Molecular Characterization of Plant Volatile Compound Interactions with Cnaphalocrocis medinalis Odorant-Binding Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Electrophysiology
2.3. Total RNA Extraction and cDNA Synthesis
2.4. Real-Time Quantitative PCR (RT-qPCR)
2.5. In Vitro CmedOBP Expression and Purification
2.6. Fluorescent Competitive Binding Assay
2.6.1. Odorant Preparation and Measurement Parameters
2.6.2. Binding Assays
2.6.3. Binding Ability Calculations
2.7. Statistical Analyses
3. Results
3.1. C. medinalis EAG Responses to a Combination of Plant Volatiles
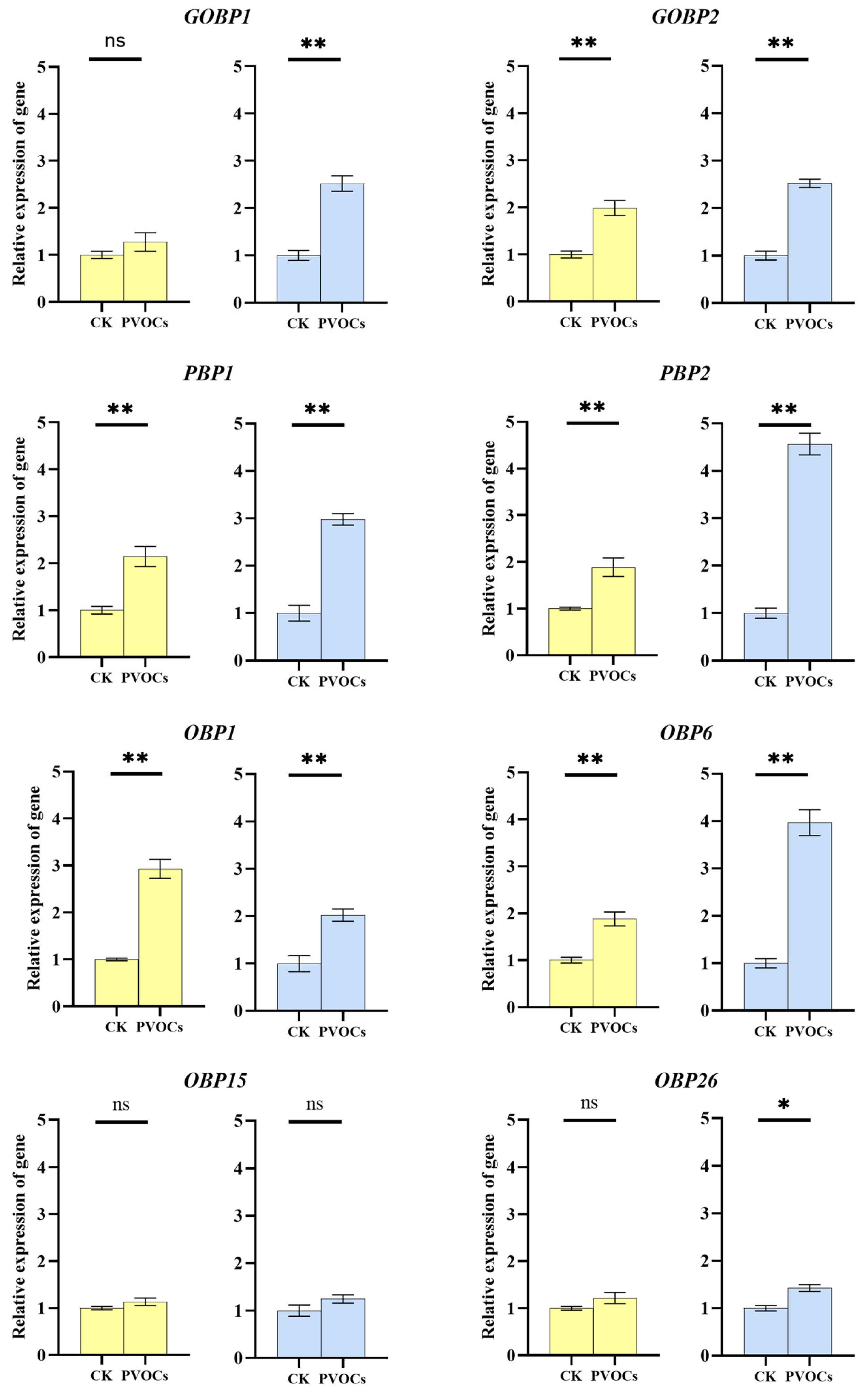
3.2. Effects of Plant Volatiles on OBP Expression in Adult C. medinalis Antennae
3.3. Temporal CmedOBP Expression in Unstimulated C. medinalis Antennae
3.4. Binding Characteristics of CmedOBPs to Plant Volatiles
4. Discussion
4.1. EAG and CmedOBP Expression Responses to Plant Volatile Exposure
4.2. Gene Expression Analysis of CmedOBPs
4.3. CmedOBP–Plant Volatile Binding Characteristics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Khan, Z.; Barrion, A.; Litsinger, J.; Castilla, N.; Joshi, R. A bibliography of rice leaffolders (Lepidoptera: Pyralidae). Insect Sci. Its Appl. 1988, 9, 129–174. [Google Scholar] [CrossRef]
- Xu, J.; Li, C.M.; Yang, Y.J.; Qi, J.H.; Zheng, X.S.; Hu, R.L.; Lu, Z.X.; Liu, Q. Growth and reproduction of artificially fed Cnaphalocrocis medinalis. Rice Sci. 2012, 19, 247–251. [Google Scholar] [CrossRef]
- Chintalapati, P.; Gururaj, K.; Vallabuni, S.; Yenumulag, P. Physiological age status of female adults and off-season survival of rice leaffolder Cnaphalocrocis medinalis in India. Rice Sci. 2015, 22, 237–244. [Google Scholar] [CrossRef]
- Khan, Z.R.; Abenes, M.L.P.; Fernandez, N.J. Suitability of graminaceous weed species as host plants for rice leaffolders, Cnaphalocrocis medinalis and Marasmia patnalis. Crop Prot. 1996, 15, 121–137. [Google Scholar] [CrossRef]
- Yang, Y.J.; Xu, H.X.; Zheng, X.S.; Tian, J.C.; Lu, Y.H.; Lu, Z.X. Progresses in management technology of rice leaffolders in China. J. Plant Prot. 2015, 42, 691–701. (In Chinese) [Google Scholar]
- Shah, S.M.A.; Hidayat-ur-Rahman; Rehman, A.; Abassi, F.M.; Khalil, I.H.; Ali, A. Characterization of wild rice species in response to leaffolder Cnaphalocrocis medinalis. Sarhad J. Agric. 2008, 24, 69–74. [Google Scholar]
- Xu, J.; Liu, Q.; Li, C.M.; Han, G.J. Field effect of Cnaphalocrocis medinalis granulovirus (CnmeGV) on the pest of rice leaffolder. J. Integr. Agric. 2019, 18, 2115–2122. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, S.; Ling, Y.; Wang, L.; Ni, H.; Guo, D.; Dong, B.; Huang, Q.; Long, L.; Zhang, S.; et al. Insecticide resistance monitoring of Cnaphalocrocis medinalis (Lepidoptera: Pyralidae) and its mechanism to chlorantraniliprole. Pest Manag. Sci. 2023, 79, 3290–3299. [Google Scholar] [CrossRef]
- Kevan, P.G.; Baker, H.G. Insects as flower visitors and pollinators. Annu. Rev. Entomol. 1983, 28, 407–453. [Google Scholar] [CrossRef]
- Breer, H.; Krieger, J.; Raming, K. A novel class of binding proteins in the antennae of the silk moth Antheraea pernyi. Insect Biochem. 1990, 20, 735–740. [Google Scholar] [CrossRef]
- Zhou, J.J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef]
- Zhou, J.J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 2018, 93, 184–200. [Google Scholar] [CrossRef]
- Hern, A.; Dorn, S. Induction of volatile emissions from ripening apple fruits infested with Cydia pomonella and the attraction of adult females. Entomol. Exp. Appl. 2002, 102, 145–151. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Fan, J.; Francis, F.; Liu, Y.; Chen, J.; Cheng, D. An overview of odorant-binding protein functions in insect peripheral olfactory reception. Genet. Mol. Res. 2011, 10, 3056–3069. [Google Scholar] [CrossRef]
- Zeng, F.F.; Zhao, Z.F.; Yan, M.J.; Zhou, W.; Zhang, Z.; Zhang, A.; Lu, Z.X.; Wang, M.Q. Identification and comparative expression profiles of chemoreception genes revealed from major chemoreception organs of the rice leaf folder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). PLoS ONE 2015, 10, e0144267. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.M.; Byers, J.A.; Manning, L.M.; Jürgens, A.; Mitchell, V.J.; Suckling, D.M. Floral scent of Canada thistle and its potential as a generic insect attractant. J. Econ. Entomol. 2008, 101, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Szanyi, S.; Nagy, A.; Szarukán, I.; Varga, Z.; Jósvai, J.K.; Tóth, M. A chemical lure for trapping both sexes of Amata phegea L. Insects 2022, 13, 1051. [Google Scholar] [CrossRef] [PubMed]
- Meagher, R.L., Jr. Trapping noctuid moths with synthetic floral volatile lures. Entomol. Exp. Appl. 2003, 103, 219–226. [Google Scholar] [CrossRef]
- Rainho, H.L.; Silva, W.D.; Gonçalves, F.G.; Savaris, M.; Bento, J.M.S. Hexanal combined with decanal mediate host location by the bamboo powderpost beetle, Dinoderus minutus. Entomol. Exp. Appl. 2022, 170, 805–811. [Google Scholar] [CrossRef]
- Zheng, X.S.; Lu, Z.X.; Xu, H.X.; Lu, Y.H.; Yang, T.J.; Tian, J.C. An Attractant for Luring Both Female and Male Moths of Cnaphalocrocis medinalis and Its Application. CN113519520A, 22 October 2021. (In Chinese). [Google Scholar]
- Zhu, A.X.; Qiu, Q.; Liu, X.D. A method for rearing the rice leaf folder (Cnaphalocrocis medinalis) using wheat seedlings. Chin. J. Appl. Entomol. 2015, 52, 883–889. (In Chinese) [Google Scholar]
- Wei, B.; Gao, H.Y.; Zheng, X.S. EAG responses of adult Cnaphalocrocis medinalis to plant volatiles. Chin. J. Appl. Entomol. 2022, 59, 988–996. (In Chinese) [Google Scholar]
- Liu, S.; Wang, W.L.; Zhang, Y.X.; Zhang, B.X.; Rao, X.J.; Liu, X.M.; Wang, D.M.; Li, S.G. Transcriptome sequencing reveals abundant olfactory genes in the antennae of the rice leaffolder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). Entomol. Sci. 2017, 20, 177–188. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zeng, F.F.; Liu, H.; Zhang, A.; Lu, Z.; Leal, W.S.; Abdelnabby, H.; Wang, M. Three chemosensory proteins from the rice leaf folder Cnaphalocrocis medinalis involved in host volatile and sex pheromone reception. Insect Mol. Biol. 2018, 27, 710–723. [Google Scholar] [CrossRef]
- Scatchard, G. The attraction of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Scala, A.; Allmann, S.; Mirabella, R.; Haring, M.A.; Schuurink, R.C. Green leaf volatiles: A plant’s multifunctional weapon against herbivores and pathogens. Int. J. Mol. Sci. 2013, 14, 17781–17811. [Google Scholar] [CrossRef]
- Fang, Y.; Zeng, R.; Lu, S.F.; Dai, L.; Wan, X. The synergistic attractiveness effect of plant volatiles to sex pheromones in a moth. J. Asia-Pac. Entomol. 2018, 21, 380–387. [Google Scholar] [CrossRef]
- Ochieng, S.A.; Park, K.C.; Baker, T.C. Host plant volatiles synergize responses of sex pheromone-specific olfactory receptor neurons in male Helicoverpa zea. J. Comp. Physiol. A 2002, 188, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.M. Current status on the functional characterization of chemosensory receptors of Cydia pomonella (Lepidoptera: Tortricidae). Front. Behav. Neurosci. 2018, 12, 189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, Z.; Cui, G.; Du, Z.; Qian, Y.; Yang, S.; Liu, M.; Guo, J. Binding properties of odorant-binding protein 4 of Tirathaba rufivena to Areca catechu volatiles. Plants 2022, 11, 167. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Duan, H.; Wang, Q.; Xiao, Y.; Wang, Q.; Xiao, Q.; Sun, L.; Zhang, Y. Key amino residues determining binding activities of the odorant binding protein AlucOBP22 to two host plant terpenoids of Apolygus lucorum. J. Agric. Food Chem. 2019, 67, 5949–5956. [Google Scholar] [CrossRef] [PubMed]
- Shields, V.D.; Hildebrand, J.G. Responses of a population of antennal olfactory receptor cells in the female moth Manduca sexta to plant-associated volatile organic compounds. J. Comp. Physiol. A 2000, 186, 1135–1151. [Google Scholar] [CrossRef] [PubMed]
- Hillier, N.K.; Kleineidam, C.; Vickers, N.J. Physiology and glomerular projections of olfactory receptor neurons on the antenna of female Heliothis virescens (Lepidoptera: Noctuidae) responsive to behaviorally relevant odors. J. Comp. Physiol. A 2006, 192, 199–219. [Google Scholar] [CrossRef]
- Kim, M.S.; Repp, A.; Smith, D.P. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics 1998, 150, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Renwick, J.A. Variable diets and changing taste in plant-insect relationships. J. Chem. Ecol. 2001, 27, 1063–1076. [Google Scholar] [CrossRef]
- Tumlinson, J.H.; Yonce, C.E.; Doolittle, R.E.; Heath, R.R.; Gentry, C.R.; Mitchell, E.R. Sex pheromones and reproductive isolation of the lesser peachtree borer and the peachtree borer. Science 1974, 185, 614–616. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, M.; Yan, S.; Wang, G.; Liu, Y. A female-biased odorant receptor from Apolygus lucorum (Meyer-Dür) tuned to some plant odors. Int. J. Mol. Sci. 2016, 17, 1165. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Liu, Y.; Ai, D.; Jiang, X.; Dong, S.; Wang, G. A pheromone antagonist regulates optimal mating time in the moth Helicoverpa armigera. Curr. Biol. 2017, 27, 1610–1615. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chen, J. The odorant binding protein, SiOBP5, mediates alarm pheromone olfactory recognition in the red imported fire ant, Solenopsis invicta. Biomolecules 2021, 11, 1595. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Mohamed, A.; Cattaneo, A.M.; Huang, X.; Keyhani, N.O.; Gu, M.; Zang, L.; Zhang, W. Odorant-binding proteins and chemosensory proteins in Spodoptera frugiperda: From genome-wide identification and developmental stage-related expression analysis to the perception of host plant odors, sex pheromones, and insecticides. Int. J. Mol. Sci. 2023, 24, 5595. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.T.; Su, Q.; Shi, L.L.; Chen, G.; Zeng, Y.; Shi, C.H.; Zhang, Y.J. Electrophysiological and behavioral responses of Bradysia odoriphaga (Diptera: Sciaridae) to volatiles from its host plant, Chinese chives (Allium tuberosum Rottler ex Spreng). J. Econ. Entomol. 2019, 112, 1638–1644. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, F.; Zhang, Y.; Yang, Y.; Hua, D. Odorant-binding protein 10 from Bradysia odoriphaga (Diptera: Sciaridae) binds volatile host plant compounds. J. Insect Sci. 2023, 23, 7. [Google Scholar] [CrossRef]
- Ullah, R.M.K.; Quershi, S.R.; Adeel, M.M.; Abdelnabby, H.; Waris, M.I.; Duan, S.G.; Wang, M.Q. An odorant binding protein (SaveOBP9) involved in chemoreception of the wheat aphid Sitobion avenae. Int. J. Mol. Sci. 2020, 21, 8331. [Google Scholar] [CrossRef]
- Khuhro, S.A.; Liao, H.; Dong, X.T.; Yu, Q.; Yan, Q.; Dong, S.L. Two general odorant binding proteins display high bindings to both host plant volatiles and sex pheromones in a pyralid moth Chilo suppressalis (Lepidoptera: Pyralidae). J. Asia-Pac. Entomol. 2017, 20, 521–528. [Google Scholar] [CrossRef]
- Leal, W.S.; Ishida, Y.; Pelletier, J.; Xu, W.; Rayo, J.; Xu, X.; Ames, J.B. Olfactory proteins mediating chemical communication in the navel orangeworm moth, Amyelois transitella. PLoS ONE 2019, 4, e7235. [Google Scholar] [CrossRef]
- Newcomb, R.D.; Sirey, T.M.; Rassam, M.; Greenwood, D.R. Pheromone binding proteins of Epiphyas postvittana (Lepidoptera: Tortricidae) are encoded at a single locus. Insect Biochem. Mol. Biol. 2002, 32, 1543–1554. [Google Scholar] [CrossRef]
- Mao, G.F.; Tian, J.; Li, T.; Fouad, H.; Ga’al, H.; Mo, J.C. Behavioral responses of Anagrus nilaparvatae to common terpenoids, aromatic compounds, and fatty acid derivatives from rice plants. Entomol. Exp. Appl. 2018, 166, 483–490. [Google Scholar] [CrossRef]
- Hinge, V.; Patil, H.; Nadaf, A. Comparative Characterization of Aroma Volatiles and Related Gene Expression Analysis at Vegetative and Mature Stages in Basmati and Non-Basmati Rice (Oryza sativa L.) Cultivars. Appl. Biochem. Biotechnol. 2016, 178, 619–639. [Google Scholar] [CrossRef] [PubMed]
- Du, H.T.; Li, Y.; Zhu, J.; Liu, F. Host-plant volatiles enhance the attraction of Cnaphalocrocis medinalis (Lepidoptera: Crambidae) to sex pheromone. Chemoecology 2022, 32, 129–138. [Google Scholar] [CrossRef]
- Paramita, B.; Chiranjit, M.; Adinpunya, M. Enzymatic production and emission of floral scent volatiles in Jasminum sambac. Plant Sci. 2017, 256, 25–38. [Google Scholar]


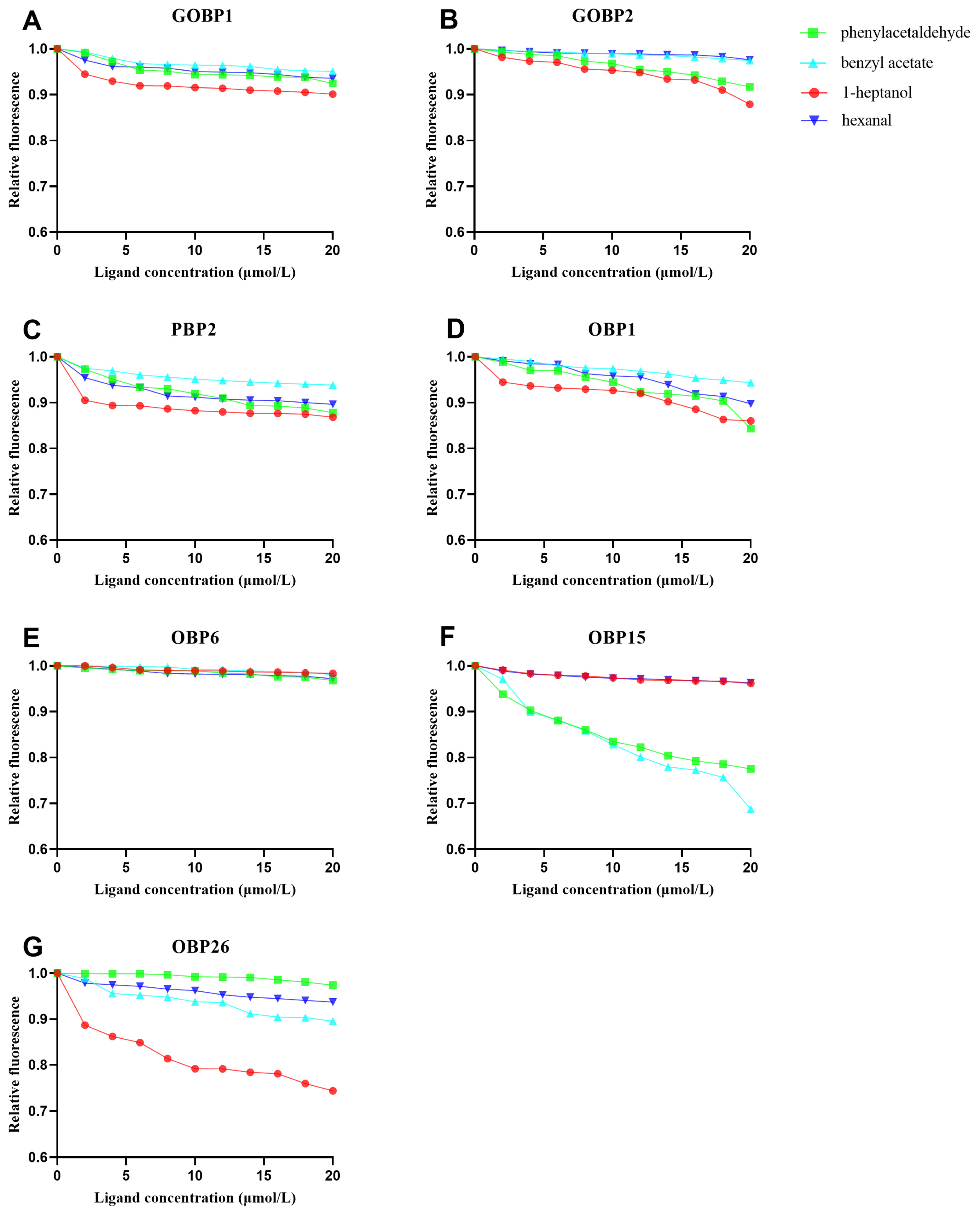
| CmedOBP | 1-Heptanol | Benzyl Acetate | Phenylacetaldehyde | Hexanal | ||||
|---|---|---|---|---|---|---|---|---|
| IC50 μmol/L | Ki μmol/L | IC50 μmol/L | Ki μmol/L | IC50 μmol/L | Ki μmol/L | IC50 μmol/L | Ki μmol/L | |
| CmedGOBP1 | 219.2 | >50 | 223.1 | >50 | 161.8 | >50 | 249 | >50 |
| CmedGOBP2 | 99.7 | >50 | 454.3 | >50 | 123.0 | >50 | 622.1 | >50 |
| CmedPBP2 | 223.9 | >50 | 235.3 | >50 | 85.8 | >50 | 154.7 | >50 |
| CmedOBP1 | 68.1 | 11.99 | 178.6 | 31.45 | 77.5 | 13.65 | 99.2 | 17.46 |
| CmedOBP6 | 623.5 | >50 | 558 | >50 | 356.8 | >50 | 382.6 | >50 |
| CmedOBP15 | 289.9 | >50 | 34.4 | 13.61 | 44.3 | 17.53 | 441.2 | >50 |
| CmedOBP26 | 52.3 | 31.89 | 96.1 | >50 | 458.1 | >50 | 199.9 | >50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Q.; Guo, X.; Wu, L.; Cui, J.; Gao, H.; Yang, Y.; Xu, H.; Lu, Z.; Zhu, P. Molecular Characterization of Plant Volatile Compound Interactions with Cnaphalocrocis medinalis Odorant-Binding Proteins. Plants 2024, 13, 479. https://doi.org/10.3390/plants13040479
Qian Q, Guo X, Wu L, Cui J, Gao H, Yang Y, Xu H, Lu Z, Zhu P. Molecular Characterization of Plant Volatile Compound Interactions with Cnaphalocrocis medinalis Odorant-Binding Proteins. Plants. 2024; 13(4):479. https://doi.org/10.3390/plants13040479
Chicago/Turabian StyleQian, Qi, Xin Guo, Lingjie Wu, Jiarong Cui, Huiying Gao, Yajun Yang, Hongxing Xu, Zhongxian Lu, and Pingyang Zhu. 2024. "Molecular Characterization of Plant Volatile Compound Interactions with Cnaphalocrocis medinalis Odorant-Binding Proteins" Plants 13, no. 4: 479. https://doi.org/10.3390/plants13040479
APA StyleQian, Q., Guo, X., Wu, L., Cui, J., Gao, H., Yang, Y., Xu, H., Lu, Z., & Zhu, P. (2024). Molecular Characterization of Plant Volatile Compound Interactions with Cnaphalocrocis medinalis Odorant-Binding Proteins. Plants, 13(4), 479. https://doi.org/10.3390/plants13040479






