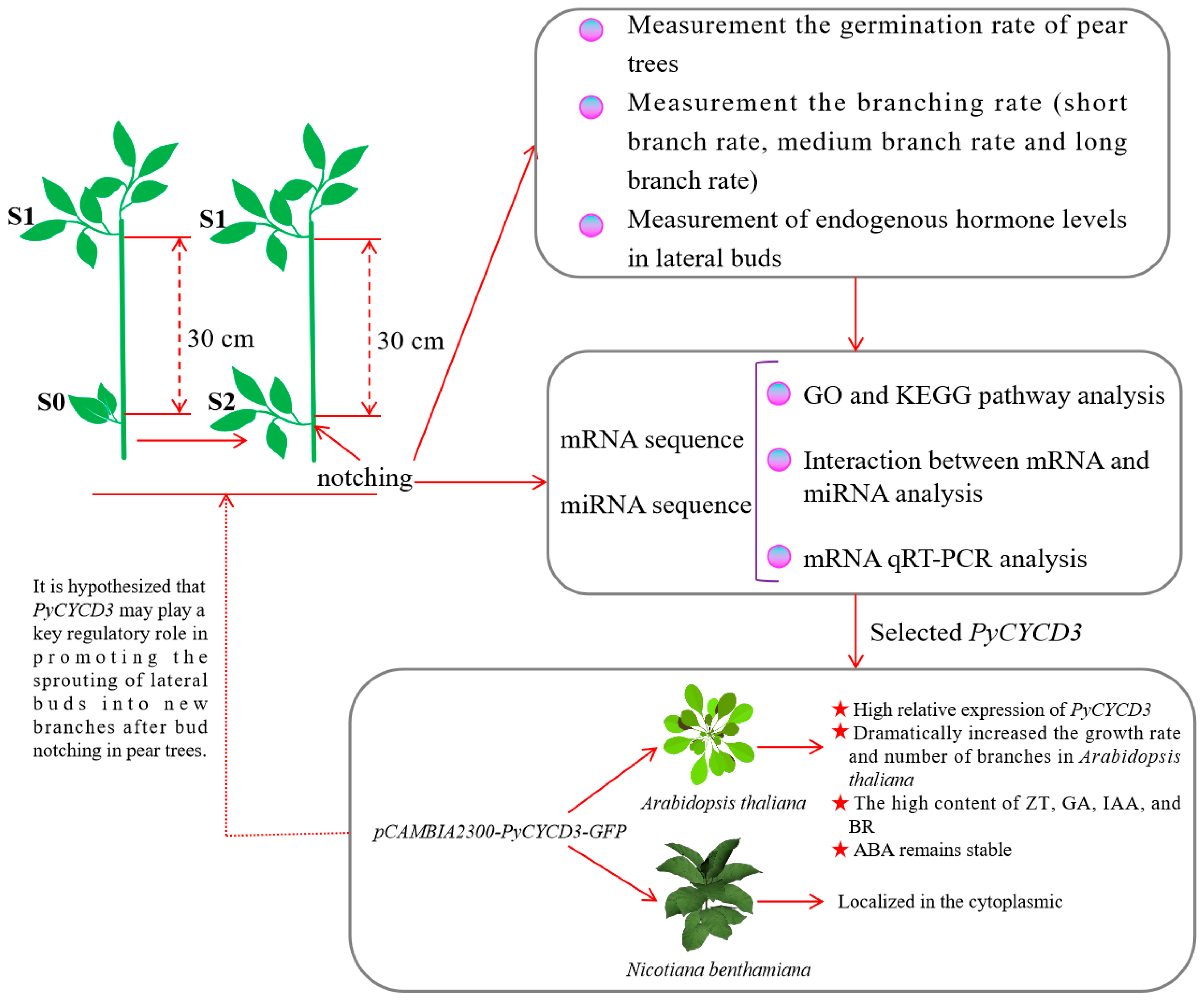
2.1. Samples Treatment and Collection
2.2. Analyzing Germination Rate, Branching Rate, and Endogenous Hormone Levels in Lateral Buds
2.3. Analysis of Sequencing Data and DEGs
2.4. Plant Hormone Signaling Pathway Analysis of mRNA DEGs
2.5. Analysis of the Interaction Network of miRNAs and Target mRNAs
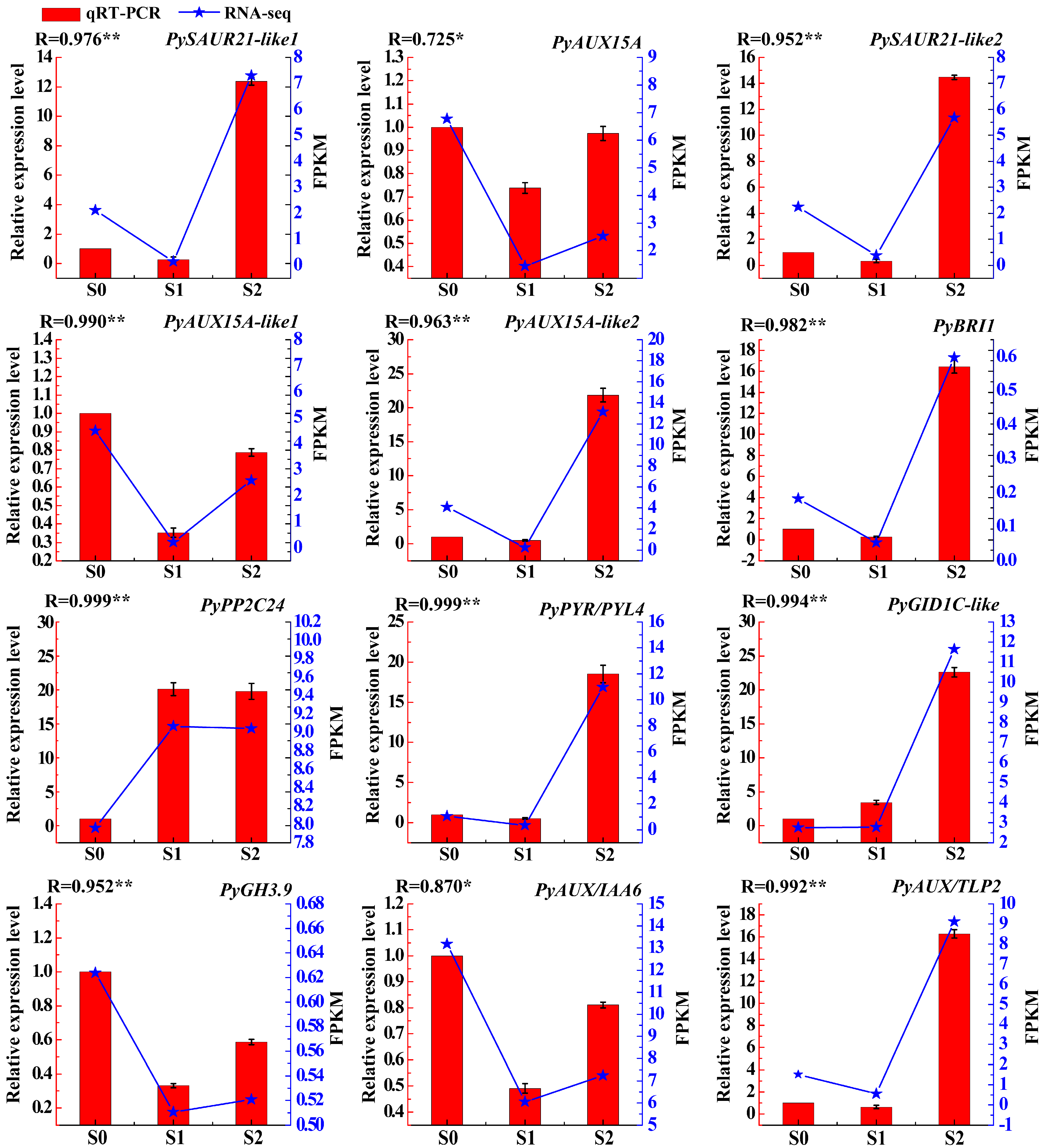
2.6. Validation of Differential Gene Expression by qRT-PCR
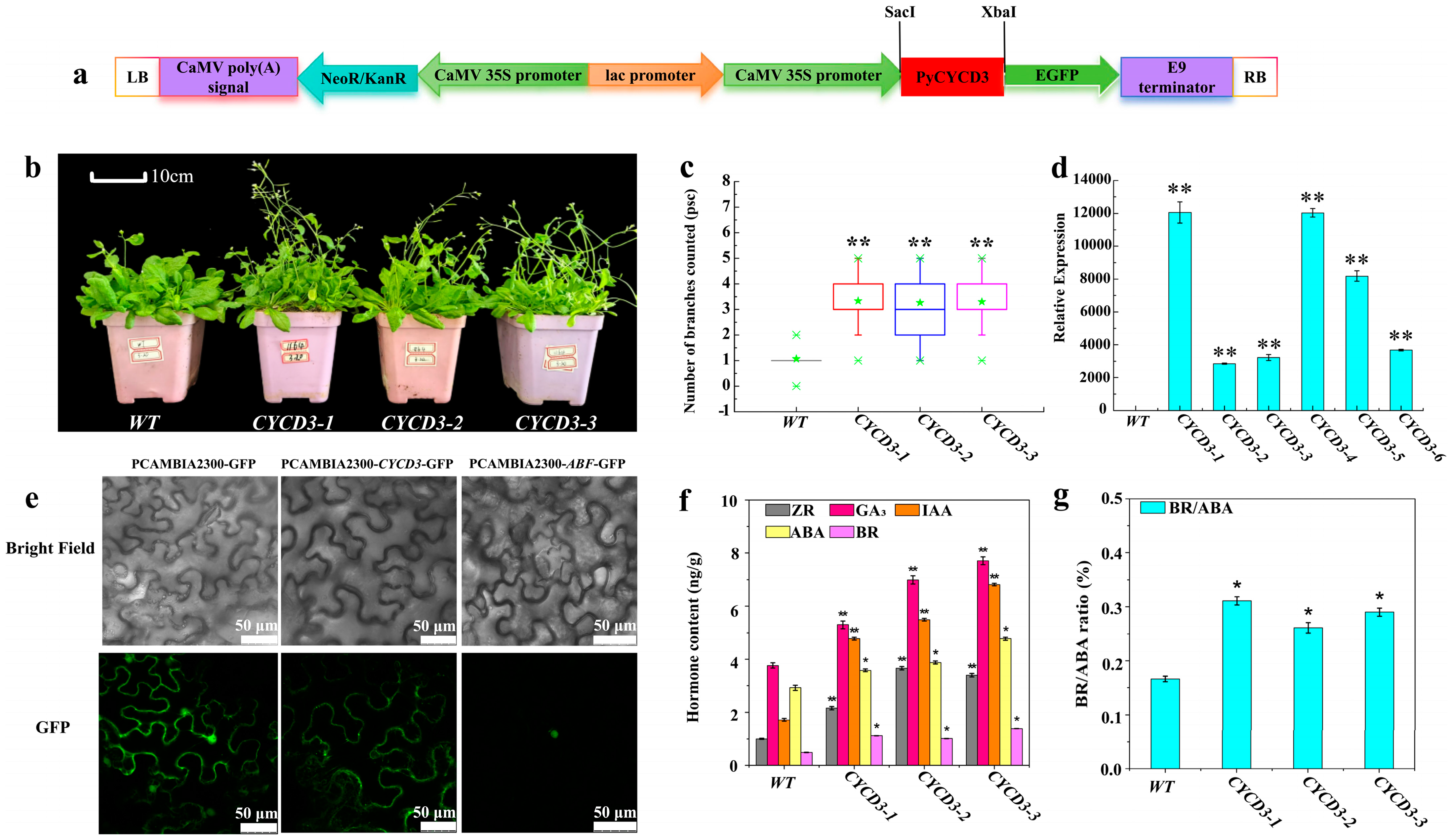
2.7. Overexpression PyCYCD3 Increased Lateral Branch Formation of Transgenic Arabidopsis
3. Discussion
3.1. The Germination, Branching Rate, and Endogenous Hormone Levels in Lateral Buds
3.2. The Genes Related to Phytohormone Signaling in Lateral Bud-Sprouting Regulation
3.3. The Interactions between mRNAs and miRNAs
3.4. The Over-Expression of PyCYCD3 in Arabidopsis
4. Materials and Methods
4.1. Plant Materials and Sample Collection
4.2. Measurement of Emergence Rate, Branching Rate, and Hormone Levels in Lateral Buds
4.3. RNA Preparation and Transcriptome Sequencing
4.4. Differential Expressed Genes (DEGs) Analysis
4.5. Quantitative Real-Time PCR (qRT-PCR) Validation
4.6. Overexpression PyCYCD3 in Arabidopsis and Subcellular Localization Analysis
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Nieminen, K.; Sanchez-Ferrero, J.C.; Rodriguez, M.L.; Chagoyen, M.; Hardtke, C.S.; Cubas, P. Strigolactone promotes degradation of DWARF14 an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 2014, 26, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Teichmann, T.; Muhr, M. Shaping plant architecture. Front. Plant Sci. 2015, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- Elfving, D.C.; Visser, D.B. Cyclanilide induces lateral branching in sweet cherry trees. Hortscience 2006, 41, 149–153. [Google Scholar] [CrossRef]
- Elfving, D.C.; Visser, D.B. Improving the efficacy of cytokinin applications for stimulation of lateral branch development in young sweet cherry trees in the orchard. Am. Soc. Hortic. Sci. 2007, 42, 251–256. [Google Scholar] [CrossRef]
- Jacyna, T. Factors influencing lateral-branch formation in woody plants. Acta Agrobot. 2002, 55, 5–25. [Google Scholar] [CrossRef]
- Jacyna, T.; Puchaa, A. Application of environment friendly branch promoting substances to advance sweet cherry tree canopy development in the orchard. J. Fruit Ornam. Plant Res. 2004, 12, 177–182. [Google Scholar]
- Zhang, L.L.; Fang, W.M.; Chen, F.D.; Song, A.P. The Role of Transcription Factors in the Regulation of Plant Shoot Branching. Plants 2022, 11, 1997. [Google Scholar] [CrossRef]
- McArtney, S.; Obermiller, J.D.J.H. Effect of notching 6-benzyladenine and 6-benzyladenine plus gibberellin A4+ A7 on budbreak and shoot development from paradormant buds on the leader of young apple trees. Hort Technol. 2015, 25, 233–237. [Google Scholar] [CrossRef]
- Aishajiang, M.; Yang, Q.; Wang, J.J.; Liu, G.J. Effects of Cutting Back, Branch-bending and Bud-notching Treatments on Endogenous Hormones in the Buds of Fuji Apple. Acta Hortic. Sin. 2013, 40, 1437–1444. [Google Scholar]
- Yang, H.H.; Zhou, K.; Wu, Q.F.; Jia, X.Y.; Wang, H.X.; Yang, W.H.; Lin, L.H.; Hu, X.M.; Pan, B.Q.; Li, P.; et al. The tomato WRKY-B transcription factor modulates lateral branching by targeting BLIND, PIN4, and IAA15. Hortic. Res. 2024, 11, uhae193. [Google Scholar] [CrossRef] [PubMed]
- Van den Ende, W. Sugars take a central position in plant growth, development and, stress responses. A focus on apical dominance. Front. Plant Sci. 2014, 5, 313. [Google Scholar] [CrossRef] [PubMed]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 741. [Google Scholar] [CrossRef]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An update on the signals controlling shoot branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef]
- Maurya, J.P.; Miskolczi, P.C.; Mishra, S.; Singh, R.K.; Bhalerao, R.P. A genetic framework for regulation and seasonal adaptation of shoot architecture in hybrid aspen. Proc. Natl. Acad. Sci. USA 2020, 117, 11523–11530. [Google Scholar] [CrossRef]
- Gonzalez-Grandio, E.; Poza-Carrion, C.; Sorzano, C.O.; Cubas, P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 2013, 25, 834–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Le Moigne, M.-A.; Bertheloot, J.; Crespel, L.; Perez-Garcia, M.-D.; Ogé, L.; Demotes-Mainard, S.; Hamama, L.; Davière, J.-M.; Sakr, S. BRANCHED1: A key hub of shoot branching. Front. Plant Sci. 2019, 10, 76. [Google Scholar] [CrossRef]
- Kovaleva, L.V.; Zakharova, E.V.; Minkina, Y.V.; Timofeeva, G.V.; Andreev, I.M. Germination and In Vitro Growth of Petunia Male Gametophyte Are Affected by Exogenous Hormones and Involve the Changes in the Endogenous Hormone Leve. Russ. J. Plant Physiol. 2005, 52, 521–526. [Google Scholar] [CrossRef]
- Zhou, Q.; Gao, B.; Li, W.F.; Mao, J.; Yang, S.J.; Li, W.; Ma, Z.H.; Zhao, X.; Chen, B.H. Effects of exogenous growth regulators and bud picking on grafting of grapevine hard branches. Sci. Hortic. 2020, 264, 109186. [Google Scholar] [CrossRef]
- Müller, D.; Leyser, O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011, 107, 1203–1212. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.R.; Hu, Y.L.; Li, P.; Hancock, J.T.; Hu, X.Y. Research Progress and Application of Plant Branching. Phyton-Int. J. Exp. Bot. 2023, 92, 680–689. [Google Scholar] [CrossRef]
- Palmer, H.J.D.; Diack, R.; Seymour, S.; Dayatilake, D.; Tustin, D.S. New approaches to the alleviation of barewood in young apple trees. Biotechnology 2005, 80, 623–627. [Google Scholar] [CrossRef]
- Tan, M.; Li, G.F.; Chen, X.L.; Xing, L.B.; Ma, J.J.; Zhang, D.; Ge, H.J.; Han, M.Y.; Sha, G.L.; An, N. Role of cytokinin strigolactone and auxin export on outgrowth of axillary buds in apple. Front. Plant Sci. 2019, 10, 616. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef]
- Anderson, J.V.; Doğramac, M.; Horvath, D.P.; Foley, M.E. Auxin and ABA act as central regulators of developmental networks associated with paradormancy in Canada thistle (Cirsium arvense). Funct. Integr. Genom. 2012, 12, 515–531. [Google Scholar] [CrossRef] [PubMed]
- González-Grandío, E.; Pajoro, A.; Franco-Zorrilla, J.M.; Tarancon, C.; Immink, R.G.H.; Cubas, P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA 2017, 114, E245–E254. [Google Scholar] [CrossRef]
- Janssen, B.J.; Drummond, R.S.; Snowden, K.C. Regulation of axillary shoot development. Curr. Opin. Plant Biol. 2014, 17, 28–35. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.; Wang, X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 2013, 27, 681–688. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, X.-Y.; Cao, D.-M.; Tang, W.; He, K.; Zhu, J.-Y.; He, J.-X.; Bai, M.-Y.; Zhu, S.; Oh, E.; et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef]
- Divi, U.K.; Rahman, T.; Krishna, P. Gene expression and functional analyses in brassinosteroid-mediated stress tolerance. Plant Biotechnol. J. 2016, 14, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; Lu, J.; Yu, J.W.; Zhang, C.Q.; He, J.X.; Liu, Q.Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2018, 1861, 561–571. [Google Scholar] [CrossRef]
- Xia, X.; Dong, H.; Yin, Y.; Song, X.; Gu, X.; Sang, K.; Zhou, J.; Shi, K.; Zhou, Y.; Foyer, C.H.; et al. Brassinosteroid signaling integrates multiple pathways to release apical dominance in tomato. Proc. Natl. Acad. Sci. USA 2021, 118, e2004384118. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Zhao, C.D.; Gao, Y.Q.; Xu, Y.; Wang, S.J.; Li, C.S.; Xie, Y.P.; Chen, P.X.; Yang, P.Z.; Yuan, L.; et al. A multifaceted module of BRI1 ETHYLMETHANE SULFONATE SUPRESSOR1 (BES1)-MYB88 in growth and stress tolerance of apple. Plant Physiol. 2021, 185, 1903–1923. [Google Scholar] [CrossRef]
- Hu, Y.X.; Bao, F.; Li, J.Y. Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. Cell Mol. 2010, 24, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wang, Z.; Mora-Garcia, S.; Li, J.M.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Zheng, L.; Ma, J.; Song, C.; Zhang, L.; Gao, C.; Zhang, D.; An, N.; Mao, J.; Han, M. Genome-wide identification and expression analysis of GRF genes regulating apple tree architecture. Tree Genet. Genomes 2018, 14, 54–59. [Google Scholar] [CrossRef]
- Zheng, L.; Zhao, C.; Mao, J.; Song, C.; Ma, J.; Zhang, D.; Han, M.; An, N. Genome-wide identification and expression analysis of brassinosteroid biosynthesis and metabolism genes regulating apple tree shoot and lateral root growth. J. Plant Physiol. 2018, 231, 68–85. [Google Scholar] [CrossRef]
- Zheng, L.W.; Gao, C.; Zhao, C.D.; Zhang, L.Z.; Han, M.Y.; An, N.; Ren, X.L. Effects of brassinosteroid associated with auxin and gibberellin on apple tree growth and gene expression patterns. Hortic. Plant J. 2019, 5, 93–108. [Google Scholar] [CrossRef]
- Dewitte, W.; Scofield, S.; Alcasabas, A.A.; Maughan, S.C.; Menges, M.; Braun, N.; Collins, C.; Nieuwland, J.; Prinsen, E.; Sundaresan, V.; et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14537–14542. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Wu, D.; Li, Y.; Tao, W.; Chao, L.; Zhang, Y. The role of auxin in nitrogen-modulated shoot branching. Plant Signal. Behav. 2021, 16, 1885888. [Google Scholar] [CrossRef]
- Prusinkiewicz, P.; Crawford, S.; Smith, R.S.; Ljung, K.; Bennett, T.; Ongaro, V.; Leyser, O. Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 2009, 106, 17431–17436. [Google Scholar] [CrossRef]
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 2005, 8, 443–449. [Google Scholar] [CrossRef]
- Gonin, M.; Salas-González, I.; Gopaulchana, D.; Frene, J.P.; Rodend, S.; Poel, B.V.D.; Salta, D.E.; Castrillo, G. Plant microbiota controls an alternative root branching regulatory mechanism in plants. Proc. Natl. Acad. Sci. USA 2023, 120, e2301054120. [Google Scholar] [CrossRef]
- Forshey, C.G. Branching responses of young apple trees to applications of 6-benzylamino purine and gibberellin A4+7. J. Am. Soc. Hortic. Sci. 1982, 107, 538–541. [Google Scholar] [CrossRef]
- Li, G.; Tan, M.; Ma, J.; Cheng, F.; Li, K.; Liu, X.; Zhao, C.; Zhang, D.; Xing, L.; Ren, X.; et al. Molecular mechanism of MdWUS2-MdTCP12 interaction in mediating cytokinin signaling to control axillary bud outgrowth. J. Exp. Bot. 2021, 72, 4822–4838. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Honma, T.; Aoyama, T.; Sato, S.; Kato, T.; Tabata, S.; Oka, A. ARR1 a transcription factor for genes immediately responsive to cytokinins. Science 2001, 294, 1519–1521. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef]
- Tajima, Y.; Imamura, A.; Kiba, T.; Amano, Y.; Yamashino, T.; Mizuno, T. Comparative Studies on the Type-B Response Regulators Revealing their Distinctive Properties in the His-to-Asp Phosphorelay Signal Transduction of Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Suzuki, T.; Yamashino, T.; Mizuno, T. Comparative Studies of the AHP Histidine-containing Phosphotransmitters Implicated in His-to-Asp Phosphorelay in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2014, 68, 462–465. [Google Scholar]
- Yamada, H.; Koizumi, N.; Nakamichi, N.; Kiba, T.; Yamashino, T.; Mizuno, T. Rapid response of Arabidopsis T87 cultured cells to cytokinin through His-to-Asp phosphorelay signal transduction. Biosci. Biotechnol. Biochem. 2004, 68, 1966–1976. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Hoad, G.V. Hormonal regulation of fruit-bud formation in fruit trees. Flower. Fruit Set Fruit Trees 1983, 149, 13–24. [Google Scholar] [CrossRef]
- Rakngan, J.; Gemma, H.; Iwahori, S. Flower bud formation in Japanese pear trees under adverse conditions and effects of some growth regulators. Jpn. J. Trop. Agric. 1995, 39, 1–6. [Google Scholar]
- Kojima, K.; Yamada, Y.; Yamamoto, M. Effects of abscisic acid injection on sugar and organic acid contents of citrus fruit. J. Jpn. Soc. Hortic. Sci. 2008, 64, 17–21. [Google Scholar] [CrossRef][Green Version]
- Kondo, S.; Gemma, H. Relationship between abscisic acid (ABA) content and maturation of the sweet cherry. J. Jpn. Soc. Hortic. Sci. 1993, 62, 63–68. [Google Scholar] [CrossRef]
- Pavicic, N.B.S.; Martina, D. Effects of combined pruning treatments on fruit quality and biennial bearing of ‘elstar’ apple (Malus domestica Borkh.). J. Food Agric. Environ. 2009, 7, 510–515. [Google Scholar]
- Swarup, R.; Parry, G.; Graham, N.; Allen, T.; Bennett, M. Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Mol. Biol. 2002, 49, 409–424. [Google Scholar] [CrossRef]
- Riou-Khamlichi, C.; Huntley, R.; Jacqmard, A.; Murray, J.A.H. Cytokinin Activation of Arabidopsis Cell Division Through a D-Type Cyclin. Science 1999, 283, 1541–1544. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.Y.; Lee, J.; Ko, H.Y.; Lee, Y.S.; Lim, D.H.; Kim, E.Y.; Cho, S.; Hong, K.S.; Ko, J.J.; Lee, S.; et al. Microinjection free delivery of miRNA inhibitor into zygotes. Sci. Rep. 2014, 4, 5417. [Google Scholar] [CrossRef]
- Dewitte, W.; Riou-Khamlichi, C.; Scofield, S.; Healy, J.M.S.; Jacqmard, A.; Kilby, N.J.; Murraya, J.A.H. Altered cell cycle distribution hyperplasia and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 2002, 15, 79–92. [Google Scholar] [CrossRef]
- Li, F.Z.; Flanary, P.L.; Altieri, D.C.; Dohlman, H.G. Cell Division regulation by BIR1 a member of the inhibitor of apoptosis family in yeast. J. Biol. Chem. 2000, 275, 6707–6711. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Mockler, T.C.; Chory, J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004, 2, E258. [Google Scholar] [CrossRef] [PubMed]
- Walcher, C.L.; Nemhauser, J.L. Bipartite promoter element required for auxin response. Plant Physiol. 2012, 158, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Xue, Y.; Jiang, P.; He, C.; Zhuge, X.; Lan, T.; Yang, H. Overexpression of PtoCYCD3;3 Promotes Growth and Causes Leaf Wrinkle and Branch Appearance in Populus. Int. J. Mol. Sci. 2021, 22, 1288. [Google Scholar] [CrossRef]
- Ohyama, A.; Tominaga, R.; Toriba, T.; Tanaka, W. D-type cyclin oscycd3;1 is involved in the maintenance of meristem activity to regulate branch formation in rice. J. Plant Physiol. 2022, 270, 153634. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Bian, W.; Duan, A.; Zhang, H. Enhancing the expression of ark1 genes in poplar leads to multiple branches and transcriptomic changes. R. Soc. Open Sci. 2020, 7, 201201. [Google Scholar] [CrossRef]
- Greene, D.W.; Autio, W.R. Notching techniques increase branching of young apple trees. J. Am. Soc. Hortic. Sci. 1994, 25, 678–682. [Google Scholar] [CrossRef]
- Xing, L.-B.; Zhang, D.; Li, Y.-M.; Shen, Y.-W.; Zhao, C.-P.; Ma, J.-J.; An, N.; Han, M.-Y. Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (Malus domestica Borkh.). Plant Cell Physiol. 2015, 55, 2052–2068. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L.J.B. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, Z.-S.; Zuo, C.-W.; Mao, J.; Ma, Z.-H.; Li, W.-F.; Chen, B.-H. Integration of mRNA-miRNA Reveals the Possible Role of PyCYCD3 in Increasing Branches Through Bud-Notching in Pear (Pyrus bretschneideri Rehd.). Plants 2024, 13, 2928. https://doi.org/10.3390/plants13202928
An Z-S, Zuo C-W, Mao J, Ma Z-H, Li W-F, Chen B-H. Integration of mRNA-miRNA Reveals the Possible Role of PyCYCD3 in Increasing Branches Through Bud-Notching in Pear (Pyrus bretschneideri Rehd.). Plants. 2024; 13(20):2928. https://doi.org/10.3390/plants13202928
Chicago/Turabian StyleAn, Ze-Shan, Cun-Wu Zuo, Juan Mao, Zong-Huan Ma, Wen-Fang Li, and Bai-Hong Chen. 2024. "Integration of mRNA-miRNA Reveals the Possible Role of PyCYCD3 in Increasing Branches Through Bud-Notching in Pear (Pyrus bretschneideri Rehd.)" Plants 13, no. 20: 2928. https://doi.org/10.3390/plants13202928
APA StyleAn, Z.-S., Zuo, C.-W., Mao, J., Ma, Z.-H., Li, W.-F., & Chen, B.-H. (2024). Integration of mRNA-miRNA Reveals the Possible Role of PyCYCD3 in Increasing Branches Through Bud-Notching in Pear (Pyrus bretschneideri Rehd.). Plants, 13(20), 2928. https://doi.org/10.3390/plants13202928




