Identification of WRKY Family Members and Characterization of the Low-Temperature-Stress-Responsive WRKY Genes in Luffa (Luffa cylindrica L.)
Abstract
:1. Introduction
2. Results
2.1. Identification of LcWRKY Genes in L. cylindrica
2.2. Chromosomal Locations and Duplication of LcWRKY Genes
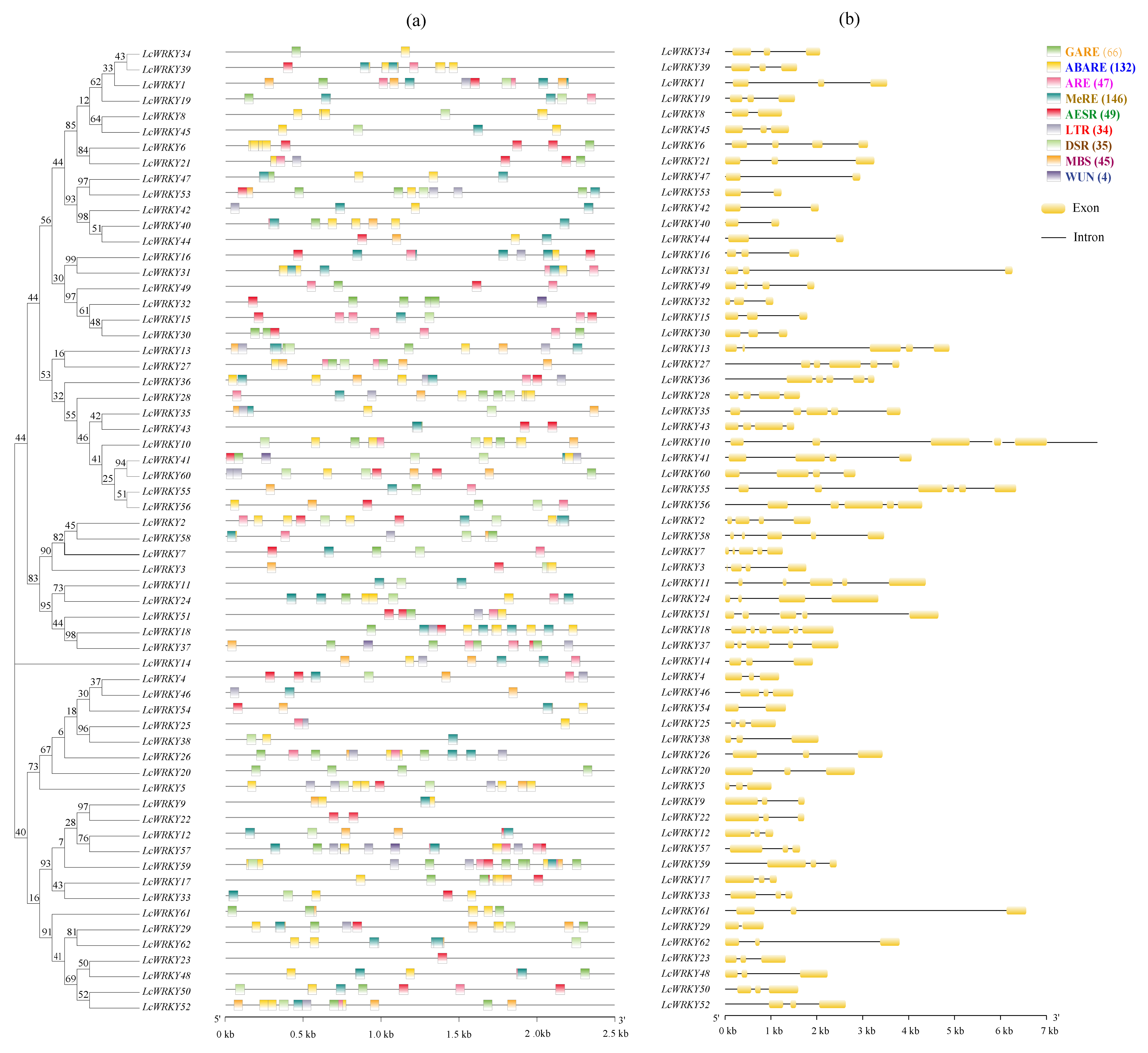
2.3. Analysis of LcWRKY Cis-Acting Elements and Gene Structures
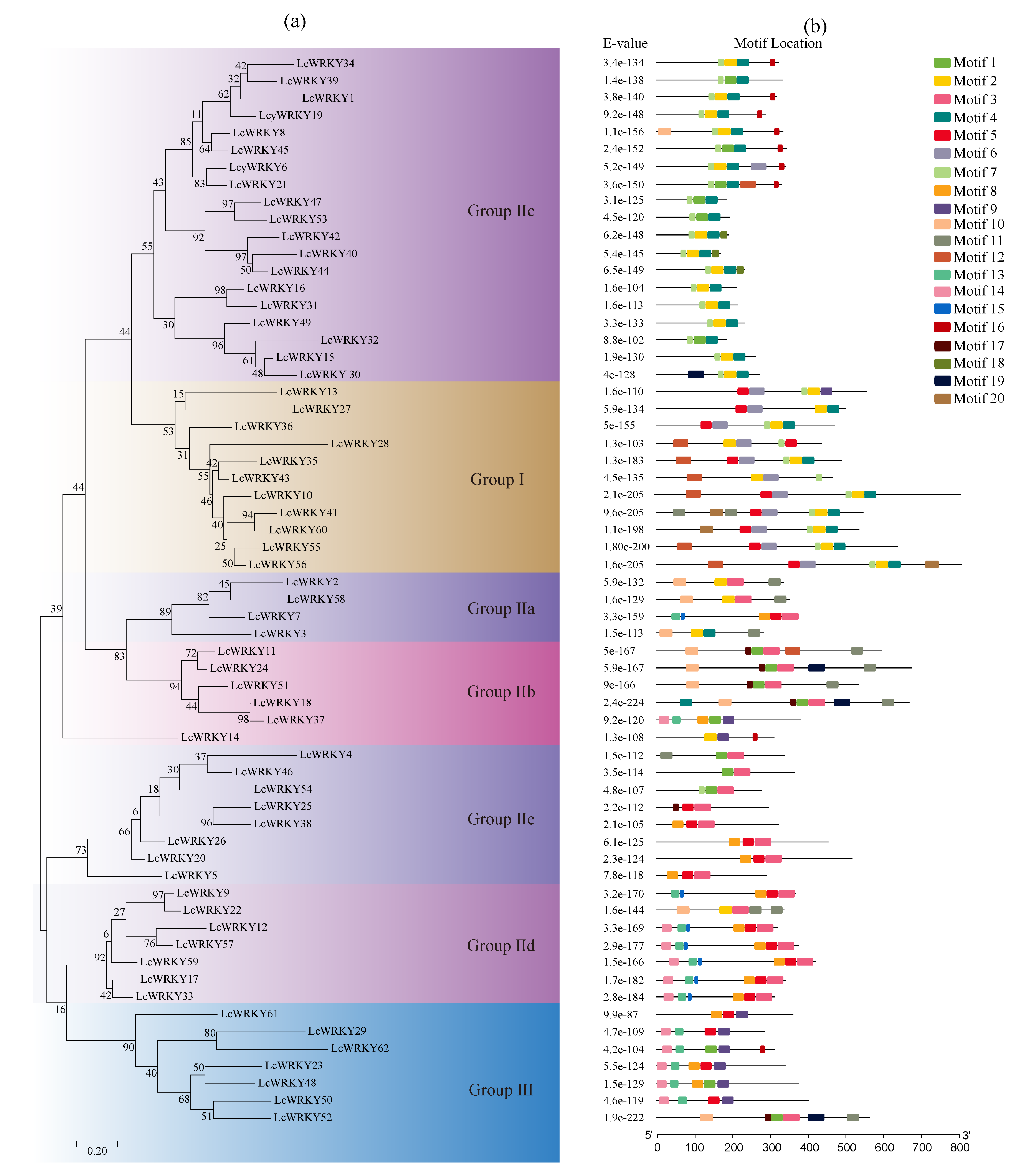
2.4. Phylogenetic Relationships and Conserved Motifs
2.5. LcWRKY Expression Profiles in Seven Tissues
2.6. Analysis of LcWRKY Expression under Low-Temperature Stress Conditions
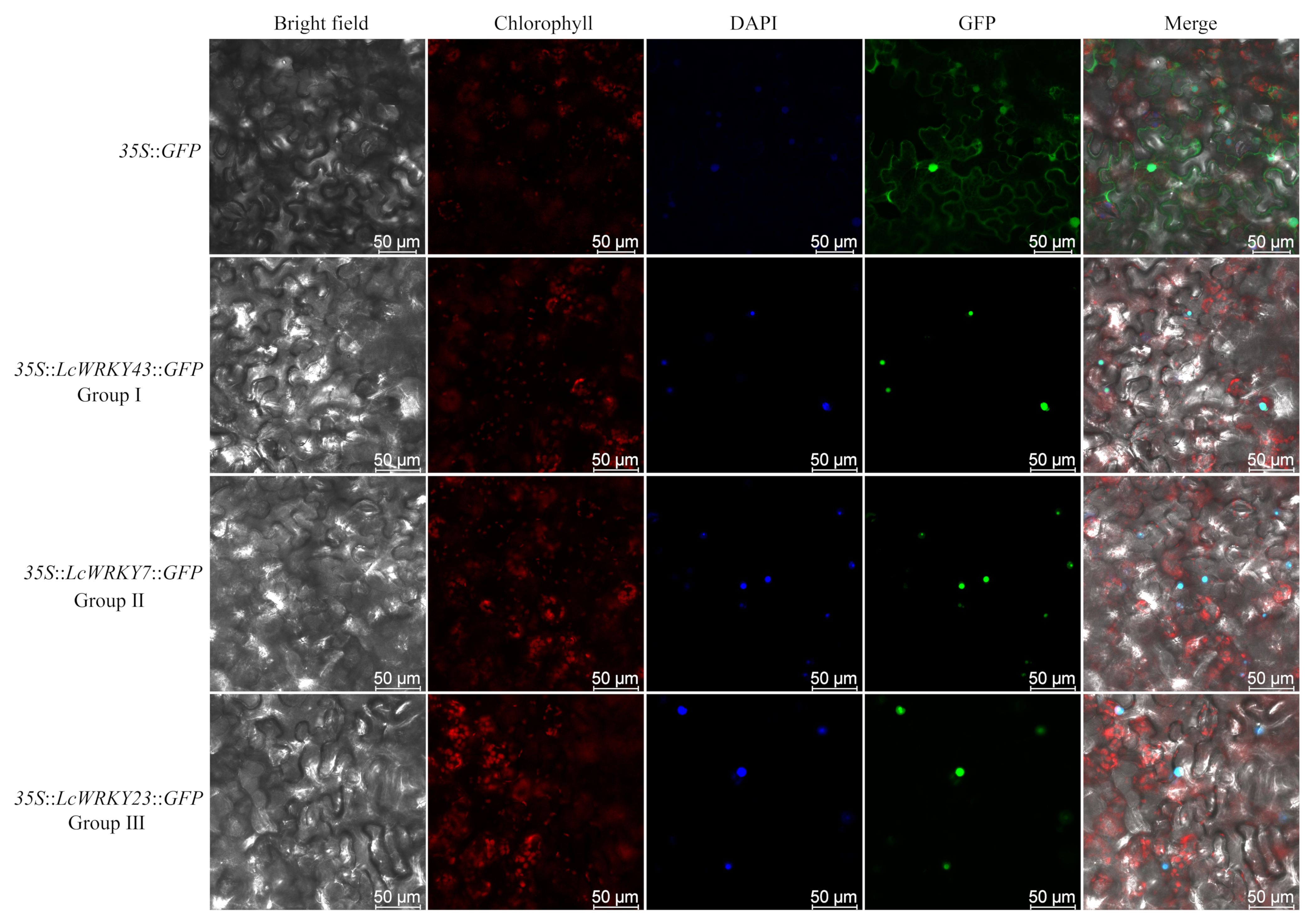
2.7. Subcellular Localization of LcWRKY Proteins
3. Discussion
3.1. Characterization of the LcWRKY Gene Family in L. cylindrica
3.2. Analysis of L. cylindrica WRKY Gene Promoters
3.3. Expression Profiles of LcWRKY Genes in L. cylindrica and the Changes Induced by Low-Temperature Stress
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatment
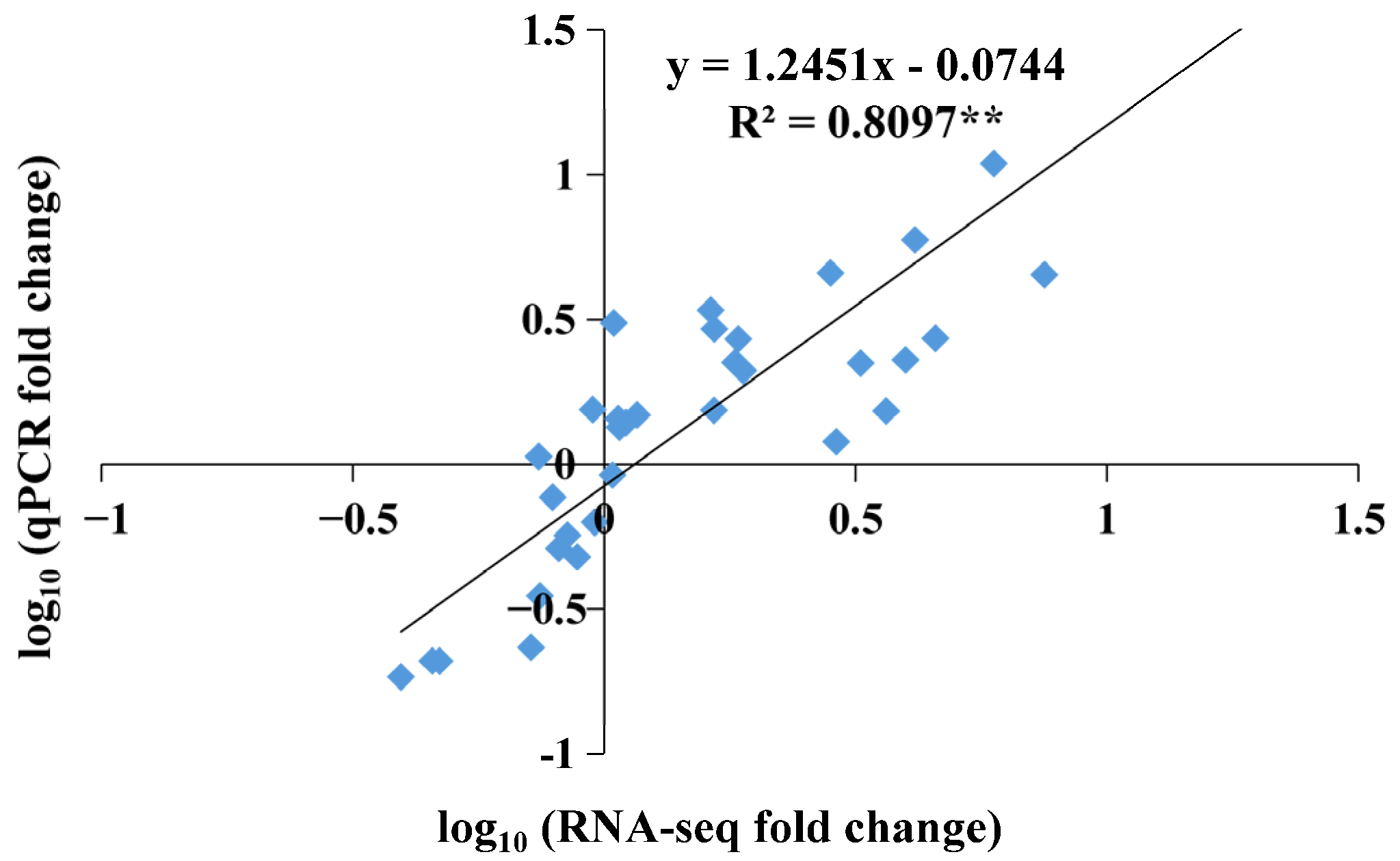
4.2. Total RNA Extraction, RNA Sequencing, and Gene Expression Analysis
4.3. Identification of LcWRKY Genes in the L. cylindrica Genome
4.4. Bioinformatics Analysis of the LcWRKY Gene Family
4.5. Subcellular Localization of LcWRKY Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Wang, H.; Xing, H.; Zhao, J.; Guo, N. GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol. 2019, 19, 598. [Google Scholar] [CrossRef]
- Chanwala, J.; Satpati, S.; Dixit, A.; Parida, A.; Giri, M.K.; Dey, N. Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genom. 2020, 21, 231. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Qiao, L.; Guo, H.; Guo, L.; Ren, F.; Bai, J.; Wang, Y. Genome-wide identification of wheat WRKY gene family reveals that TaWRKY75-A is referred to drought and salt resistances. Front. Plant Sci. 2021, 12, 663118. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, X.; Song, X.; Wen, B.; Xie, N.; Wang, K.; Huang, J.; Liu, Z. Identification of a WRKY transcriptional activator from Camellia sinensis that regulates methylated EGCG biosynthesis. Hortic. Res. 2022, 19, uhac024. [Google Scholar] [CrossRef]
- Sun, W.; Ma, Z.; Chen, H.; Liu, M. Genome-wide investigation of WRKY transcription factors in Tartary buckwheat (Fagopyrum tataricum) and their potential roles in regulating growth and development. PeerJ 2020, 8, e8727. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Cai, W.; Shen, L.; Cao, J.; Liu, C.; Hu, J.; Guan, D.; He, S. A CaCDPK29–CaWRKY27b module promotes CaWRKY40-mediated thermotolerance and immunity to Ralstonia solanacearum in pepper. New Phytol. 2022, 233, 1843–1863. [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Ding, A.; Cheng, T.; Wang, J.; Zhang, Q. Genome-wide analysis of members of the WRKY gene family and their cold stress response in Prunus mume. Genes 2019, 10, 911. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wu, Z.; Teng, R.; Xu, S.; Cao, X.; Yuan, G.; Zhang, D.; Teng, N. LlWRKY39 is involved in thermotolerance by activating LlMBF1c and interacting with LlCaM3 in lily (Lilium longiflorum). Hortic. Res. 2021, 8, 36. [Google Scholar] [CrossRef]
- Yuan, Y.; Ren, S.; Liu, X.; Su, L.; Wu, Y.; Zhang, W.; Li, Y.; Jiang, Y.; Wang, H.; Fu, R. SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. 2022, 234, 164–178. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Zou, C.; Jiang, W.; Yu, D. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 2010, 61, 3901–3914. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Ao, C.; Wang, X.; Wu, Y.; Du, X. GhWRKY1-like, a WRKY transcription factor, mediates drought tolerance in Arabidopsis via modulating ABA biosynthesis. BMC Plant Biol. 2021, 21, 458. [Google Scholar] [CrossRef] [PubMed]
- Negi, N.; Khurana, P. A salicylic acid inducible mulberry WRKY transcription factor, Mi WRKY53 is involved in plant defence response. Plant Cell Rep. 2021, 40, 2151–2171. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, X.; Liu, H.; Wang, Y.; van Nocker, S.; Tu, M.; Fang, J.; Guo, J.; Li, Z.; Wang, X. Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Hortic. Res. 2022, 19, uhab064. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.; Yeap, W.; Appleton, D.; Ho, C.; Kulaveerasingam, H. Identification of drought responsive Elaeis guineensis WRKY transcription factors with sensitivity to other abiotic stresses and hormone treatments. BMC Genom. 2022, 23, 164. [Google Scholar] [CrossRef] [PubMed]
- Goyal, P.; Manzoor, M.M.; Vishwakarma, R.A.; Sharma, D.; Dhar, M.K.; Gupta, S. A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in Glycyrrhiza glabra. Sci. Rep. 2020, 10, 373. [Google Scholar] [CrossRef]
- Govardhana, M.; Kumudini, B.S. In-silico analysis of cucumber (Cucumis sativus L.) Genome for WRKY transcription factors and cis-acting elements. Comput. Biol. Chem. 2020, 85, 107212. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Y.; Zhang, Z.; Liu, X.; Hsu, C.-C.; Du, Y.; Sang, T.; Zhu, C.; Wang, Y.; Satheesh, V. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 2020, 11, 613. [Google Scholar] [CrossRef]
- Huang, X.; Cao, L.; Fan, J.; Ma, G.; Chen, L. CdWRKY2-mediated sucrose biosynthesis and CBF-signalling pathways coordinately contribute to cold tolerance in bermudagrass. Plant Biotechnol. J. 2021, 20, 660–675. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T. The OsWRKY63–OsWRKY76–OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zheng, W.; Liu, B.; Zheng, J.; Dong, F.; Liu, Z.; Wen, Z.; Yang, F.; Wang, H.; Xu, Z. Characterizing the role of TaWRKY13 in salt tolerance. Int. J. Mol. Sci. 2019, 20, 5712. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Tian, A.; Zou, H.; Xie, Z.; Lei, G.; Jian, H.; Wang, C.; Wang, H.; Zhang, J.; Chen, S. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 05, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Nai, G.; Liang, G.; Ma, W.; Lu, S.; Li, Y.; Gou, H.; Guo, L.; Chen, B.; Mao, J. Overexpression VaPYL9 improves cold tolerance in tomato by regulating key genes in hormone signaling and antioxidant enzyme. BMC Plant Biol. 2022, 22, 344. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Bioch. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Fei, J.; Wang, Y.; Cheng, H.; Su, Y.; Zhong, Y.; Zheng, L. The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 2022, 22, 274. [Google Scholar] [CrossRef]
- Wei, Z.; Ye, J.; Zhou, Z.; Chen, G.; Meng, F.; Liu, Y. Isolation and characterization of PoWRKY, an abiotic stress-related WRKY transcription factor from Polygonatum odoratum. Physiol. Mol. Biol. Plants 2021, 27, 1–9. [Google Scholar] [CrossRef]
- Ye, Y.J.; Xiao, Y.Y.; Han, Y.C.; Shan, W.; Fan, Z.Q.; Xu, Q.G.; Kuang, J.F.; Lu, W.J.; Lakshmanan, P.; Chen, J.Y. Banana fruit VQ motif-containing protein5 represses cold-responsive transcription factor MaWRKY26 involved in the regulation of JA biosynthetic genes. Sci. Rep. 2016, 6, 23632. [Google Scholar] [CrossRef]
- Liu, Y.; Xiong, Y.; Zhao, J.; Bai, S.; Li, D.; Chen, L.; Feng, J.; Li, Y.; Ma, X.; Zhang, J. Molecular mechanism of cold tolerance of centipedegrass based on the transcriptome. Int. J. Mol. Sci. 2023, 24, 1265. [Google Scholar] [CrossRef]
- Liu, S.; Kracher, B.; Ziegler, J.; Birkenbihl, R.P.; Somssich, I.E. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife 2015, 4, e07295. [Google Scholar] [CrossRef]
- Khan, I.; Zhang, Y.; Liu, Z.; Hu, J.; Liu, C.; Sheng, Y.; Husssain, A.; Ashraf, M.F.; Noman, A.; Shen, L.; et al. CaWRKY40b in Pepper Acts as a Negative Regulator in Response to Ralstonia solanacearum by Directly Modulating Defense Genes Including CaWRKY40. Int. J. Mol. Sci. 2018, 19, 1403. [Google Scholar] [CrossRef]
- Zhou, L.; Huang, Y.; Wang, Q.; Guo, D. AaHY5 ChIP-seq based on transient expression system reveals the role of AaWRKY14 in artemisinin biosynthetic gene regulation. Plant Physiol. Biochem. 2021, 168, 321–328. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, L.; Yang, S.; Qiu, S.; Ma, X.; Cai, J.; Guan, D.; Wang, Z.; He, S. A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol. Plant Pathol. 2020, 22, 3–18. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Martí-Gómez, C.; Ferriol, M.; Gómez, P.; Jamilena, M.; Mueller, L. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018, 16, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Xu, Y.; Wang, J.; Liu, W.; Zhou, Q.; Luo, S.; Huang, W.; He, X.; Li, Q.; Peng, Q.; et al. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat. Commun. 2019, 10, 5158. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef]
- Saensuk, C.; Ruangnam, S.; Pitaloka, M.K.; Dumhai, R.; Mahatheeranont, S.; de Hoop, S.J.; Balatero, C.; Riangwong, K.; Ruanjaichon, V.; Toojinda, T.; et al. A SNP of betaine aldehyde dehydrogenase (BADH) enhances an aroma (2-acetyl-1-pyrroline) in sponge gourd (Luffa cylindrica) and ridge gourd (Luffa acutangula). Sci. Rep. 2022, 12, 3718. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhao, G.; Gong, H.; Li, J.; Luo, C.; He, X.; Luo, S.; Zheng, X.; Liu, X.; Guo, J.; et al. A high-quality sponge gourd (Luffa cylindrica) genome. Hortic. Res. 2020, 7, 128. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Xu, L.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Hao, Z.; Lu, Y.; Yang, L. Genome-wide identification of the Liriodendron chinense WRKY gene family and its diverse roles in response to multiple abiotic stress. BMC Plant Biol. 2022, 22, 25. [Google Scholar] [CrossRef]
- Huang, J.; Chen, G.; Ahmad, S.; Hao, Y.; Chen, J.; Zhou, Y.; Lan, S.; Liu, Z.; Peng, D. Genome-Wide Identification and Characterization of the GRF Gene Family in Melastoma dodecandrum. Int. J. Mol. Sci. 2023, 24, 1261. [Google Scholar] [CrossRef]
- Guo, J.; Xu, W.; Hu, Y.; Huang, J.; Zhao, Y.; Zhang, L.; Huang, C.; Ma, H. Phylotranscriptomics in Cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Mol. Plant 2020, 13, 1117–1133. [Google Scholar] [CrossRef]
- Barrera-Redondo, J.; Lira-Saade, R.; Eguiarte, L.E. Gourds and tendrils of cucurbitaceae: How their shape diversity, molecular and morphological novelties evolved via whole-genome duplications. Mol. Plant 2020, 13, 1108–1110. [Google Scholar] [CrossRef]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef]
- Luo, D.; Ba, L.; Shan, W.; Kuang, J.; Lu, W.; Chen, J. Involvement of WRKY transcription factors in abscisic-acid-induced cold tolerance of banana fruit. J. Agric. Food Chem. 2017, 65, 3627–3635. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Zhang, R.; Wu, B.; Xiao, G. Expression identification of three OsWRKY genes in response to abiotic stress and hormone treatments in rice. Plant Signal Behav. 2023, 18, 2292844. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Song, Z.; Wei, L.; Li, L. Molecular characterization and expression analysis of WRKY family genes in Dendrobium officinale. Genes Genom. 2018, 40, 265–279. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Wei, J.; Cai, K.; Zhang, H.; Ge, L.; Ren, Z.; Zhao, C.; Zhao, X. Genome-wide identification and analysis of the WRKY gene family and cold stress response in Acer truncatum. Genes 2021, 12, 1867. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, Y.; Xu, S.; Zhan, Z.; Xu, Y.; Zhang, J.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, B.; Li, Y.; Huang, L.; Zhang, Q.; Zhu, H.; Wen, Q. RNA sequencing analysis of low temperature and low light intensity-responsive transcriptomes of zucchini (Cucurbita pepo L.). Sci. Hortic. 2020, 265, 109263. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, J. Genomic analysis of Dof transcription factors in Hevea brasiliensis, a rubber-producing tree. Ind. Crop Prod. 2019, 134, 271–283. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Peng, L.; Cao, C.; Bai, C.; Wang, Y.; Li, Z.; Zhu, H.; Wen, Q.; He, S. Identification of WRKY Family Members and Characterization of the Low-Temperature-Stress-Responsive WRKY Genes in Luffa (Luffa cylindrica L.). Plants 2024, 13, 676. https://doi.org/10.3390/plants13050676
Liu J, Peng L, Cao C, Bai C, Wang Y, Li Z, Zhu H, Wen Q, He S. Identification of WRKY Family Members and Characterization of the Low-Temperature-Stress-Responsive WRKY Genes in Luffa (Luffa cylindrica L.). Plants. 2024; 13(5):676. https://doi.org/10.3390/plants13050676
Chicago/Turabian StyleLiu, Jianting, Lijuan Peng, Chengjuan Cao, Changhui Bai, Yuqian Wang, Zuliang Li, Haisheng Zhu, Qingfang Wen, and Shuilin He. 2024. "Identification of WRKY Family Members and Characterization of the Low-Temperature-Stress-Responsive WRKY Genes in Luffa (Luffa cylindrica L.)" Plants 13, no. 5: 676. https://doi.org/10.3390/plants13050676
APA StyleLiu, J., Peng, L., Cao, C., Bai, C., Wang, Y., Li, Z., Zhu, H., Wen, Q., & He, S. (2024). Identification of WRKY Family Members and Characterization of the Low-Temperature-Stress-Responsive WRKY Genes in Luffa (Luffa cylindrica L.). Plants, 13(5), 676. https://doi.org/10.3390/plants13050676





