Identification of QTLs and Key Genes Enhancing Lodging Resistance in Soybean Through Chemical and Physical Trait Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials Collections and Storage
2.2. Soybean Seeding Growing Conditions
2.3. Field and Pot Soybean Planting and Sampling Surveys
2.4. Hormone Treatment of Soybean Seedlings
2.5. Determination of Cell Wall Component Content
2.5.1. Determination of Lignin and Cellulose Content
2.5.2. Determination of the Total Pectin Content
2.6. RNA Extraction
2.7. RT–qPCR Analysis of Candidate Genes
2.8. QTL Analysis via Euclidean Distance (ED)
2.9. Function Analysis of the Candidate Genes
2.10. Genotypes of Soybean Varieties Using a Soybean SNP Array
3. Results
3.1. Impact of Planting Density on Phenotypic Traits Related to Lodging Resistance
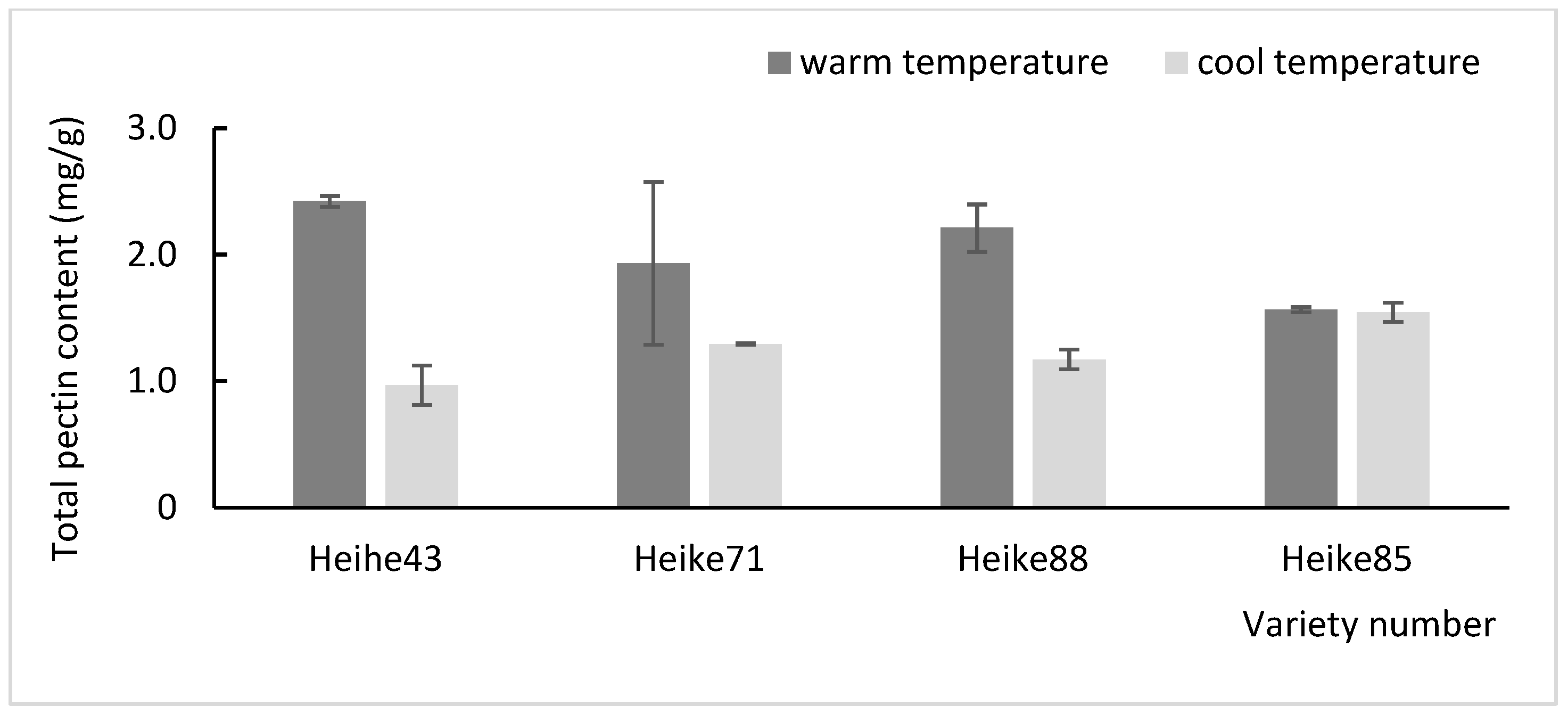
3.2. Temperature Effects on Soybean Cell Wall Composition for Environmental Adaptation
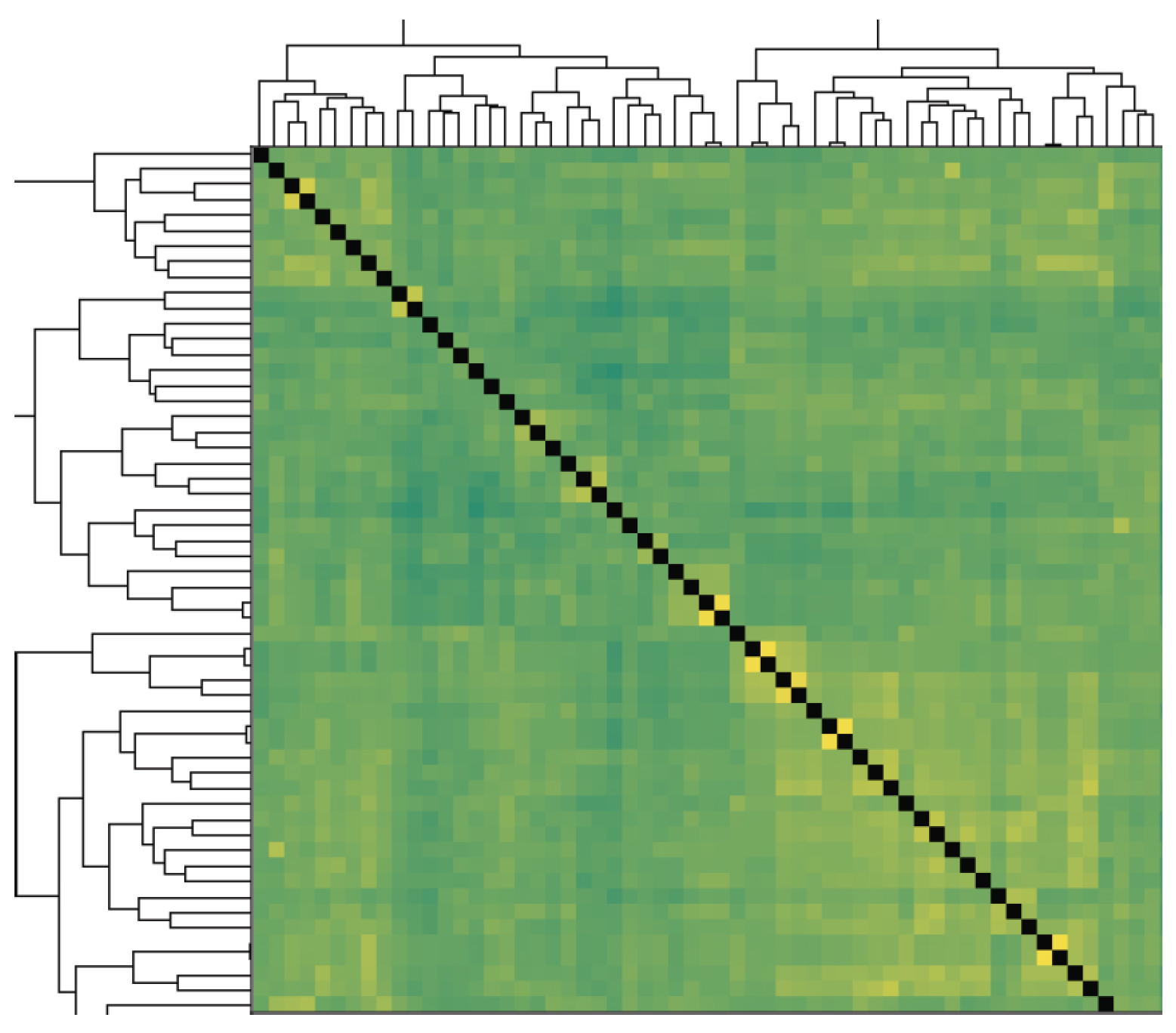
3.3. Genetic Analysis and Phenotypic Variation of Lodging Resistance in Soybean Varieties
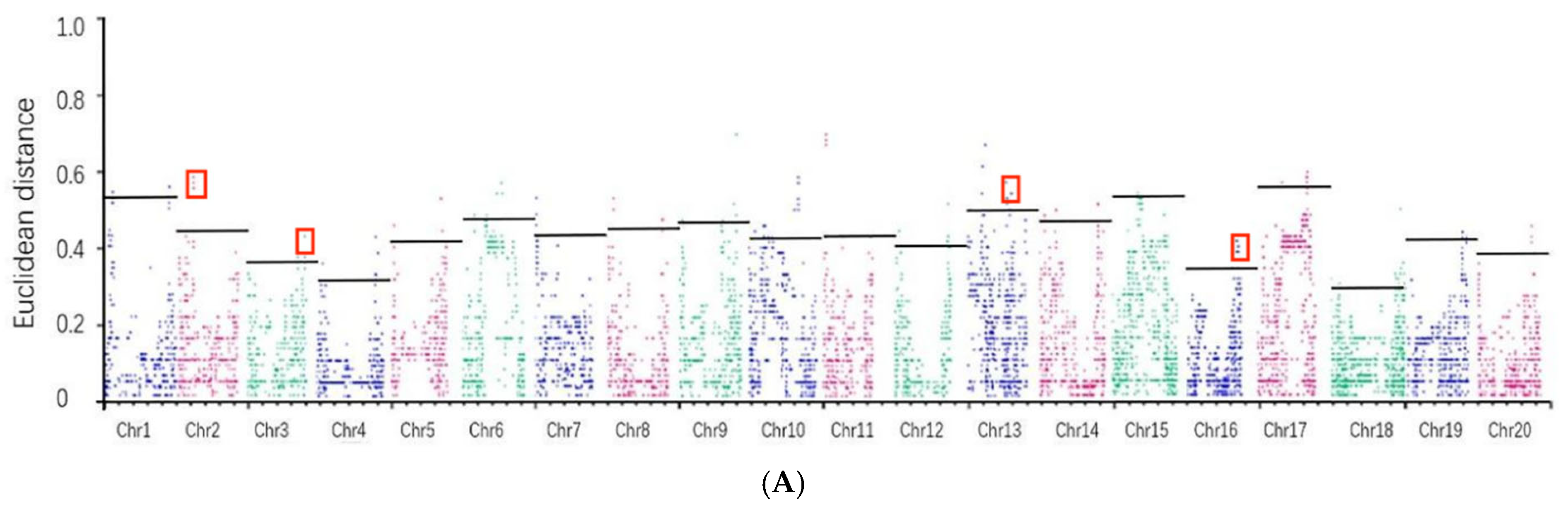
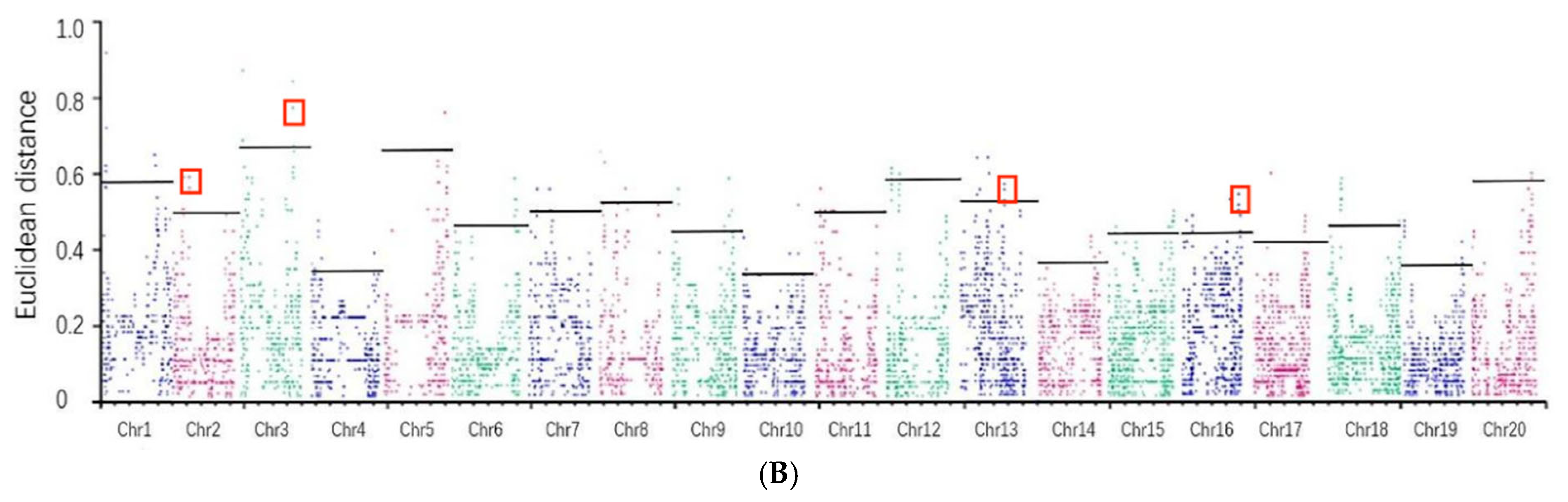
3.4. Identification of Candidate Genomic Regions Associated with Target Traits in Soybean
3.5. Screening Genes Related to Lodging Resistance
3.6. Candidate Gene Verification Using RT–qPCR Data
3.7. Functional Analysis of Candidate Genes
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, C.H.; Chang, Y.L.; Luo, Y.L.; Li, W.Q.; Jin, M.; Wang, Y.Y.; Cui, H.X.; Sun, S.F.; Li, Y.; Wang, Z.L. Nitrogen regulates stem lodging resistance by breaking the balance of photosynthetic carbon allocation in wheat. Field Crop Res. 2023, 296, 108908. [Google Scholar] [CrossRef]
- Liu, Y.; Nie, C.W.; Zhang, Z.; Wang, Z.X.; Ming, B.; Xue, J.; Yang, H.Y.; Xu, H.G.; Meng, L.; Cui, N.B.; et al. Evaluating how lodging affects maize yield estimation based on UAV observations. Front. Plant Sci. 2023, 13, 979103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Gu, S.C.; Wang, Y.Y.; Xu, C.C.; Zhao, Y.T.; Liu, X.L.; Wang, P.; Huang, S.B. The relationships between maize (Zea mays L.) lodging resistance and yield formation depend on dry matter allocation to ear and stem. Crop J. 2023, 11, 258–268. [Google Scholar] [CrossRef]
- Feng, S.W.; Jiang, X.L.; Ding, W.H.; Niu, L.Y.; Song, X.M.; Ru, Z.G. Studies of wheat lodging resistance on the basis of a new method. Acta Agric. Boreali-Sinica 2015, 30, 69–72. [Google Scholar]
- Peng, J.Y.; Lu, L.; Noor, M.A.; Li, S.Y.; Ma, W.; Wang, J. Mid-season lodging modulates photosynthesis, evapotranspiration, and dry matter accumulation and distribution simulated by the optimized model in maize. Front. Ecol. Evol. 2023, 11, 1178609. [Google Scholar] [CrossRef]
- Yang, Y.S.; Guo, X.X.; Hou, P.; Xue, J.; Liu, G.Z.; Liu, W.M.; Wang, Y.H.; Zhao, R.L.; Ming, B.; Xie, R.Z.; et al. Quantitative effects of solar radiation on maize lodging resistance mechanical properties. Field Crop Res. 2020, 255, 107906. [Google Scholar] [CrossRef]
- Xue, J.; Gao, S.; Fan, Y.H.; Li, L.L.; Ming, B.; Wang, K.R.; Xie, R.Z.; Hou, P.; Li, S.K. Traits of plant morphology, stalk mechanical strength, and biomass accumulation in the selection of lodging-resistant maize cultivars. Eur. J. Agron. 2020, 117, 126073. [Google Scholar] [CrossRef]
- Kashiwagi, T. Novel QTL for Lodging Resistance, PRL4, Improves Physical Properties with High Non-Structural Carbohydrate Accumulation of Basal Culms in Rice (Oryza sativa, L.). Euphytica 2022, 218, 83. [Google Scholar] [CrossRef]
- Yano, K.; Ookawa, T.; Aya, K.; Ochiai, Y.; Hirasawa, T.; Ebitani, T.; Takarada, T.; Yano, M.; Yamamoto, T.; Fukuoka, S.; et al. Isolation of a Novel Lodging Resistance QTL Gene Involved in Strigolactone Signaling and Its Pyramiding with a QTL Gene Involved in Another Mechanism. Mol. Plant. 2015, 8, 303–314. [Google Scholar] [CrossRef]
- Piñera-Chávez, F.J.; Berry, P.M.; Foulkes, M.J.; Molero, G.; Reynolds, M.P. Avoiding lodging in irrigated spring wheat. II. Genetic variation of stem and root structural properties. Field Crop Res. 2016, 196, 64–74. [Google Scholar] [CrossRef]
- Sajjad, M.; Ahmad, A.; Riaz, M.W.; Hussain, Q.; Yasir, M.; Lu, M.Z. Recent genome resequencing paraded COBRA-Like gene family roles in abiotic stress and wood formation in Poplar. Front. Plant Sci. 2023, 14, 1242836. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, J.; Li, N.; Pei, Y.F.; Peng, J.; Wang, Z. An integrated strategy coordinating endogenous and exogenous approaches to alleviate crop lodging. Plant Stress 2023, 9, 100197. [Google Scholar] [CrossRef]
- Hu, D.; Liu, X.B.; She, H.Z.; Gao, Z.; Ruan, R.W.; Wu, D.Q.; Yi, Z.L. The lignin synthesis related genes and lodging resistance of Fagopyrum esculentum. Biol. Plant 2017, 61, 138–146. [Google Scholar] [CrossRef]
- Ahmad, I.; Meng, X.P.; Kamran, M.; Ali, S.; Ahmad, S.; Liu, T.N.; Cai, T.; Han, Q.F. Effects of uniconazole with or without micronutrient on the lignin biosynthesis, lodging resistance, and winter wheat production in semiarid regions. J. Integr. Agric. 2020, 19, 62–77. [Google Scholar] [CrossRef]
- Miao, W.; Li, F.C.; Lu, J.C.; Wang, D.L.; Chen, M.K.; Tang, L.; Xu, Z.J.; Chen, W.F. Biochar application enhanced rice biomass production and lodging resistance via promoting co-deposition of silica with hemicellulose and lignin. Sci. Total Environ. 2023, 855, 158818. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.X.; Guo, Y.L.; Huang, G.M.; Liu, Y.R.; Li, Z.H.; Zhou, Y.Y.; Duan, L.S. Effects of a novel plant growth regulator B2 on stalk quality and grain yield of winter wheat in North China. Plant Soil. 2023, 487, 437–453. [Google Scholar] [CrossRef]
- Hussain, S.; Iqbal, N.; Rahman, T.; Liu, T.; Brestic, M.; Safdar, M.E.; Asghar, M.A.; Farooq, M.U.; Shafiq, I.; Ali, A. Shade effect on carbohydrates dynamics and stem strength of soybean genotypes. Environ. Exp. Bot. 2019, 162, 374–382. [Google Scholar] [CrossRef]
- Chen, X.G.; Wang, J.; Wang, Z.L.; Li, W.Y.; Wang, C.Y.; Yan, S.H.; Li, H.M.; Zhang, A.J.; Tang, Z.H.; Wei, M. Optimized nitrogen fertilizer application mode increased culms lignin accumulation and lodging resistance in culms of winter wheat. Field Crops Res. 2018, 228, 31–38. [Google Scholar] [CrossRef]
- Ke, S.W.; Luan, X.; Liang, J.Y.; Hung, Y.H.; Hsieh, T.F.; Zhang, X.Q. Rice OsPEX1, an extensin-like protein, affects lignin biosynthesis and plant growth. Plant Mol. Biol. 2019, 100, 151–161. [Google Scholar] [CrossRef]
- Li, Q.; Fu, C.F.; Liang, C.L.; Ni, X.J.; Zhao, X.H.; Chen, M.; Ou, L.J. Crop lodging and the roles of lignin, cellulose, and hemicellulose in lodging resistance. Agronomy 2022, 12, 1795. [Google Scholar] [CrossRef]
- Ma, Q.H. Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 2010, 61, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Asghar, M.A.; Javed, H.H.; Ullah, A.; Cheng, B.; Xu, M.; Wang, W.Y.; Liu, C.Y.; Rahman, A.; Iqbal, T.; et al. Optimum nitrogen improved stem breaking resistance of intercropped soybean by modifying the stem anatomical structure and lignin metabolism. Plant Physiol. Biochem. 2023, 199, 107720. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.J.; Qi, F.Y.; Qin, L.; Zhang, M.N.; Sun, Z.Q.; Li, H.Y.; Cui, M.J.; Zhang, M.Y.; Li, C.Y.; Li, X.N.; et al. Mapping of a QTL associated with sucrose content in peanut kernels using BSA–seq. Front. Genet. 2023, 13, 1089389. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.H.; Yan, L.; Ma, X.W.; Chen, Y.P.; Wu, L.M.; Ma, T.T.; Zhao, L.; Yi, B.; Ma, C.Z.; Tu, J.X.; et al. Combined BSA-seq based mapping and RNA-seq profiling reveal candidate genes associated with plant architecture in Brassica napus. Int. J. Mol. Sci. 2022, 23, 2472. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.; Johar, P.; Raina, A.; Salgotra, R.K.; Feng, X.; Bhat, J.A. Harnessing the potential of bulk segregant analysis sequencing and its related approaches in crop breeding. Front. Genet. 2022, 13, 944501. [Google Scholar] [CrossRef]
- Liu, X.M.; Bi, B.; Xu, X.; Li, B.H.; Tian, S.M.; Wang, J.P.; Zhang, H.; Wang, G.Q.; Han, Y.J.; McElroy, J.S. Rapid identification of a candidate nicosulfuron sensitivity gene (Nss) in maize (Zea mays L.) via combining bulked segregant analysis and RNA-seq. Threo Appl. Genet. 2019, 132, 1351–1361. [Google Scholar] [CrossRef]
- Zhu, T.; Wu, L.R.; He, H.G.; Song, J.C.; Jia, M.S.; Liu, L.C.; Wang, X.L.; Han, R.; Niu, L.P.; Du, W.X.; et al. Bulked segregant RNA—Seq reveals distinct expression profiling in Chinese wheat cultivar Jimai 23 responding to powdery mildew. Front. Genet. 2020, 11, 474. [Google Scholar] [CrossRef]
- Lin, M.M.; Sun, S.H.; Fang, J.B.; Qi, X.J.; Sun, L.M.; Zhong, Y.P.; Sun, Y.X.; Hong, G.; Wang, R.; Li, Y.K. BSR-Seq analysis provides insights into the cold stress response of Actinidia arguta F1 populations. BMC Genom. 2021, 22, 72. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, D.; Wang, T.; Liang, X.; Cui, Y.; Liu, Z.; Li, W. Concentration difference of auxin involved in stem development in soybean. J. Integr. Agr. 2020, 19, 953–964. [Google Scholar] [CrossRef]
- Fehr, W.R.; Caviness, C.E. Stages of Soybean Development. J. Stages of Soybean Development. (Iowa Agriculture and Economics Experiment Station Special Report 80); Iowa State University: Ames, IA, USA, 1977. [Google Scholar]
- Foster, C.E.; Martin, T.M.; Pauly, M. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: Lignin. J. Vis. Exp. 2010, 37, e1745. [Google Scholar] [CrossRef]
- Foster, C.E.; Martin, T.M.; Pauly, M. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: Carbohydrates. J. Vis. Exp. 2010, 37, e1837. [Google Scholar] [CrossRef]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.C.; Yost, H.J. MMAPPR: Mutation Mapping Analysis Pipeline for Pooled RNA–seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Chen, Z.; Li, X.; Song, G.; Wu, Y.; Jin, J.; Cui, F.; Yuan, J.; Qi, H.; Wang, J.; et al. Optimization of fermentation conditions for Bacillus velezensis TCS001 and evaluation of its growth promotion and disease prevention effects on strawberries. Biol. Control 2024, 198, 105632. [Google Scholar] [CrossRef]
- Zhang, N.; Xie, Y.D.; Guo, H.J.; Zhao, L.S.; Xiong, H.C.; Gu, J.Y.; Li, J.H.; Kong, F.Q.; Sui, L.; Zhao, Z.W.; et al. Gibberellins regulate the stem elongation rate without affecting the mature plant height of a quick development mutant of winter wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2016, 107, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Zou, M.; Li, J. Basally distributed actin array drives embryonic hypocotyl elongation during the seed-to-seedling transition in Arabidopsis. New Phytol. 2023, 240, 191–206. [Google Scholar] [CrossRef]
- Resentini, F.; Cyprys, P.; Steffen, J.G.; Alter, S.; Morandini, P.; Mizzotti, C.; Lloyd, A.; Drews, G.N.; Dresselhaus, T.; Colombo, L.; et al. SUPPRESSOR OF FRIGIDA (SUF4) supports gamete fusion via regulating Arabidopsis EC1 gene expression. Plant Physiol. 2017, 173, 155–166. [Google Scholar] [CrossRef]
- Muto, H.; Yabe, N.; Asami, T.; Hasunuma, K.; Yamamoto, K.T. Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol. 2004, 136, 3124–3133. [Google Scholar] [CrossRef]
- Fristedt, R. Chloroplast function revealed through analysis of GreenCut2 genes. J. Exp. Bot. 2017, 68, 2111–2120. [Google Scholar] [CrossRef]
- Guo, R.K.; Wen, X.; Zhang, W.; Huang, L.; Peng, Y.; Jin, L.; Han, H.H.; Zhang, L.L.; Li, W.Y.; Guo, H.W. Arabidopsis EIN2 represses ABA responses during germination and early seedling growth by inactivating HLS1 protein independently of the canonical ethylene pathway. Plant J. 2023, 115, 1514–1527. [Google Scholar] [CrossRef]
- Ge, Y.H.; Yan, F.L.; Zourelidou, M.; Wang, M.L.; Ljung, K.; Fastner, A.; Hammes, U.Z.; Donato, M.D.; Geisler, M.; Schwechheimer, C.; et al. SHADE AVOIDANCE 4 is required for proper auxin distribution in the hypocotyl. Plant Physiol. 2017, 173, 788–800. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Song, L.H.; Hegie, A.; Suzuki, N.; Shulaev, E.; Luo, X.Z.; Cenariu, D.; Ma, V.; Kao, S.; Lim, J.; Gunay, M.B. Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plant. 2013, 148, 322–333. [Google Scholar] [CrossRef]
- Sandoval, F.J.; Roje, S. An FMN hydrolase is fused to a riboflavin kinase homolog in plants. J. Biol. Chem. 2005, 280, 38337–38345. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Fu, Q.T.; Huang, W.D.; Yu, D.Q. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009, 28, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Botwright, T.L.; Rebetzke, G.J.; Condon, A.G.; Richards, R.A. Influence of the gibberellin-sensitive Rht8 dwarfing gene on leaf epidermal cell dimensions and early vigour in wheat (Triticum aestivum L.). Ann. Bot. 2005, 95, 631–639. [Google Scholar] [CrossRef]
- Zhang, W.H.; Li, J.; Li, H.Y.; Zhang, D.D.; Zhu, B.C.; Yuan, H.L.; Gao, T.G. Transcriptomic analysis of humic acid in relieving the inhibitory effect of high nitrogen on soybean nodulation. Front. Plant Sci. 2023, 14, 1196939. [Google Scholar] [CrossRef]
- Noor, R.B.M.; Caviness, C.E. Influence of induced lodging on pod distribution and seed yield in Soybeans. Agronomy 1980, 72, 904–906. [Google Scholar] [CrossRef]
- Tian, J.Y.; Li, S.P.; Cheng, S.; Liu, Q.Y.; Zhou, L.; Tao, Y.; Xing, Z.P.; Hu, Y.J.; Guo, B.W.; Wei, H.Y.; et al. Increasing the appropriate seedling density for higher yield in dry direct-seeded rice sown by a multifunctional seeder after wheat-straw return. J. Integr. Agric. 2023, 22, 400–416. [Google Scholar] [CrossRef]
- Kirkby, E.A.; Nikolic, M.; White, P.J.; Xu, G. Mineral nutrition, yield, and source-sink relationships. In Marschner’s Mineral Nutrition of Plants; Academic Press: Cambridge, MA, USA, 2023; pp. 131–200. [Google Scholar] [CrossRef]
- Shao, H.; Shi, D.F.; Shi, W.J.; Ban, X.B.; Chen, Y.C.; Ren, W.; Chen, F.J.; Mi, G.H. The impact of high plant density on dry matter remobilization and stalk lodging in maize genotypes with a different stay-green degree. Arch. Agron. Soil. Sci. 2021, 67, 504–518. [Google Scholar] [CrossRef]
- Xiang, D.B.; Zhao, G.; Wan, Y.; Tan, M.L.; Song, C.; Song, Y. Effect of planting density on lodging-related morphology, lodging rate, and yield of tartary buckwheat (Fagopyrum tataricum). Plant Prod. Sci. 2016, 19, 479–488. [Google Scholar] [CrossRef]
- Zhang, P.; Yan, Y.; Gu, S.C.; Wang, Y.Y.; Xu, C.L.; Sheng, D.C.; Li, Y.B.; Wang, P.; Huang, S.B. Lodging resistance in maize: A function of root-shoot interactions. Eur. J. Agron. 2022, 132, 126393. [Google Scholar] [CrossRef]
- Gao, H.W.; Yang, M.Y.; Yan, L.; Hu, X.Z.; Hong, H.L.; Zhang, X.; Sun, R.J.; Wang, H.R.; Wang, X.B.; Liu, L.K.; et al. Identification of tolerance to high density and lodging in short petiolate germplasm M657 and the effect of density on yield–related phenotypes of soybean. J. Integr. Agr. 2023, 22, 434–446. [Google Scholar] [CrossRef]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. Root lodging is a physical stress that changes gene expression from sucrose accumulation to degradation in sorghum. BMC Plant Biol. 2018, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Liu, T.; Iqbal, N.; Brestic, M.; Pang, T.; Mumtaz, M.; Shafiq, I.; Li, S.X.; Wang, L.; Gao, Y.; et al. Effects of lignin, cellulose, hemicellulose, sucrose and monosaccharaides carbohydrates on soybean physical stem strength and yield in intercropping. Photochem. Photobiol. Sci. 2020, 19, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef]
- Ogden, M.; Hoefgen, R.; Roessner, U.; Persson, S.; Khan, G.A. Feeding the walls: How does nutrient availability regulate cell wall composition? Int. J. Mol. Sci. 2018, 19, 2691. [Google Scholar] [CrossRef]
- Manga-Robles, A.; Santiago, R.; Malvar, R.A.; Moreno-González, V.; Fornalé, S.; López, I.; Centeno, M.L.; Acebes, J.L.; Álvarez, J.M.; Caparros-Ruiz, D.; et al. Elucidating compositional factors of maize cell walls contributing to stalk strength and lodging resistance. Plant Sci. 2021, 307, 110882. [Google Scholar] [CrossRef]
- Liu, S.T.; Tang, Y.J.; Ruan, N.; Dang, Z.J.; Huang, Y.W.; Miao, W.; Xu, Z.J.; Li, F.C. The rice BZ1 locus is required for glycosylation of arabinogalactan proteins and galactolipid and plays a role in both mechanical strength and leaf color. Rice 2020, 13, 41. [Google Scholar] [CrossRef]
- Wang, X.Q.; Shi, Z.; Zhang, R.Y.; Sun, X.; Wang, J.D.; Wang, S.; Zhang, Y.; Zhao, Y.X.; Su, A.G.; Li, C.H.; et al. Stalk architecture, cell wall composition, and QTL underlying high stalk flexibility for improved lodging resistance in maize. BMC Plant Biol. 2020, 20, 515. [Google Scholar] [CrossRef]
- Zhou, C.Y.; Xiong, H.C.; Li, Y.T.; Guo, H.J.; Xie, Y.D.; Zhao, L.S.; Gu, J.Y.; Zhao, S.R.; Ding, Y.P.; Song, X.Y. Genetic analysis and QTL mapping of a novel reduced height gene in common wheat (Triticum aestivum L.). J. Integr. Agric. 2020, 19, 1721–1730. [Google Scholar] [CrossRef]
- Jiang, J.Y.; Liao, X.L.; Jin, X.Y.; Tan, L.; Lu, Q.F.; Yuan, C.L.; Xue, Y.F.; Yin, N.W.; Lin, N.; Chai, Y.R. MYB43 in oilseed rape (Brassica napus) positively regulates vascular lignification, plant morphology and yield potential but negatively affects resistance to Sclerotinia sclerotiorum. Genes 2020, 11, 581. [Google Scholar] [CrossRef]
- Brulé, V.; Rafsanjani, A.; Pasini, D.; Western, T.L. Hierarchies of plant stiffness. Plant Sci. 2016, 250, 79–96. [Google Scholar] [CrossRef]
- Shah, D.U.; Reynolds, T.P.; Ramage, M.H. The strength of plants: Theory and experimental methods to measure the mechanical properties of stems. J. Exp. Bot. 2017, 68, 4497–4516. [Google Scholar] [CrossRef]
- Liao, P.; Bell, S.M.; Chen, L.; Huang, S.; Wang, H.Y.; Miao, J.H.; Qi, Y.M.; Sun, Y.N.; Liao, B.; Zeng, Y.J.; et al. Improving rice grain yield and reducing lodging risk simultaneously: A meta-analysis. Eur. J. Agron. 2023, 143, 126709. [Google Scholar] [CrossRef]
- Luo, X.Y.; Wu, Z.F.; Fu, L.; Dan, Z.W.; Long, W.X.; Yuan, Z.Q.; Liang, T.; Zhu, R.S.; Hu, Z.L.; Wu, X.T. Responses of the lodging resistance of indica rice cultivars to temperature and solar radiation under field conditions. Agronomy 2022, 12, 2603. [Google Scholar] [CrossRef]
- Wang, T.T.; Jin, Y.; Deng, L.X.; Li, F.; Wang, Z.Y.; Zhu, Y.Y.; Wu, Y.F.; Qu, H.Y.; Zhang, S.N.; Liu, Y.; et al. The transcription factor MYB110 regulates plant height, lodging resistance, and grain yield in rice. Plant Cell 2023, 36, 298–323. [Google Scholar] [CrossRef]
- Hwang, S.; Lee, T.G. Integration of lodging resistance QTL in soybean. Sci. Rep. 2019, 9, 6540. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Li, Y.H.; Li, D.L.; Zhang, X.R.; Kong, L.P.; Zhou, Y.G.; Lyu, X.G.; Ji, R.H.; Wei, X.Z.; Cheng, Q.C.; et al. PH13 improves soybean shade traits and enhances yield for high-density planting at high latitudes. Nat. Commun. 2023, 14, 6813. [Google Scholar] [CrossRef]
- Wang, X.; Han, L.Q.; Li, J.; Shang, X.Y.; Liu, Q.; Li, L.; Zhang, H.W. Next-generation bulked segregant analysis for Breeding 4.0. Cell Rep. 2023, 42, 113039. [Google Scholar] [CrossRef]
- Shah, L.; Yahya, M.; Shah, S.M.A.; Nadeem, M.; Ali, A.; Ali, A.; Wang, J.; Riaz, M.W.; Reman, S.; Wu, W.X.; et al. Improving lodging resistance: Using wheat and rice as classical examples. Int. J. Mol. Sci. 2019, 20, 4211. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Slafer, G.A.; Davies, W.J.; Berry, P.M.; Sylvester–Bradley, R.; Martre, P.; Calderini, D.F.; Griffiths, S.; Reynolds, M.P. Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. J. Exp. Bot. 2011, 62, 469–486. [Google Scholar] [CrossRef] [PubMed]




| Gene ID | Primer Sequence (5′-3′) | |
|---|---|---|
| Glyma.02G138700 | F: AATCCCAAATGCTCTGTTCCG | R: GGTAAGGTTCTTTTCCGTCGTG |
| Glyma.02G139500 | F: GACCGCAAACCCCACGATAA | R: CACCAGAAGAACCTGTTGGAGAAA |
| Glyma.03G181800 | F: CAGGTCTTTGCGACATTCTACG | R: AACATCGGTGGCTTCCTCTTC |
| Glyma.03G182400 | F: TGCAATTCCCAGTTTGTCCC | R: TCACTTCTGTCACCCATTCCCTA |
| Glyma.02G139600 | F: CAAGCTCGTCGAGGATTTTGA | R: AACCCAGGTAACAGCACCAACTAT |
| Glyma.03G183600 | F: GAGCACCGTTGTATCCCTCATT | R: CCTATCTGCTGCCTCAACCATC |
| Glyma.03G184500 | F: CAAACAATGAAGCACATCCCG | R: TTGGCACATCATCCACTAGGAA |
| Glyma.03G184600 | F: CAATAACGGCGAGCATCACA | R: CACATCCTTCAACGAGCATTTCAC |
| Glyma.03G184200 | F: TTGGTGTTCGTTTGGCTCCT | R: CCACAATTCAGTTATGCCCTCA |
| Glyma.16G196200 | F: ATCTGAGCCGTTGGATTGCC | R: CACCCTTTGATTTGTCCTGTGAC |
| Glyma.16G196000 | F: AAACCAAGGCAACTCCGTCTC | R: CTGGTTGTTGGACCCATTGAA |
| Glyma.03G181400 | F: GAATGGTGGAGCCGATGAGGAT | R: CCGCAATTTGGAACCGAAGA |
| Stage | Index | Plant Height/cm | Diameter/mm | |||||
|---|---|---|---|---|---|---|---|---|
| Vn | R5 | R8 | Vn | R5 | R8 | |||
| Variety | ||||||||
| Heike71 (L) | 14.20 ± 0.20 | 41.60 ± 0.63 | 50.10 ± 0.23 | 4.71 ± 0.46 | 6.65 ± 0.71 | 6.32 ± 0.17 | ||
| Heike71 (H) | 22.07 ± 0.03 | 54.00 ± 0.30 | 59.50 ± 0.41 | 4.43 ± 0.23 | 5.00 ± 0.02 | 5.42 ± 0.34 | ||
| Heihe43 (L) | 21.78 ± 0.88 | 54.58 ± 0.28 | 59.90 ± 0.43 | 5.19 ± 0.24 | 5.66 ± 0.06 | 7.22 ± 0.34 | ||
| Heihe43 (H) | 27.31 ± 0.30 | 66.50 ± 0.21 | 79.77 ± 0.31 | 4.12 ± 0.22 | 4.84 ± 0.37 | 6.16 ± 0.35 | ||
| Heike88 (L) | 21.80 ± 0.25 | 64.92 ± 0.18 | 65.70 ± 0.64 | 5.33 ± 0.27 | 5.37 ± 0.24 | 6.86 ± 0.53 | ||
| Heike88 (H) | 24.16 ± 0.52 | 73.00 ± 0.19 | 80.40 ± 0.40 | 4.12 ± 0.19 | 5.08 ± 0.16 | 5.47 ± 0.60 | ||
| Heike85 (L) | 20.85 ± 0.57 | 49.60 ± 0.13 | 53.50 ± 0.11 | 4.64 ± 0.49 | 5.41 ± 0.29 | 5.52 ± 0.49 | ||
| Heike85 (H) | 25.69 ± 0.27 | 61.83 ± 0.40 | 69.70 ± 0.50 | 4.01 ± 0.28 | 4.61 ± 0.29 | 4.71 ± 0.24 | ||
| Stage | Index | Hardness/N | Resilience/N | |||||
|---|---|---|---|---|---|---|---|---|
| R5 | R8 | R8 (Test Field) | R5 | R8 | R8 (Test Field) | |||
| Variety | ||||||||
| Heike71 (L) | 3887.58 ± 733 | 3613.83 ± 965 | 5226.28 ± 984 | 57.50 ± 0.38 | 57.50 ± 0.38 | 57.50 ± 0.38 | ||
| Heike71 (H) | 1333.81 ± 140 | 2144.71 ± 425 | 4678.51 ± 771 | 58.59 ± 1.46 | 58.59 ± 1.46 | 58.59 ± 1.46 | ||
| Heihe43 (L) | 3604.26 ± 80 | 5482.83 ± 486 | 11,053.16 ± 106 | 56.93 ± 0.43 | 56.93 ± 0.43 | 56.93 ± 0.43 | ||
| Heihe43 (H) | 1611.39 ± 504 | 2417.05 ± 359 | 7184.96 ± 1143 | 60.81 ± 0.09 | 60.81 ± 0.09 | 60.81 ± 0.09 | ||
| Heike88 (L) | 2070.06 ± 311 | 5402.55 ± 556 | 11,475.91 ± 219 | 54.21 ± 0.65 | 54.21 ± 0.65 | 54.21 ± 0.65 | ||
| Heike88 (H) | 1964.84 ± 597 | 2082.91 ± 220 | 5221.12 ± 1201 | 58.20 ± 1.23 | 58.20 ± 1.23 | 58.20 ± 1.23 | ||
| Heike85 (L) | 2472.86 ± 509 | 4424.22 ± 1564 | 5576.78 ± 1097 | 56.71 ± 0.70 | 56.71 ± 0.70 | 56.71 ± 0.70 | ||
| Heike85 (H) | 1020.08 ± 351 | 1822.26 ± 291 | 3572.91 ± 600 | 62.35 ± 0.91 | 62.35 ± 0.91 | 62.35 ± 0.91 | ||
| Year | Max/mm | Min/mm | Mean/mm | Standard Deviation | Kutosis | Skewness |
|---|---|---|---|---|---|---|
| 2020 | 12.95 | 2.57 | 7.38 | 1.91 | 0.52 | 0.38 |
| 2021 | 12.93 | 5.77 | 8.41 | 1.38 | 0.69 | 0.71 |
| Chromosome | ED Association Analysis of Natural Populations in 2020 | ED Association Analysis of Natural Populations in 2021 |
|---|---|---|
| Chr2 | 0.4525 | 0.5091 |
| Chr3 | 0.3688 | 0.6788 |
| Chr13 | 0.5091 | 0.5374 |
| Chr16 | 0.3538 | 0.4534 |
| Chromosome | Start Location | End Location | Size/Mb |
|---|---|---|---|
| Chr2 | 14333783 | 14628947 | 0.2952 |
| Chr3 | 41117720 | 41517255 | 0.3995 |
| Chr13 | 32018780 | 32072589 | 0.0538 |
| Chr16 | 35982944 | 36098060 | 0.1151 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Zeng, D.; Zhao, C.; Han, D.; Li, S.; Wen, M.; Liang, X.; Zhang, X.; Liu, Z.; Ali, S.; et al. Identification of QTLs and Key Genes Enhancing Lodging Resistance in Soybean Through Chemical and Physical Trait Analysis. Plants 2024, 13, 3470. https://doi.org/10.3390/plants13243470
Zhao W, Zeng D, Zhao C, Han D, Li S, Wen M, Liang X, Zhang X, Liu Z, Ali S, et al. Identification of QTLs and Key Genes Enhancing Lodging Resistance in Soybean Through Chemical and Physical Trait Analysis. Plants. 2024; 13(24):3470. https://doi.org/10.3390/plants13243470
Chicago/Turabian StyleZhao, Wanying, Depeng Zeng, Caitong Zhao, Dezhi Han, Shuo Li, Mingxing Wen, Xuefeng Liang, Xianfeng Zhang, Zhihua Liu, Shahid Ali, and et al. 2024. "Identification of QTLs and Key Genes Enhancing Lodging Resistance in Soybean Through Chemical and Physical Trait Analysis" Plants 13, no. 24: 3470. https://doi.org/10.3390/plants13243470
APA StyleZhao, W., Zeng, D., Zhao, C., Han, D., Li, S., Wen, M., Liang, X., Zhang, X., Liu, Z., Ali, S., & Jiang, Z. (2024). Identification of QTLs and Key Genes Enhancing Lodging Resistance in Soybean Through Chemical and Physical Trait Analysis. Plants, 13(24), 3470. https://doi.org/10.3390/plants13243470






