Unraveling the Role of AtSRT2 in Energy Metabolism, Stress Responses, and Gene Expression during Osmotic Stress in Arabidopsis thaliana
Abstract
1. Introduction
2. Results and Discussion
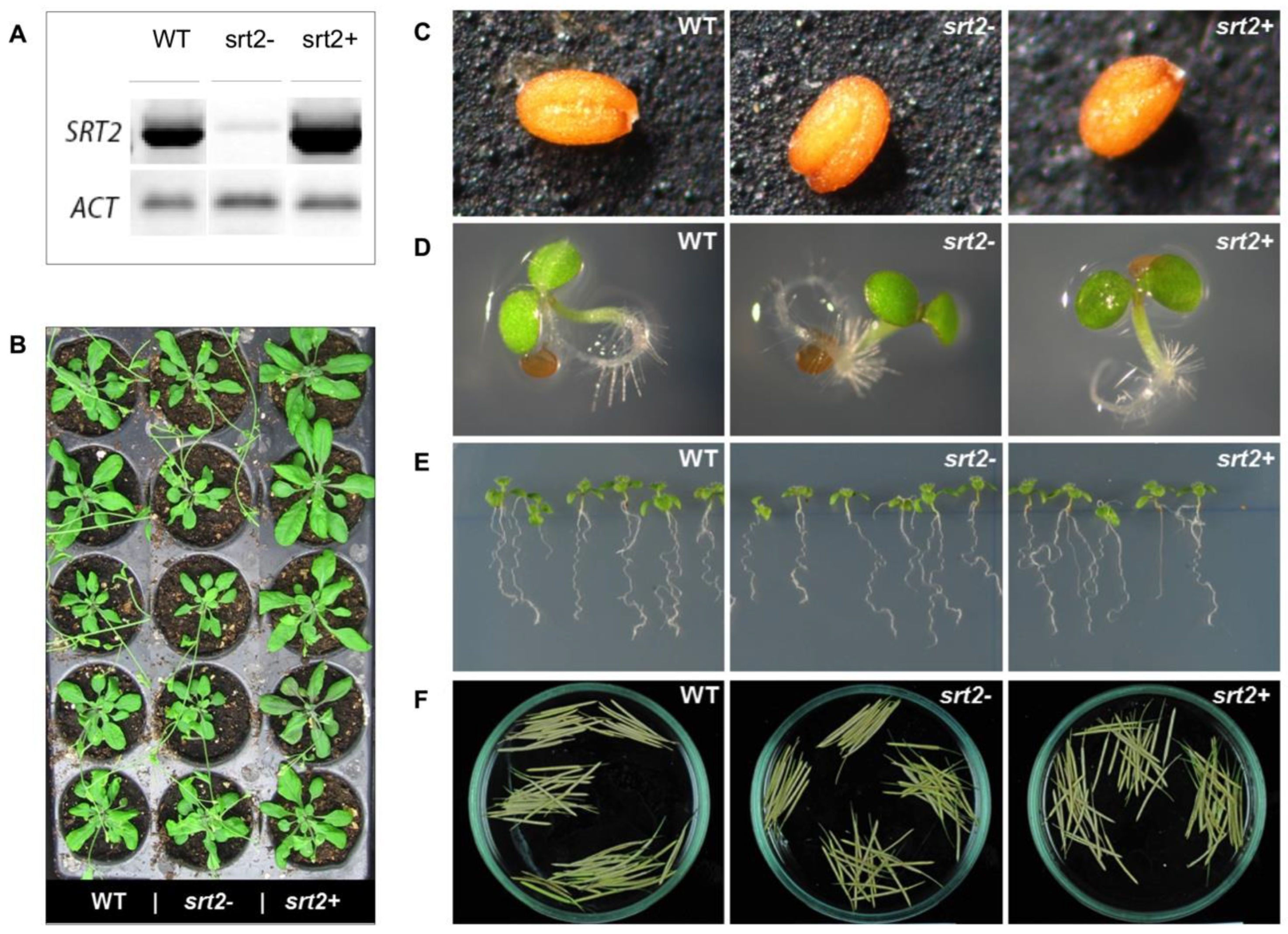
2.1. Effect of SRT2 on the Development of A. thaliana
2.2. Root Length under Stress Conditions
2.3. Membrane Lipoperoxidation
2.4. SRT2 Is Critical for Activating Osmotic Stress Response Genes
2.5. Nicotinamide’s Contribution to Stress Signaling
3. Materials and Methods
3.1. Plant Material, Growth Conditions, and Stress Treatment
3.2. Greenhouse Growth Conditions
3.3. Measurement of Leaf Properties
3.4. Germination Assay
3.5. Lipid Peroxidation
3.6. RNA Extraction and First Strand cDNA Synthesis
3.7. qRT-PCR Analysis
3.8. Molecular Markers of Abiotic Stress Response
3.9. Statistical Analyses
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Grubisha, O.; Smith, B.C.; Denu, J.M. Small molecule regulation of Sir2 protein deacetylases. FEBS J. 2005, 272, 4607–4616. [Google Scholar] [CrossRef]
- Zhang, Q.; Piston, D.W.; Goodman, R.H. Regulation of corepressor function by nuclear NADH. Science 2002, 295, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Pétriacq, P.; de Bont, L.; Tcherkez, G.; Gakière, B. NAD: Not just a pawn on the board of plant-pathogen interactions. Plant Signal. Behav. 2013, 8, e22477. [Google Scholar] [CrossRef]
- Guarente, L. Sirtuins, aging, and metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Mostoslavsky, R. Fine tuning our cellular factories: Sirtuins in mitochondrial biology. Cell Metab. 2011, 13, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Canto, C.; Auwerx, J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Verdin, E. NAD(+) in aging, metabolism, and neurodegeneration. Science 2015, 350, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Berglund, T.; Ohlsson, A.B.; Rydström, J.; Jordan, B.R.; Strid, Å. Effect of nicotinamide on gene expression and glutathione levels in tissue cultures of Pisum sativum. J. Plant Physiol. 1993, 142, 676–684. [Google Scholar] [CrossRef]
- Berglund, T.; Wallström, A.; Nguyen, T.V.; Laurell, C.; Ohlsson, A.B. Nicotinamide; antioxidative and DNA hypomethylation effects in plant cells. Plant Physiol. Biochem. 2017, 118, 551–560. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Chao, E.D.; Blackwell, H.E.; Moazed, D.; Schreiber, S.L. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 2001, 276, 38837–38843. [Google Scholar] [CrossRef]
- Tanny, J.C.; Moazed, D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 2001, 98, 415–420. [Google Scholar] [CrossRef]
- Berger, F.; Ramirez-Hernandez, M.H.; Ziegler, M. The new life of a centenarian: Signalling functions of NAD(P). Trends Biochem. Sci. 2004, 29, 111–118. [Google Scholar] [CrossRef]
- Adams, J.D.; Klaidman, L.K., Jr. Sirtuins, Nicotinamide and Aging, A Critical Review. Lett. Drug Des. Discov. 2007, 4, 44–48. [Google Scholar] [CrossRef]
- Martínez-Redondo, P.; Vaquero, A. The Diversity of Histone Versus Nonhistone Sirtuin Substrates. Genes Cancer 2013, 4, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Szucko, I. Sirtuins: Not only animal proteins. Acta Physiol. Plant 2016, 38, 237. [Google Scholar] [CrossRef]
- Gardner, J.G.; Escalante-Semerena, J.C. In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase. J. Bacteriol. 2009, 191, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.D.; Botting, C.H.; Wardleworth, B.N.; Jackson, S.P.; White, M.F. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 2002, 296, 148–151. [Google Scholar] [CrossRef]
- Lu, S.P.; Lin, S.J. Regulation of yeast sirtuins by NAD+ metabolism and calorie restriction. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Wirth, M.; Paap, F.; Fischle, W.; Wenzel, D.; Agafonov, D.E.; Samatov, T.R.; Wisniewski, J.R.; Jedrusik-Bode, M. HIS-24 linker histone and SIR-2.1 deacetylase induce H3K27me3 in the Caenorhabditis elegans germ line. Mol. Cell Biol. 2009, 29, 3700–3709. [Google Scholar] [CrossRef]
- Whitaker, R.; Faulkner, S.; Miyokawa, R.; Burhenn, L.; Henriksen, M.; Wood, J.G.; Helfand, S.L. Increased expression of Drosophila sir2 extends life span in a dose dependent manner. Aging 2013, 5, 682–691. [Google Scholar] [CrossRef]
- Frye, R.A. Characterization of Five Human cDNAs with Homology to the Yeast SIR2 Gene: Sir2-like Proteins (Sirtuins) Metabolize NAD and May Have Protein ADP-Ribosyltransferase Activity. Biochem. Biophys. Res. Commun. 1999, 260, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Zhang, H.; Zhao, Y.; Sun, Q.; Hu, Y.; Peng, H.; Zhou, D.X. The rice NAD(+)-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements. PLoS ONE 2013, 8, e66807. [Google Scholar] [CrossRef] [PubMed]
- König, A.C.; Hartl, M.; Pham, P.A.; Laxa, M.; Boersema, P.J.; Orwat, A.; Kalitventseva, I.; Plöchinger, M.; Braun, H.P.; Leister, D.; et al. The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 2014, 164, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Finkemeier, I.; Laxa, M.; Miguet, L.; Howden, A.J.; Sweetlove, L.J. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 2011, 155, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Oh, M.H.; Schwarz, E.M.; Larue, C.T.; Sivaguru, M.; Imai, B.S.; Yau, P.M.; Ort, D.R.; Huber, S.C. Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 2011, 155, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lu, Y.; Zhao, Y.; Zhou, D.X. OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression. Plant Sci. 2016, 248, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Busconi, M.; Reggi, S.; Fogher, C.; Bavaresco, L. Evidence of a sirtuin gene family in grapevine (Vitis vinifera L.). Plant Physiol Biochem. 2009, 7, 650–652. [Google Scholar] [CrossRef] [PubMed]
- Cucurachi, M.; Busconi, M.; Morreale, G.; Zanetti, A.; Bavaresco, L.; Fogher, C. Characterization and differential expression analysis of complete coding sequences of Vitis vinifera L. sirtuin genes. Plant Physiol. Biochem. 2012, 54, 123–132. [Google Scholar] [CrossRef]
- Aiese Cigliano, R.; Sanseverino, W.; Cremona, G.; Ercolano, M.R.; Conicella, C.; Consiglio, F.M. Genome-wide analysis of histone modifiers in tomato: Gaining an insight into their developmental roles. BMC Genom. 2013, 14, 5. [Google Scholar] [CrossRef]
- Zhao, L.; Lu, J.; Zhang, J.; Wu, P.Y.; Yang, S.; Wu, K. Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front. Plant Sci. 2015, 5, 760. [Google Scholar] [CrossRef]
- Huang, L.; Sun, Q.; Qin, F.; Li, C.; Zhao, Y.; Zhou, D.X. Down-Regulation of a Silent Information regulator2-Related Histone Deacetylase Gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007, 144, 1508–1519. [Google Scholar] [CrossRef]
- Wang, C.; Gao, F.; Wu, J.; Dai, J.; Wei, C.; Li, Y. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5, and SID2 expression. Plant Cell Physiol. 2010, 51, 1291–1299. [Google Scholar] [CrossRef]
- Shen, Y.; Issakidis-Bourguet, E.; Zhou, D. Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 2016, 67, 5291–5300. [Google Scholar] [CrossRef] [PubMed]
- Kupis, W.; Pałyga, J.; Tomal, E.; Niewiadomska, E. The role of sirtuins in cellular homeostasis. J. Physiol. Biochem. 2016, 72, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Lebon, G.; Brun, O.; Magné, C.; Clément, C. Photosynthesis of the grapevine (Vitis vinifera) inflorescence. Tree Physiol. 2005, 25, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Aquea, F.; Timmermann, T.; Arce-Johnson, P. Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol. Biochem. 2010, 48, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Finkemeier, I. Plant mitochondrial retrograde signaling: Post-translational modifications enter the stage. Front. Plant Sci. 2012, 253, 253. [Google Scholar] [CrossRef]
- Soto-Burgos, J.; Bassham, D.C. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS ONE 2017, 12, e0182591. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Ryabova, L.A. Recent Discoveries on the Role of TOR (Target of Rapamycin) Signaling in Translation in Plants. Plant Physiol. 2018, 176, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, A.; Yadav, S.; Gautam, V.; Singh, A.; Sarkar, A.K. Sirtinol, a Sir2 protein inhibitor, affects stem cell maintenance and root development in Arabidopsis thaliana by modulating auxin-cytokinin signaling components. Sci. Rep. 2017, 14, 42450. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, H.G. Plant root development: Is the classical theory for auxin-regulated root growth false? Protoplasma 2022, 259, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Lv, S.; Zhang, C.; Yang, C. Histone deacetylases and their functions in plants. Plant Cell Rep. 2013, 32, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sheen, J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014, 164, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Hohl, M.; Schopfer, P. Water relations of growing maize coleoptiles. Comparison between mannitol and polyethylene glycol 6000 as external osmotica for adjusting turgor pressure. Plant Physiol. 1991, 95, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Deprost, D.; Yao, L.; Sormani, R.; Moreau, M.; Leterreux, G.; Nicolaï, M.; Bedu, M.; Robaglia, C.; Meyer, C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007, 9, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Armengaud, P.; Zambaux, K.; Hills, A.; Sulpice, R.; Pattison, R.J.; Blatt, M.R.; Amtmann, A. EZ-Rhizo: Integrated software for the fast and accurate measurement of root system architecture. Plant J. 2009, 57, 945–956. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Hoefsloot, H.C.; Mol, S.; Feron, R.; de Boer, G.J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Gul, B.; Weber, D.J. Effect of dormancy relieving compounds on the seed germination of non-dormant Allenrolfea occidentalis under salinity stress. Ann. Bot. 1998, 82, 555–560. [Google Scholar] [CrossRef]
- Hodges, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Shi, H.T.; Li, R.J.; Cai, W.; Liu, W.; Wang, C.L.; Lu, Y.T. Increasing nitric oxide content in Arabidopsis thaliana by expressing rat neuronal nitric oxide synthase resulted in enhanced stress tolerance. Plant Cell Physiol. 2011, 53, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Luo, X.; Sun, M.; Chen, C.; Ding, X.; Wang, X.; Yang, S.; Yu, Q.; Jia, B.; Ji, W.; et al. A Glycine Soja 14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 99–118. [Google Scholar] [CrossRef]
- Levin, D.E.; Hammond, C.I.; Ralston, R.O.; Bishop, J.M. Two yeast genes that encode unusual protein kinases. Proc. Natl. Acad. Sci. USA 1987, 84, 6035–6039. [Google Scholar] [CrossRef]
- Kurkela, S.; Borg-Franck, M. Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 1992, 19, 689–692. [Google Scholar] [CrossRef]
- Wang, H.; Datla, R.; Georges, F.; Loewen, M.; Cutler, A.J. Promoters from kin1 and cor6.6, two homologous Arabidopsis thaliana genes: Transcriptional regulation and gene expression induced by low temperature, ABA, osmoticum and dehydration. Plant Mol. Biol. 1995, 28, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008, 11, 2991–3007. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 14, 4731–4748. [Google Scholar] [CrossRef]
- Jia, W.; Zhang, L.; Wu, D.; Liu, S.; Gong, X.; Cui, Z.; Cui, N.; Cao, H.; Rao, L.; Wang, C. Sucrose transporter AtSUC9 mediated by a low sucrose level is involved in Arabidopsis abiotic stress resistance by regulating sucrose distribution and ABA accumulation. Plant Cell Physiol. 2015, 56, 1574–1587. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol. Gen. Genet 1993, 238, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Hou, X.; Xie, K.; Xiong, L. Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses. Planta 2009, 230, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [CrossRef]
- Matus, J.T.; Aquea, F.; Espinoza, C.; Vega, A.; Cavallini, E.; Santo, S.D.; Cañón, P.; de la Guardia, A.R.H.; Serrano, J.; Tornielli, G.B.; et al. Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses. PLoS ONE 2014, 9, e110372. [Google Scholar] [CrossRef]
- Chen, T.; Li, W.; Hu, X.; Guo, J.; Liu, A.; Zhang, B. A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015, 56, 917–929. [Google Scholar] [CrossRef]







| Genotype | WT | srt2- | srt2+ |
|---|---|---|---|
| Feature | Mean ± SD | Mean ± SD | Mean ± SD |
| Silique length (mm) | 13.93 ± 0.16 | 13.53 ± 0.12 * | 13.01 ± 0.12 * |
| Seed number per silique | 48.67 ± 0.95 | 44.49 ± 0.82 * | 46.63 ± 0.89 |
| Siliques number per plant | 90.21 ± 4.21 | 82.41 ± 4.92 | 87.042 ± 4.03 |
| Seed width (mm) | 0.40 ± 0.03 | 0.45 ± 0.02 | 0.44 ± 0.02 |
| Seed length (mm) | 0.72 ± 0.03 | 0.75 ± 0.01 | 0.73 ± 0.03 |
| Seed area (mm2) | 0.23 ± 0.04 | 0.26 ± 0.03 | 0.25 ± 0.02 |
| Oligonucleotide | Sequence |
|---|---|
| SALK_N649295-PCR1-D | 5′-CGC AGA GAG AGA ACA AAA TCG-3′ |
| SALK_N649295-PCR1-R | 5′-TTC CAC ATT CTG TGC TAA CCC-3′ |
| SALK_N649295-PCR2-D | 5′-TGG TTC ACG TAG TGG GCC ATC G-3′ |
| SALK_N649295-PCR2-D | 5′-TTC CAC ATT CTG TGC TAA CCC-3′ |
| SALK_N58811-PCR1-D | 5′-AGA GTT CAT AAA AAC AAT GAA TCA AG-3′ |
| SALK_N58811-PCR1-R | 5′-AAC TCA TTG CAT TTG CAT AGG-3′ |
| SALK_N58811-PCR2-D | 5′-AAC TCA TTG CAT TTG CAT AGG-3′ |
| SALK_N58811-PCR2-R | 5′-TGG TTC ACG TAG TGG GCC ATC G-3′ |
| SALK_N631994-PCR1-D | 5′-TTT AAG GCA TTT TCA AGG CTG-3′ |
| SALK_N631994-PCR1-R | 5′-GGA GTT TAC TCG ATC AAG CCG-3′ |
| SALK_N631994-PCR1-D | 5′-GGA GTT TAC TCG ATC AAG CCG-3′ |
| SALK_N631994-PCR1-R | 5′-TGG TTC ACG TAG TGG GCC ATC G-3′ |
| Oligonucleotide | Sequence |
|---|---|
| AtSRT2-CDS3-LP | 5′-AGAAGCCGCACTCTAAGCAC-3′ |
| AtSRT2-CDS3-RP | 5′-AGTTACGCTGGATGGAGGAG-3′ |
| Gene | Gene Code | Primer Sequence (Forward/Reverse) | Amplicon Length (bp) | Tm °C |
|---|---|---|---|---|
| Act2 | At3g18780 | 5′-TGCCAATCTACGAGGGTTTC-3′ | 203 | 57.3 |
| 5′-TGAGGTTTCCATCTCCTGCT-3′ | 57.3 | |||
| Kin2-LP | At5g15970 | 5′-ACCAACAAGAATGCCTTCCA-3′ | 146 | 55.2 |
| 5′-ACTGCCGCATCCGATATACT-3′ | 57.3 | |||
| RD22-LP | At5g25610 | 5′-GGGCTGTTTCCACTGAGGT-3′ | 160 | 58.8 |
| 5′-GTCGTCATCATCGCCTTGT-3′ | 56.7 | |||
| SRT2-LP | At5g09230 | 5′-AGTTACGCTGGATGGAGGAG-3′ | 177 | 59.4 |
| 5′-AGAAGCCGCACTCTAAGCAC-3′ | 59.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obrecht, A.; Paneque, M. Unraveling the Role of AtSRT2 in Energy Metabolism, Stress Responses, and Gene Expression during Osmotic Stress in Arabidopsis thaliana. Plants 2024, 13, 711. https://doi.org/10.3390/plants13050711
Obrecht A, Paneque M. Unraveling the Role of AtSRT2 in Energy Metabolism, Stress Responses, and Gene Expression during Osmotic Stress in Arabidopsis thaliana. Plants. 2024; 13(5):711. https://doi.org/10.3390/plants13050711
Chicago/Turabian StyleObrecht, Alberto, and Manuel Paneque. 2024. "Unraveling the Role of AtSRT2 in Energy Metabolism, Stress Responses, and Gene Expression during Osmotic Stress in Arabidopsis thaliana" Plants 13, no. 5: 711. https://doi.org/10.3390/plants13050711
APA StyleObrecht, A., & Paneque, M. (2024). Unraveling the Role of AtSRT2 in Energy Metabolism, Stress Responses, and Gene Expression during Osmotic Stress in Arabidopsis thaliana. Plants, 13(5), 711. https://doi.org/10.3390/plants13050711







