Assessment of Genetic Diversity and Population Structure of Exotic Sugar Beet (Beta vulgaris L.) Varieties Using Three Molecular Markers
Abstract
:1. Introduction
2. Results
2.1. Analysis of Genetic Diversity
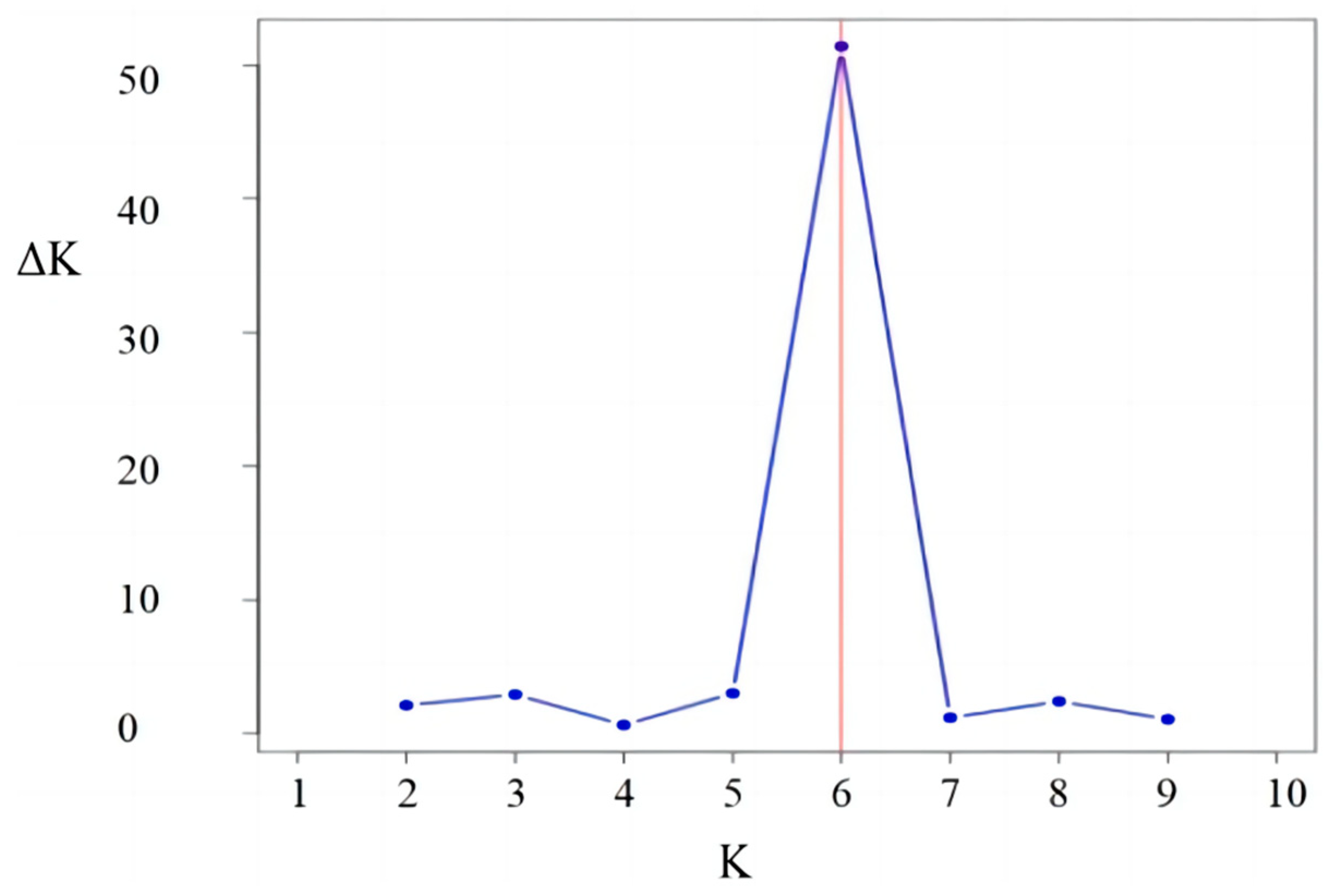
2.2. Analysis of Population Structure
2.3. Genetic Distance and Cluster Analysis
2.4. Analysis of Molecular Variance
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Extraction of Sugar Beet Varieties
4.3. Primer Information Used in the Experiment
4.4. PCR Amplification Reaction System and Procedure
4.5. Statistical Analysis of Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Romeiras, M.M.; Vieira, A.; Silva, D.N.; Moura, M.; Santos-Guerra, A.; Batista, D.; Duarte, M.C.; Paulo, O.S. Evolutionary and Biogeographic Insights on the Macaronesian Beta-Patellifolia Species (Amaranthaceae) from a Time-Scaled Molecular Phylogeny. PLoS ONE 2016, 11, e0152456. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Schulz, B.; Stich, B. Population structure and genetic diversity in elite sugar beet germplasm investigated with SSR markers. Euphytica 2010, 175, 35–42. [Google Scholar] [CrossRef]
- Deng, D.D.; Shen, L.; Li, M.; Guo, J.X.; Liu, X. Overview of the World Sugar Market in the 2021/22 Cruising Season. Sugarcane Canesugar 2022, 51, 91–99. [Google Scholar]
- Xu, Q.L.; Lu, B.F. Review and Prospect of Mechanization Development of Sugar Beet Production. Sugar Crops China 2016, 38, 73–75+78. [Google Scholar]
- Stevanato, P.; Chiodi, C.; Broccanello, C.; Concheri, G.; Biancardi, E.; Pavli, O.; Skaracis, G. Sustainability of the Sugar Beet Crop. Sugar Tech 2019, 21, 703–716. [Google Scholar] [CrossRef]
- Schwichtenberg, K.; Wenke, T.; Zakrzewski, F.; Seibt, K.M.; Minoche, A.; Dohm, J.C.; Weisshaar, B.; Himmelbauer, H.; Schmidt, T. Diversification, evolution and methylation of short interspersed nuclear element families in sugar beet and related Amaranthaceae species. Plant J. 2016, 85, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; An, Z.; Mao, Z.; Ma, L.; Lu, Z. Enhanced Heavy Metal Tolerance and Accumulation by Transgenic Sugar Beets Expressing Streptococcus thermophilus StGCS-GS in the Presence of Cd, Zn and Cu Alone or in Combination. PLoS ONE 2015, 10, e0128824. [Google Scholar] [CrossRef]
- Craig, S.A.S. Betaine in human nutrition. Am. J. Clin. Nutr. 2004, 80, 539–549. [Google Scholar] [CrossRef]
- Curcic, Z.; Taski-Ajdukovic, K.; Nagl, N. Relationship between hybrid performance and genetic variation in self-fertile and self-sterile sugar beet pollinators as estimated by SSR markers. Euphytica 2017, 213, 108. [Google Scholar] [CrossRef]
- Wang, Z.H.; Wu, Z.D.; Wang, X.W.; Fang, Z.Y. Analysis of the Genetic Diversity in Different Types of Sugar Beets by SRAP and SSR Markers. Acta Agron. Sin. 2008, 1, 37–46. [Google Scholar] [CrossRef]
- Witzel, K.; Kurina, A.B.; Artemyeva, A.M. Opening the Treasure Chest: The Current Status of Research on Brassica oleracea and B. rapa Vegetables from ex situ Germplasm Collections. Front. Plant Sci. 2021, 12, 643047. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.T.; Ni, H.B.; Zhang, F.S. Application of Molecular Marker Technology in Sugarbeet Breeding. Chin. Agric. Sci. Bull. 2016, 32, 132–137. [Google Scholar]
- McGrath, J.M. Assisted Breeding in Sugar Beets. Sugar Tech 2010, 12, 187–193. [Google Scholar] [CrossRef]
- Vieira, E.A.; de Carvalho, F.I.F.; Bertan, I.; Kopp, M.M.; Zimmer, P.D.; Benin, G.; da Silva, J.A.G.; Hartwig, I.; Malone, G.; de Oliveira, A.C. Association between genetic distances in wheat (Triticum aestivum L.) as estimated by AFLP and morphological markers. Genet. Mol. Biol. 2007, 30, 392–399. [Google Scholar] [CrossRef]
- Zhang, Y.G.; Yuan, X.Y.; Zhang, G.F.; Li, Y.J.; Yin, J.H.; Lin, J.X.; Li, X.J. The Application of Click Chemistry Reactions in Plant Cell Labeling. Chin. Bull. Bot. 2023, 58, 956–965. [Google Scholar]
- Hannani, M.T.; Thudium, C.S.; Karsdal, M.A.; Ladel, C.; Mobasheri, A.; Uebelhoer, M.; Larkin, J.; Bacardit, J.; Struglics, A.; Bay-Jensen, A.C. From biochemical markers to molecular endotypes of osteoarthritis: A review on validated biomarkers. Expert Rev. Mol. Diagn. 2024, 24, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Bakooie, M.; Pourjam, E.; Mahmoudi, S.B.; Safaie, N.; Naderpour, M. Development of an SNP Marker for Sugar Beet Resistance/Susceptible Genotyping to Root-Knot Nematode. J. Agric. Sci. Technol. 2015, 17, 443–454. [Google Scholar]
- Kritsiriwuthinan, K.; Ngrenngarmlert, W.; Patrapuvich, R.; Phuagthong, S.; Choosang, K. Distinct Allelic Diversity of Plasmodium vivax Merozoite Surface Protein 3-Alpha (PvMSP-3α) Gene in Thailand Using PCR-RFLP. J. Trop. Med. 2023, 2023, 8855171. [Google Scholar] [CrossRef]
- Mousa, H.M.; Abd Al-Abbas, M.J. Random Amplified Polymorphic DNA for identical Streptococcus salivarius strains isolated from tongue of peoples before and after Listerine In vivo. J. Popul. Ther. Clin. Pharmacol. 2023, 30, E9–E13. [Google Scholar]
- Rini, D.S.; Budiyanti, Y.; Valentine, M.; Permana, R. ISSR and SRAP for assessing genetic variability of Indonesian local rice genotypes (Oryza sativa L.). Crop Breed. Appl. Biotechnol. 2023, 23, e448923411. [Google Scholar] [CrossRef]
- Sathapondecha, P.; Suksri, P.; Nuanpirom, J.; Nakkanong, K.; Nualsri, C.; Whankaew, S. Development of Gene-Based InDel Markers on Putative Drought Stress-Responsive Genes and Genetic Diversity of Durian (Durio zibethinus). Biochem. Genet. 2024. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Hu, Y.; Li, F.; Liu, L.; Cui, L. Development of genomic SSR markers and genetic diversity of Sphaerulina musiva in China. J. Phytopathol. 2024, 172, e13253. [Google Scholar] [CrossRef]
- Srinivas, P.R. Introduction to Protein Electrophoresis. Methods Mol. Biol. (Clifton N.J.) 2012, 869, 23–28. [Google Scholar]
- Jander, G.; Norris, S.R.; Rounsley, S.D.; Bush, D.F.; Levin, I.M.; Last, R.L. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002, 129, 440–450. [Google Scholar] [CrossRef]
- Peng, F.; Pi, Z.; Li, S.N.; Wu, Z.D. Genetic Diversity and Population Structure Analysis of Excellent Sugar Beet (Beta vulgaris L.) Germplasm Resources. Horticulturae 2024, 10, 120. [Google Scholar] [CrossRef]
- Liang, X.M.; Pi, Z.; Wu, Z.D.; Li, S.N. Constructing DNA Fingerprinting and Evaluating Genetic Diversity Among Sugar Beet (Beta vulgaris L.) Varieties Based on Four Molecular Markers. Sugar Tech 2023, 25, 1361–1373. [Google Scholar] [CrossRef]
- Patel, S.; Ravikiran, R.; Chakraborty, S.; Macwana, S.; Sasidharan, N.; Trivedi, R.; Aher, B. Genetic diversity analysis of colored and white rice genotypes using Microsatellite (SSR) and Insertion-Deletion (INDEL) markers. Emir. J. Food Agric. 2014, 26, 497–507. [Google Scholar] [CrossRef]
- Chen, M.; He, X.; Huang, X.; Lu, T.; Zhang, Y.; Zhu, J.; Yu, H.; Luo, C. Cis-element amplified polymorphism (CEAP), a novel promoter- and gene-targeted molecular marker of plants. Physiol. Mol. Biol. Plants 2022, 28, 1407–1419. [Google Scholar] [CrossRef]
- Chen, M.Y. A Novel Cis-Element Amplified Polymorphism (CEAP), and Its Application in the Analysis of Mango Germplasm Resources. Bachelor’s Thesis, Guangxi University, Guangxi, China, 2022. [Google Scholar]
- Liu, Z.; Li, J.; Fa, X.; Htwe, N.M.P.S.; Wang, S.; Huang, W.; Yang, J.; Xing, L.; Chen, L.; Li, Y. Assessing the numbers of SNPs needed to establish molecular IDs and characterize the genetic diversity of soybean cultivars derived from Tokachi nagaha. Crop J. 2017, 5, 326–336. [Google Scholar] [CrossRef]
- Pathaichindachote, W.; Panyawut, N.; Sikaewtung, K.; Patarapuwadol, S.; Muangprom, A. Genetic Diversity and Allelic Frequency of Selected Thai and Exotic Rice Germplasm Using SSR Markers. Rice Sci. 2019, 26, 393–403. [Google Scholar] [CrossRef]
- Mikel, M.A. Genetic diversity and improvement of contemporary proprietary North American dent corn. Crop Sci. 2008, 48, 1686–1695. [Google Scholar] [CrossRef]
- Zhao, J.P.; Quan, B.Q.; Ren, J.C.; Guo, P.Y.; Xu, Y. DNA Molecular Marker Technology and its Application in Wheat Genetic Breeding. Barley Cereal Sci. 2024, 41, 9–13. [Google Scholar]
- Zhang, J.; Ge, X.; Zhao, Z.; Zheng, X.; Lu, C.; Jiang, N.; Liu, Y. Population genetic diversity of tomato spotted wilt orthotospovirus isolates from tobacco in Yunnan Province, China. Physiol. Mol. Plant Pathol. 2024, 130, 102228. [Google Scholar] [CrossRef]
- Babarinde, S.; Burlakoti, R.R.; Peters, R.D.; Al-Mughrabi, K.; Novinscak, A.; Sapkota, S.; Prithiviraj, B. Genetic structure and population diversity of Phytophthora infestans strains in Pacific western Canada. Appl. Microbiol. Biotechnol. 2024, 108, 237. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Wu, Z.D.; Zhang, W.B. The evolution of the world’s major sugar beet breeding companies and their enlightenment to the development of China’s sugar beet industry. Sugar Crops China 2014, 3, 82–84+86. [Google Scholar]
- Liu, R.; Liu, N.X.; Wu, Y.M.; Wu, Z.D.; Xing, W.; Wang, Q.H. Genetic Diversity Analysis of Red Beet: Based on SSR and Indel Molecular Markers. Chin. Agric. Sci. Bull. 2019, 35, 47–52. [Google Scholar]
- Tehseen, M.M.; Poore, R.C.; Fugate, K.K.; Bolton, M.D.; Ramachandran, V.; Wyatt, N.A.; Li, X.H.; Chu, C.G. Potential of publicly available Beta vulgaris germplasm for sustainable sugarbeet improvement indicated by combining analysis of genetic diversity and historic resistance evaluation. Crop Sci. 2023, 4, 2255–2273. [Google Scholar] [CrossRef]
- Wu, C.H.; Yang, L.; Xiang, S.Z.; Hu, Q.Q.; Zhou, Z.X.; Zheng, B.; Shi, X.P.; Liu, M. Study on Genomic DNA Extraction Methods for the Rare and Endangered Tree Species Dipteronia sinensis Oliv. J. Anhui Agric. Sci. 2024, 52, 91–94. [Google Scholar]
- Smulders, M.J.M.; Esselink, G.D.; Everaert, I.; De Riek, J.; Vosman, B. Characterisation of sugar beet (Beta vulgari L. ssp vulgaris) varieties using microsatellite markers. BMC Genet. 2010, 11, 41. [Google Scholar] [CrossRef]
- Fugate, K.K.; Fajardo, D.; Schlautman, B.; Ferrareze, J.P.; Bolton, M.D.; Campbell, L.G.; Wiesman, E.; Zalapa, J. Generation and Characterization of a Sugarbeet Transcriptome and Transcript-Based SSR Markers. Plant Genome 2014, 7, 1–13. [Google Scholar] [CrossRef]
- Plomion, C.; Liu, B.H.; O’Malley, D.M. Genetic analysis using trans-dominant linked markers in an F2 family. Theor. Appl. Genet. 1996, 93, 1083–1089. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1996, 129, 157. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Weir, B.S. Genetic Data Analysis II: Methods for Discrete Population Genetic Data; Sinauer Associates: Sunderland, MA, USA, 1996. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Ge, X.J.; Yu, Y.; Yuan, Y.M.; Huang, H.W.; Yan, C. Genetic diversity and geographic differentiation in endangered Ammopiptanthus (Leguminosae) populations in desert regions of northwest China as revealed by ISSR analysis. Ann. Bot. 2005, 95, 843–851. [Google Scholar] [CrossRef]


| Number | Primers | Na | Ne | I | Ho | He | Nei’s | PIC |
|---|---|---|---|---|---|---|---|---|
| 1 | 27,906 | 10 | 8.252 | 2.206 | 0.985 | 0.882 | 0.879 | 0.869 |
| 2 | 2305 | 13 | 6.585 | 2.115 | 0.930 | 0.851 | 0.848 | 0.840 |
| 3 | 11,965 | 15 | 7.023 | 2.321 | 0.879 | 0.861 | 0.858 | 0.847 |
| 4 | 57,236 | 9 | 6.269 | 1.959 | 0.791 | 0.844 | 0.840 | 0.847 |
| 5 | D17 | 17 | 9.126 | 2.420 | 0.814 | 0.894 | 0.890 | 0.886 |
| 6 | D31 | 15 | 8.047 | 2.344 | 0.830 | 0.879 | 0.876 | 0.871 |
| 7 | D32 | 19 | 9.300 | 2.451 | 0.651 | 0.896 | 0.893 | 0.892 |
| 8 | TGAC9 | 8 | 4.811 | 1.738 | 0.898 | 0.795 | 0.792 | 0.781 |
| 9 | TGAC10 | 11 | 7.640 | 2.186 | 0.931 | 0.873 | 0.869 | 0.860 |
| 10 | TCAC26 | 17 | 10.579 | 2.549 | 0.906 | 0.909 | 0.906 | 0.907 |
| 11 | TCAC27 | 10 | 7.077 | 2.096 | 0.605 | 0.864 | 0.859 | 0.801 |
| 12 | TGAC28 | 15 | 11.035 | 2.515 | 0.954 | 0.913 | 0.909 | 0.906 |
| 13 | ACGTG4 | 9 | 5.483 | 1.828 | 0.207 | 0.822 | 0.818 | 0.781 |
| 14 | GATAA1 | 12 | 7.818 | 2.202 | 0.754 | 0.876 | 0.872 | 0.876 |
| 15 | GATAA2 | 11 | 4.001 | 1.806 | 0.487 | 0.753 | 0.750 | 0.773 |
| 16 | TGAC23 | 6 | 3.591 | 1.470 | 1.000 | 0.724 | 0.722 | 0.682 |
| 17 | TGAC19 | 9 | 7.189 | 2.071 | 0.887 | 0.865 | 0.861 | 0.866 |
| 18 | TGAC18 | 9 | 6.833 | 2.015 | 0.418 | 0.860 | 0.854 | 0.700 |
| 19 | TGAC12 | 7 | 4.048 | 1.559 | 0.858 | 0.756 | 0.753 | 0.737 |
| 20 | TGAC6 | 11 | 6.154 | 2.017 | 0.714 | 0.795 | 0.838 | 0.781 |
| 21 | TGAC7 | 8 | 5.701 | 1.846 | 0.857 | 0.843 | 0.825 | 0.817 |
| 22 | TGAC20 | 11 | 8.220 | 2.193 | 0.780 | 0.883 | 0.878 | 0.859 |
| 23 | TGAC21 | 10 | 6.410 | 1.995 | 0.954 | 0.847 | 0.844 | 0.831 |
| 24 | TGAC22 | 10 | 6.013 | 1.950 | 0.930 | 0.837 | 0.834 | 0.824 |
| 25 | ACGTG1 | 11 | 5.473 | 1.948 | 0.930 | 0.821 | 0.817 | 0.808 |
| 26 | ACGTG3 | 10 | 6.593 | 2.007 | 0.902 | 0.852 | 0.848 | 0.830 |
| 27 | AAAG28 | 5 | 3.155 | 1.368 | 0.221 | 0.688 | 0.683 | 0.639 |
| Total | 298 | 182.426 | 55.175 | 21.073 | 22.683 | 22.616 | 22.111 | |
| Mean | 11 | 6.757 | 2.044 | 0.780 | 0.840 | 0.838 | 0.819 |
| Source | df | SS | MS | PV% | p | Est. Var | Fst | Nm |
|---|---|---|---|---|---|---|---|---|
| Among populations | 5 | 61.699 | 12.340 | 5% | <0.001 | 0.215 | ||
| Within populations | 258 | 910.956 | 3.739 | 95% | <0.001 | 3.739 | ||
| Total | 263 | 972.655 | 100% | 3.954 | 0.057 * | 4.98 |
| Number | Variety Name | Breeding Company | Number | Variety Name | Breeding Company |
|---|---|---|---|---|---|
| 1 | KWS126 | KWS SAAT SE | 67 | Ma1 | Maribohilleshög ApS |
| 2 | KWS127 | KWS SAAT SE | 68 | Ma2 | Maribohilleshög ApS |
| 3 | KWS128 | KWS SAAT SE | 69 | Ma3 | Maribohilleshög ApS |
| 4 | KWS129 | KWS SAAT SE | 70 | Ma4 | Maribohilleshög ApS |
| 5 | KWS130 | KWS SAAT SE | 71 | Ma7 | Maribohilleshög ApS |
| 6 | KWS131 | KWS SAAT SE | 72 | Ma8 | Maribohilleshög ApS |
| 7 | KWS132 | KWS SAAT SE | 73 | Ma9 | Maribohilleshög ApS |
| 8 | KWS133 | KWS SAAT SE | 74 | Ma10 | Maribohilleshög ApS |
| 9 | KWS134 | KWS SAAT SE | 75 | Ma11 | Maribohilleshög ApS |
| 10 | KWS136 | KWS SAAT SE | 76 | Ma12 | Maribohilleshög ApS |
| 11 | KWS137 | KWS SAAT SE | 77 | Ma14 | Maribohilleshög ApS |
| 12 | KWS138 | KWS SAAT SE | 78 | Ma15 | Maribohilleshög ApS |
| 13 | KWS139 | KWS SAAT SE | 79 | Ma16 | Maribohilleshög ApS |
| 14 | KWS140 | KWS SAAT SE | 80 | Ma17 | Maribohilleshög ApS |
| 15 | KWS141 | KWS SAAT SE | 81 | Ma18 | Maribohilleshög ApS |
| 16 | KWS158 | KWS SAAT SE | 82 | Ma19 | Maribohilleshög ApS |
| 17 | KWS0023 | KWS SAAT SE | 83 | Ma20 | Maribohilleshög ApS |
| 18 | KWS1130 | KWS SAAT SE | 84 | MA22 | Maribohilleshög ApS |
| 19 | KWS1131 | KWS SAAT SE | 85 | MA23 | Maribohilleshög ApS |
| 20 | KWS2407 | KWS SAAT SE | 86 | 23MH1 | Maribohilleshög ApS |
| 21 | KWS2408 | KWS SAAT SE | 87 | 23MH2 | Maribohilleshög ApS |
| 22 | KWS3473 | KWS SAAT SE | 88 | 23MH3 | Maribohilleshög ApS |
| 23 | KWS3504 | KWS SAAT SE | 89 | 23MH4 | Maribohilleshög ApS |
| 24 | KWS3505 | KWS SAAT SE | 90 | 23MH6 | Maribohilleshög ApS |
| 25 | KWS6637 | KWS SAAT SE | 91 | 23MH7 | Maribohilleshög ApS |
| 26 | KWS6653 | KWS SAAT SE | 92 | 23MH8 | Maribohilleshög ApS |
| 27 | KWS7748 | KWS SAAT SE | 93 | 23MH9 | Maribohilleshög ApS |
| 28 | KWS7772 | KWS SAAT SE | 94 | 23MH10 | Maribohilleshög ApS |
| 29 | KWS8805 | KWS SAAT SE | 95 | ST12528 | STRUBE |
| 30 | KWS9147 | KWS SAAT SE | 96 | ST12655 | STRUBE |
| 31 | KWS9898 | KWS SAAT SE | 97 | ST12763 | STRUBE |
| 32 | KWS9962 | KWS SAAT SE | 98 | ST12764 | STRUBE |
| 33 | SX1535 | SES VanderHave | 99 | ST12816 | STRUBE |
| 34 | SX1537 | SES VanderHave | 100 | ST12817 | STRUBE |
| 35 | SV2427 | SES VanderHave | 101 | ST12846 | STRUBE |
| 36 | SV2538 | SES VanderHave | 102 | ST12908 | STRUBE |
| 37 | SV2674 | SES VanderHave | 103 | ST12909 | STRUBE |
| 38 | SV2675 | SES VanderHave | 104 | ST13103 | STRUBE |
| 39 | SV2676 | SES VanderHave | 105 | ST13112 | STRUBE |
| 40 | SV2761 | SES VanderHave | 106 | ST13237 | STRUBE |
| 41 | SV2762 | SES VanderHave | 107 | ST13527 | STRUBE |
| 42 | SV2763 | SES VanderHave | 108 | ST13528 | STRUBE |
| 43 | MK4185 | SES VanderHave | 109 | ST13529 | STRUBE |
| 44 | MK4205 | SES VanderHave | 110 | ST13790 | STRUBE |
| 45 | MK4241 | SES VanderHave | 111 | ST13832 | STRUBE |
| 46 | MK4245 | SES VanderHave | 112 | ST13903 | STRUBE |
| 47 | MK4256 | SES VanderHave | 113 | ST13915 | STRUBE |
| 48 | MK4257 | SES VanderHave | 114 | ST13943 | STRUBE |
| 49 | SR23001 | SES VanderHave | 115 | ST15216 | STRUBE |
| 50 | SR230010 | SES VanderHave | 116 | ST15217 | STRUBE |
| 51 | SR230011 | SES VanderHave | 117 | L2301 | Lion Seeds Ltd. |
| 52 | SR230012 | SES VanderHave | 118 | L2302 | Lion Seeds Ltd. |
| 53 | SR230013 | SES VanderHave | 119 | L2305 | Lion Seeds Ltd. |
| 54 | SR230015 | SES VanderHave | 120 | L2306 | Lion Seeds Ltd. |
| 55 | SR230016 | SES VanderHave | 121 | L2307 | Lion Seeds Ltd. |
| 56 | SR230017 | SES VanderHave | 122 | LN001 | Lion Seeds Ltd. |
| 57 | SR230018 | SES VanderHave | 123 | LN002 | Lion Seeds Ltd. |
| 58 | SR230019 | SES VanderHave | 124 | LN003 | Lion Seeds Ltd. |
| 59 | SR23002 | SES VanderHave | 125 | Bts1714 | BETASEED |
| 60 | SR230020 | SES VanderHave | 126 | Bts1715 | BETASEED |
| 61 | SR23004 | SES VanderHave | 127 | Bts1730 | BETASEED |
| 62 | SR23005 | SES VanderHave | 128 | Bts3880 | BETASEED |
| 63 | SR23006 | SES VanderHave | 129 | Bts5940 | BETASEED |
| 64 | SR23007 | SES VanderHave | 130 | Bts6870 | BETASEED |
| 65 | SR23008 | SES VanderHave | 131 | Bts6871 | BETASEED |
| 66 | SR23009 | SES VanderHave | 132 | Bts7715 | BETASEED |
| Primer Type | Primer Name | Primer Sequences (5′-3′) | Annealing Temperature |
|---|---|---|---|
| SSR | 27906 | F GAGCAGCAAACATGATAAGA | 57 °C |
| R GAAAACAGTGAGTATGGGTCTA | |||
| 2305 | F TACTAAAACCCTACGAACTCCA | 55 °C | |
| R TACACCTGTGATTGTCAGAAGA | |||
| 11965 | F TTGAGTATTTTCGTCGGC | 57 °C | |
| R CATCTACATCAGTTTTCCCTTC | |||
| 57236 | F TTGGAGAGAGAAAAGAGAGAAG | 57 °C | |
| R ATCCCTTGACAGTAGAACTCC | |||
| InDel | D17 | F GATGGGGGAGATCCCAAC | Touch down |
| R GCTTGACCCAGTGCCATC | |||
| D31 | F CGCAGAGTGGTGTGTTGG | Touch down | |
| R TGGAGAATGGGTGTGCTG | |||
| D32 | F GGGGGAGAGCAGTGGGTA | Touch down | |
| R AGCAGAGGAGGTGTGTGTGA | |||
| CEAP | TGAC9 | GCAGCTGAGAGTTGACGA | Touch down |
| TGAC10 | GCAGCTGAGAGTTGACGT | Touch down | |
| TCAC26 | GCAGCTGAGGTTGACCAG | Touch down | |
| TGAC27 | GCAGCTGAGGTTGACCTC | Touch down | |
| TGAC28 | GCAGCTGAGGTTGACCGA | Touch down | |
| ACGTG4 | GCAGTCAGATCACGTGAC | Touch down | |
| GATAA1 | GCAGCTGCGTGGATAAAT | Touch down | |
| GATAA2 | GCAGCTCGCTGGATAAAG | Touch down | |
| TGAC23 | GCAGCTGAGGTTGACGAC | Touch down | |
| TGAC19 | GCAGCTGAGGTTGACTAG | Touch down | |
| TGAC18 | GCAGCTGAGGTTGACACA | Touch down | |
| TGAC12 | GCAGCTGAGAGTTGACGG | Touch down | |
| TGAC6 | GCAGCTGAGAGTTGACTT | Touch down | |
| TGAC7 | GCAGCTGAGAGTTGACTG | Touch down | |
| TGAC20 | GCAGCTGAGGTTGACTCA | Touch down | |
| TGAC21 | GCAGCTGAGGTTGACTGT | Touch down | |
| TGAC22 | GCAGCTGAGGTTGACTTC | Touch down | |
| ACGTG1 | GCAGTCAGATCACGTGAA | Touch down | |
| ACGTG3 | GCAGTCAGATCACGTGAG | Touch down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, B.; Li, S.; Pi, Z.; Wu, Z.; Wang, R. Assessment of Genetic Diversity and Population Structure of Exotic Sugar Beet (Beta vulgaris L.) Varieties Using Three Molecular Markers. Plants 2024, 13, 2954. https://doi.org/10.3390/plants13212954
Sun B, Li S, Pi Z, Wu Z, Wang R. Assessment of Genetic Diversity and Population Structure of Exotic Sugar Beet (Beta vulgaris L.) Varieties Using Three Molecular Markers. Plants. 2024; 13(21):2954. https://doi.org/10.3390/plants13212954
Chicago/Turabian StyleSun, Bowei, Shengnan Li, Zhi Pi, Zedong Wu, and Ronghua Wang. 2024. "Assessment of Genetic Diversity and Population Structure of Exotic Sugar Beet (Beta vulgaris L.) Varieties Using Three Molecular Markers" Plants 13, no. 21: 2954. https://doi.org/10.3390/plants13212954
APA StyleSun, B., Li, S., Pi, Z., Wu, Z., & Wang, R. (2024). Assessment of Genetic Diversity and Population Structure of Exotic Sugar Beet (Beta vulgaris L.) Varieties Using Three Molecular Markers. Plants, 13(21), 2954. https://doi.org/10.3390/plants13212954






