Analysis of the UDP-Glucosyltransferase (UGT) Gene Family and Its Functional Involvement in Drought and Salt Stress Tolerance in Phoebe bournei
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Analysis of PbUGT Gene Members
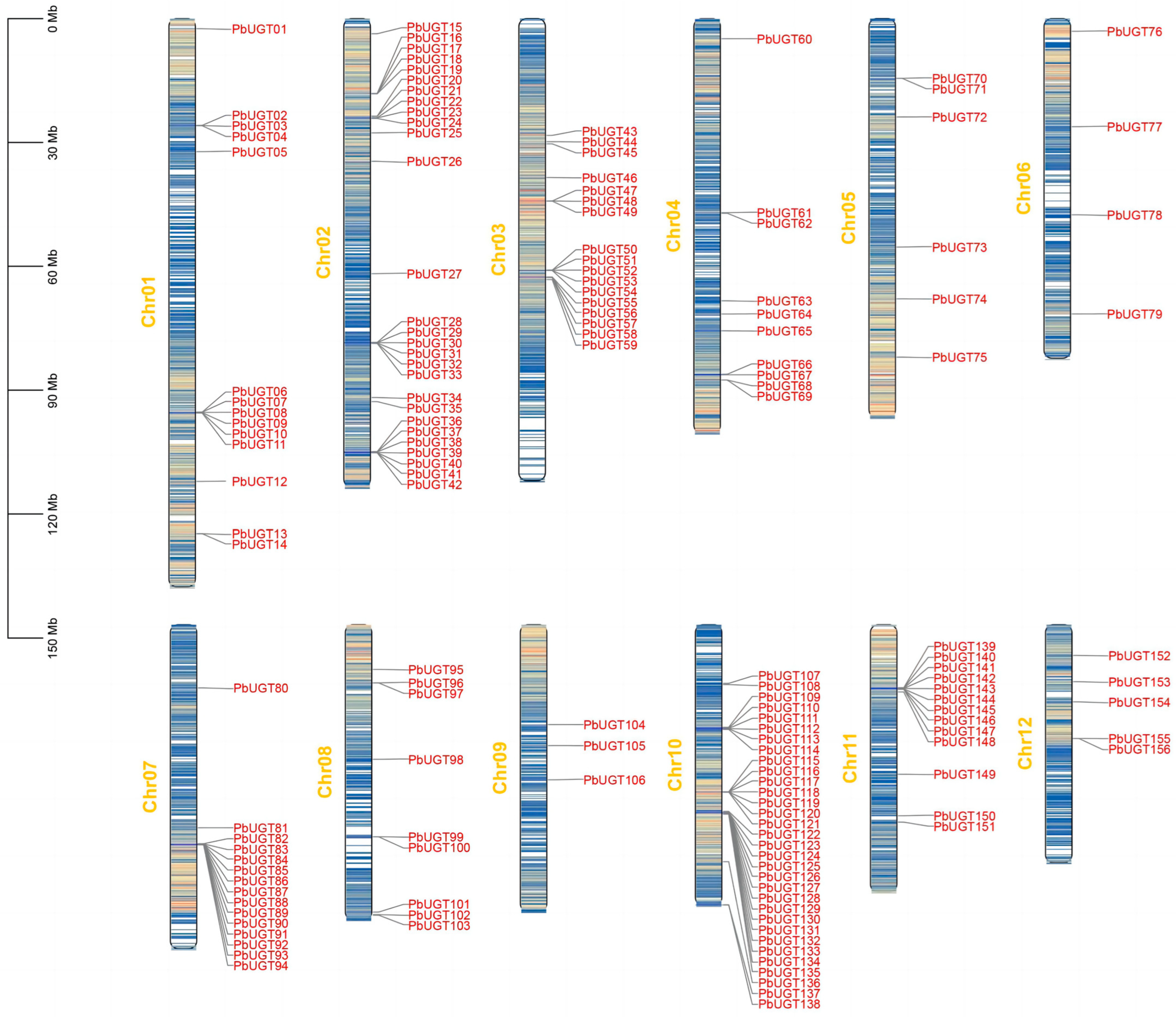
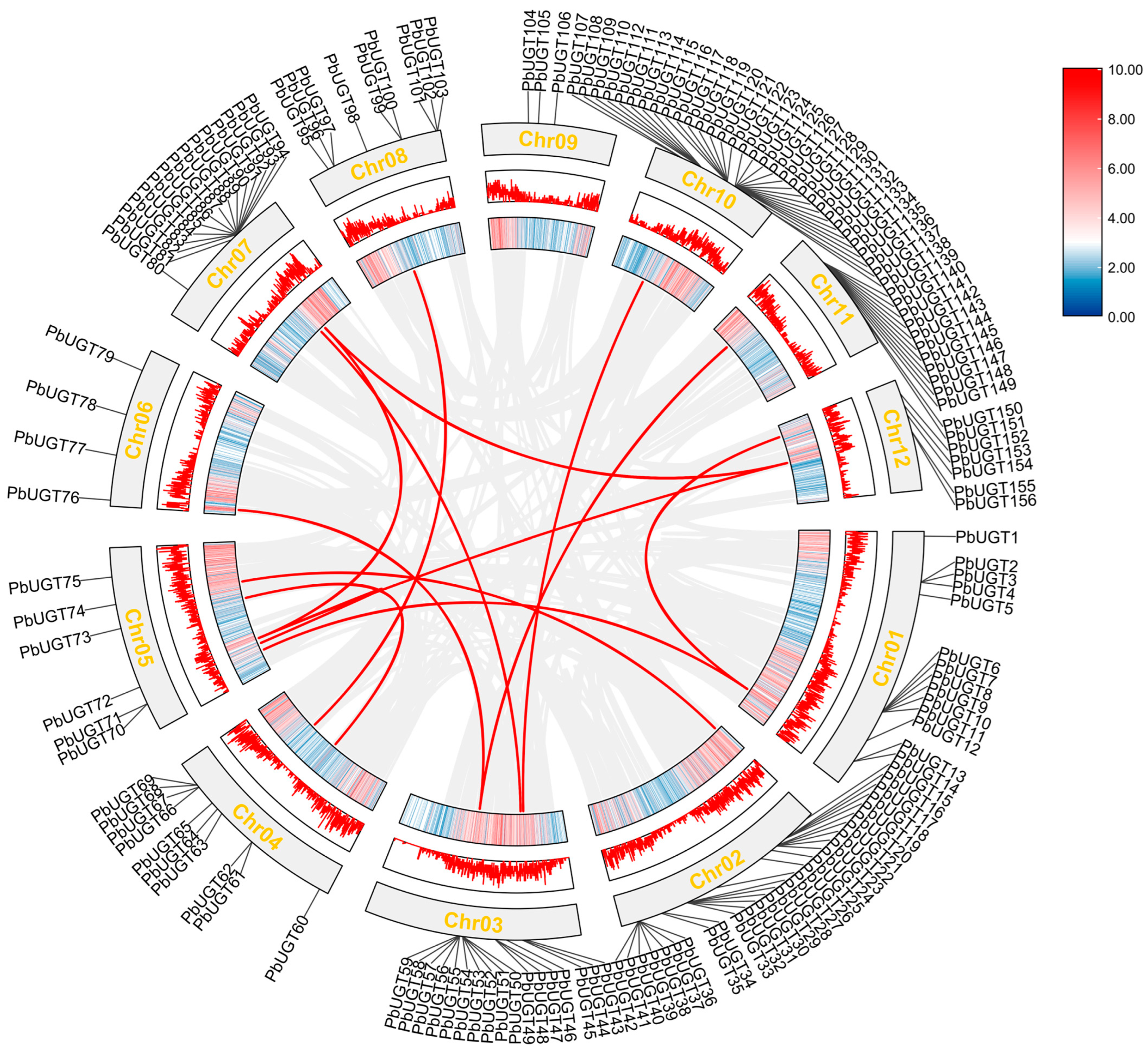
2.2. Chromosome Localization Analysis of the PbUGT Gene Family
2.3. PbUGT Protein Conserved Motif Analysis
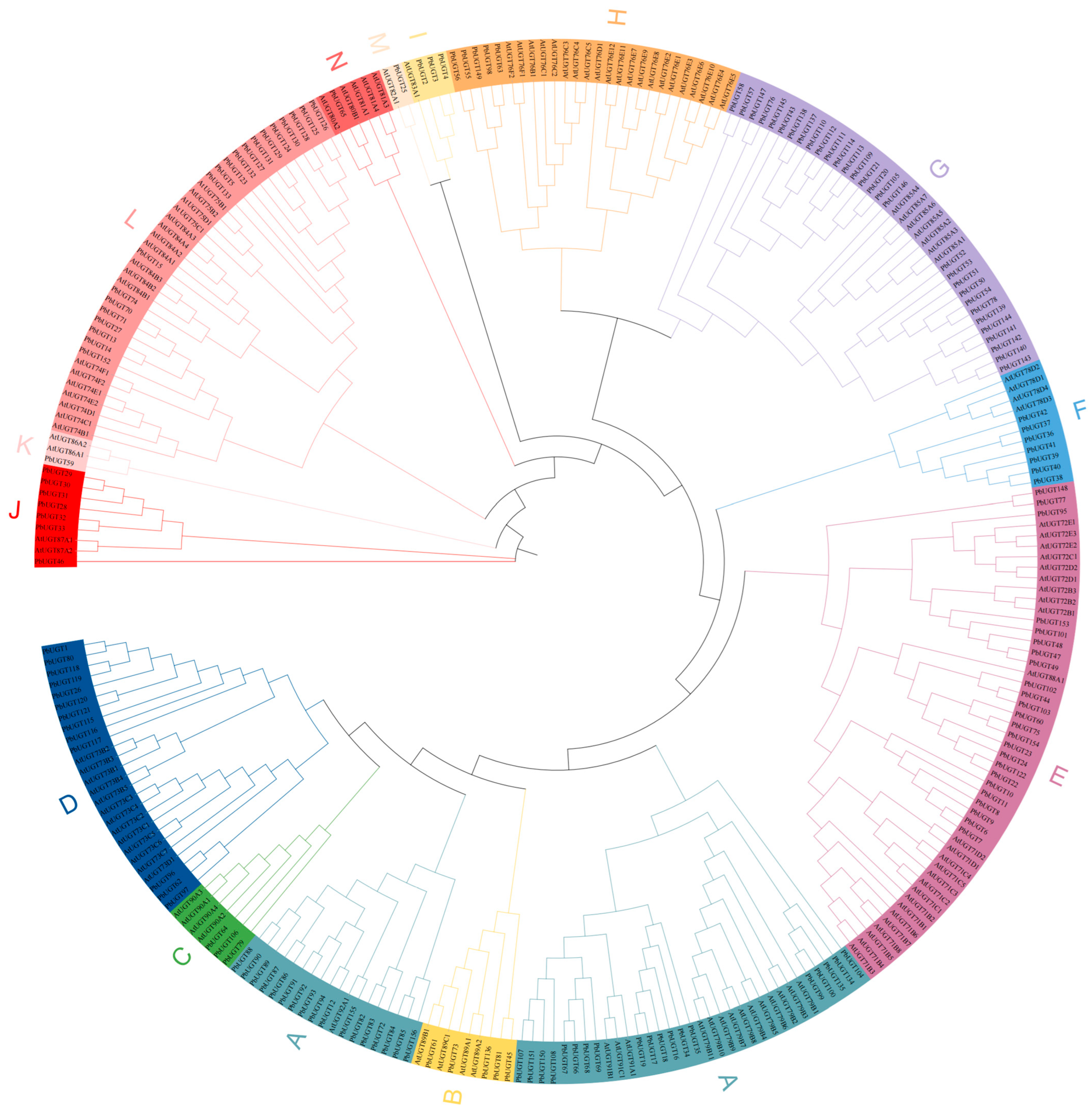
2.4. Phylogenetic Analysis of the PbUGT Gene Family
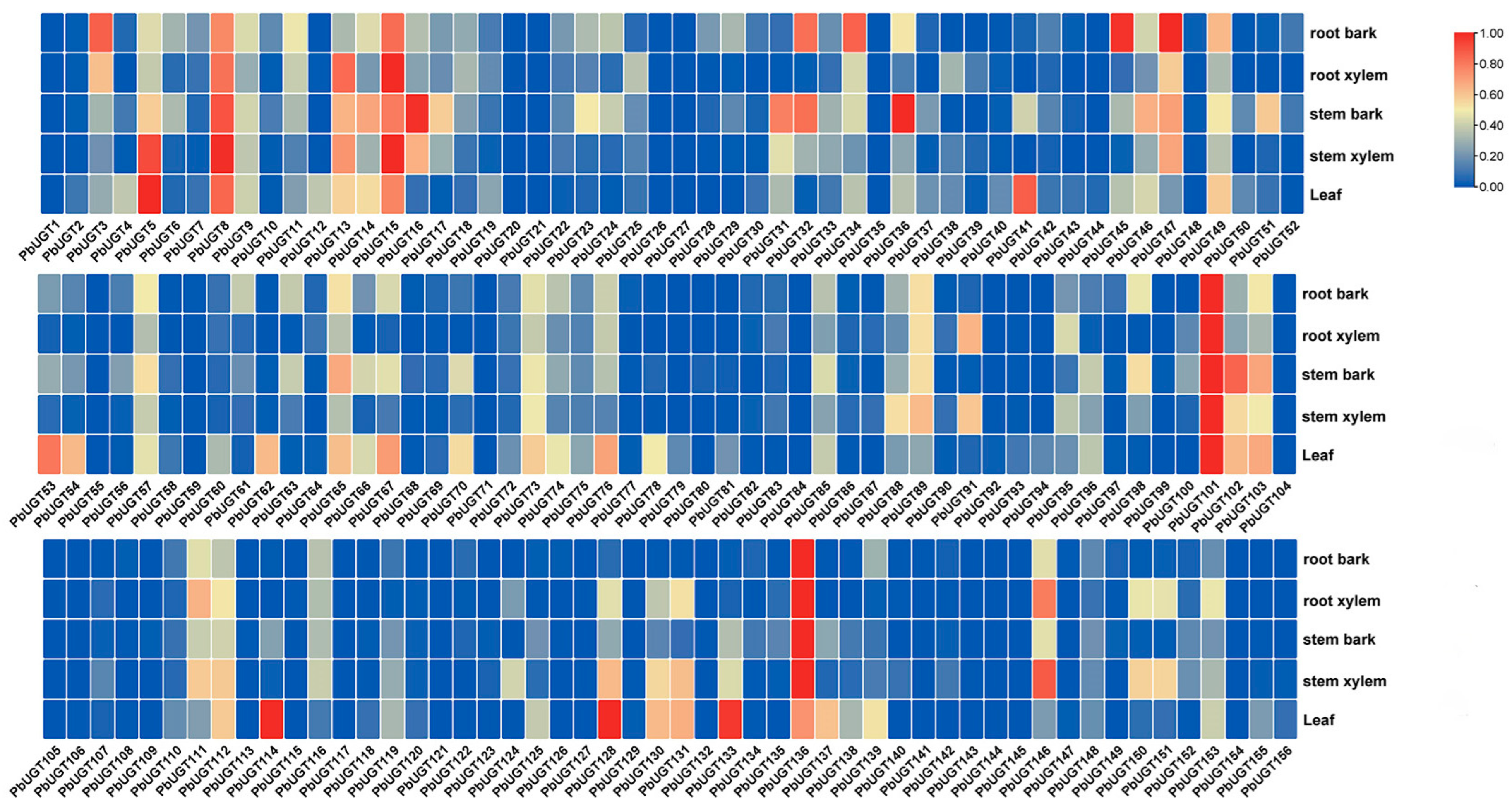
2.5. Organ and Tissue Expression Profile Analysis of the PbUGT Gene Family
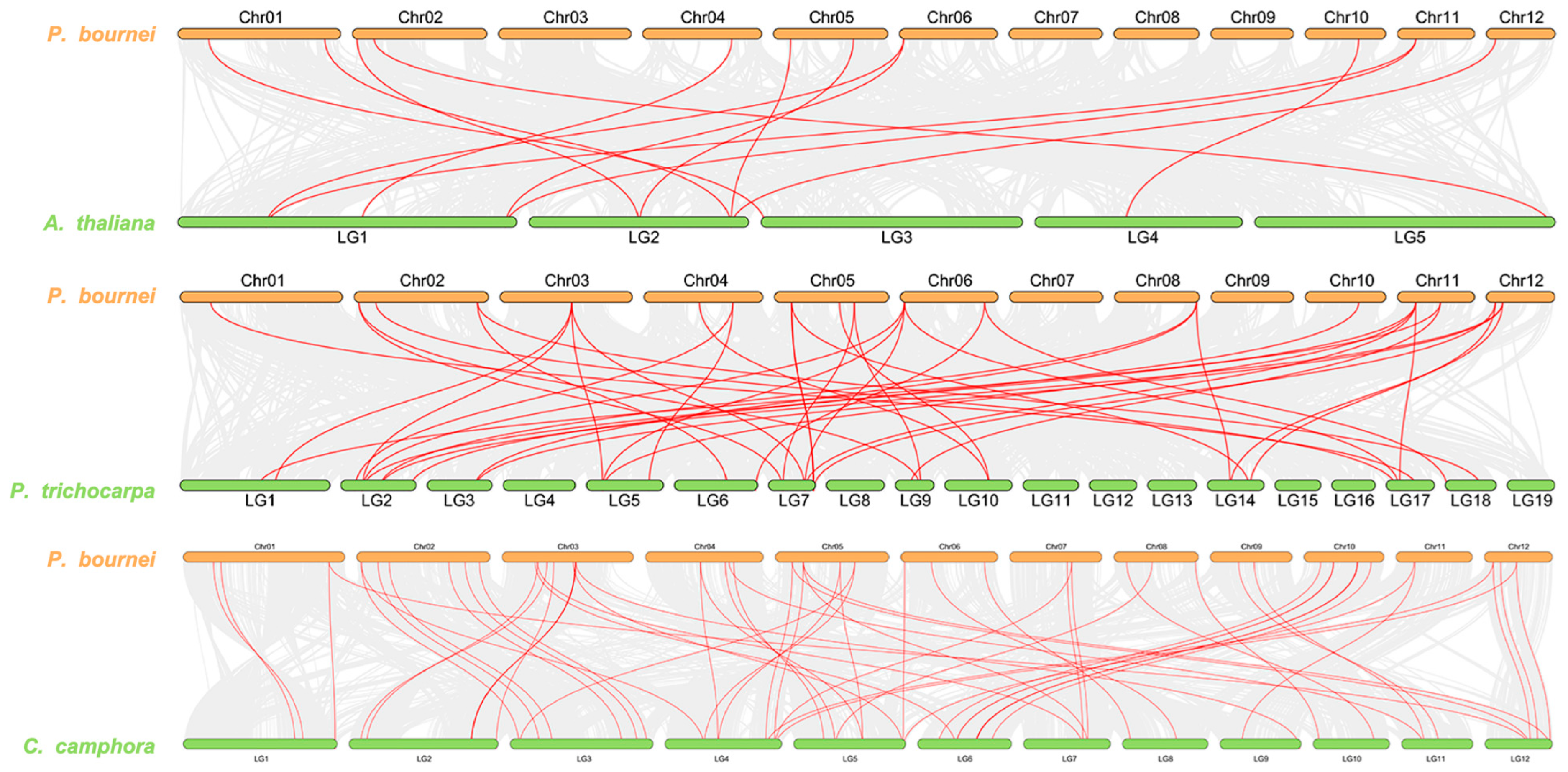
2.6. Synteny Analysis of Homologous UGT Genes among Phoebe bournei, Arabidopsis thaliana, Populus trichocarpa, and Cinnamomum camphora
2.7. Duplication Analysis of PbUGT Genes in Chromosomal Replication
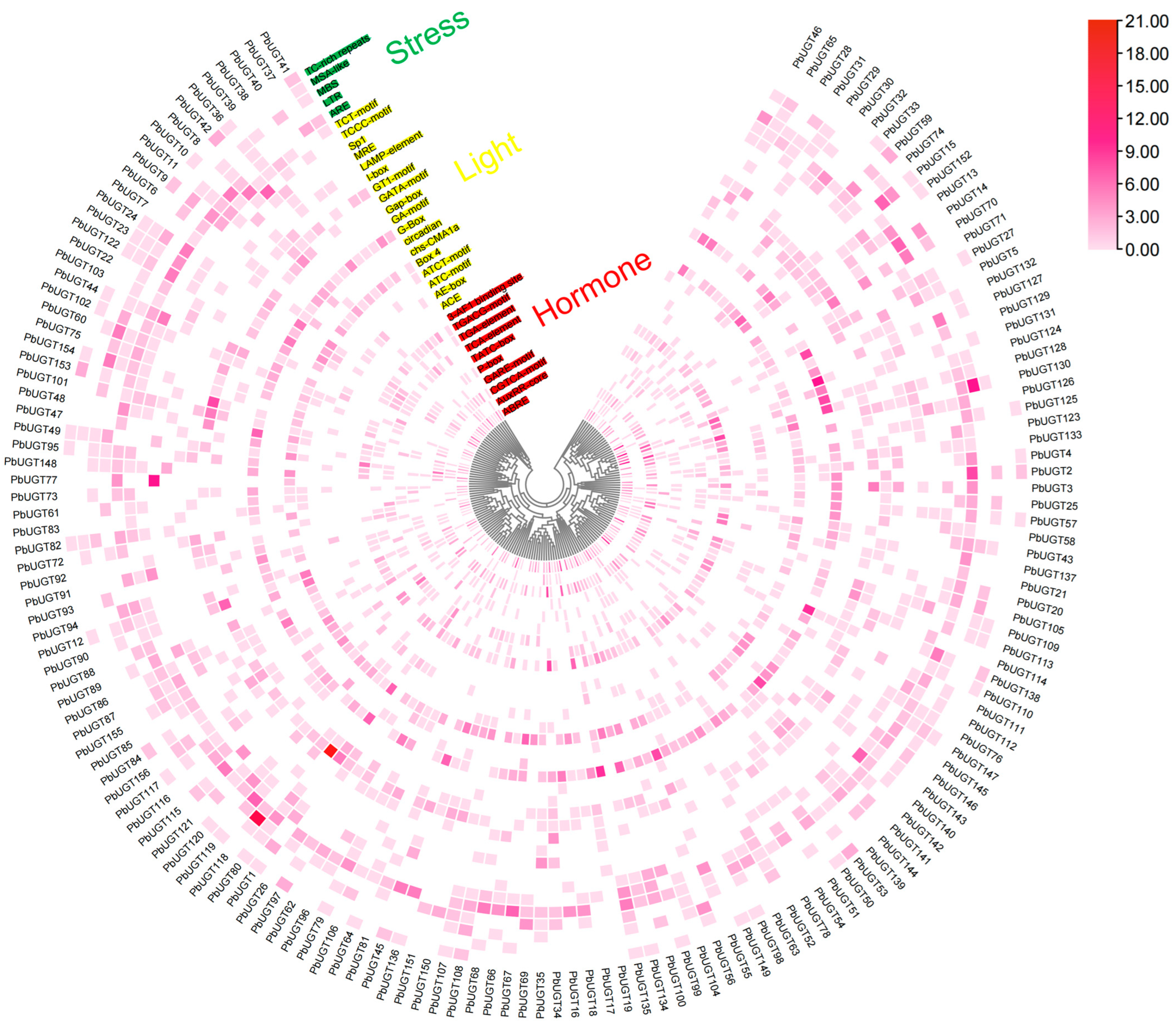
2.8. Enrichment Analysis of Stress-, Hormone-, and Light-Responsive Cis-Elements in the Promoter Region of Phoebe bournei UGT Genes
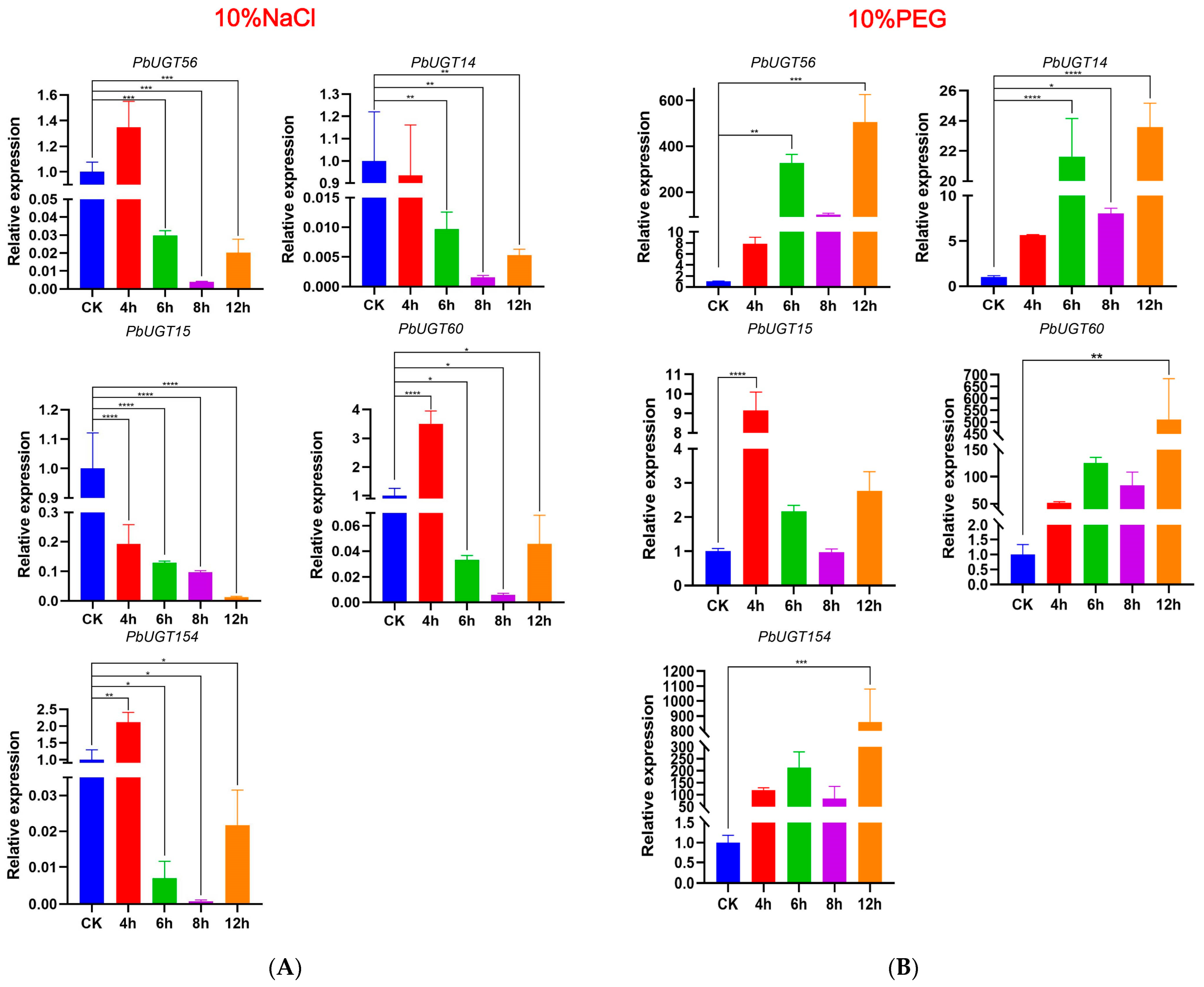
2.9. Screening and Verification of Stress Resistance Genes in PbUGT Genes
3. Discussion
4. Materials and Methods
4.1. Identification, Chromosomal Localization, and Analysis of Protein Physicochemical Properties
4.2. Gene Structure, Protein Conserved Motifs, and Exon–Intron Analysis
4.3. Phylogenetic Analysis and Construction of the Phylogenetic Tree
4.4. Analysis of Tissue Expression Profiles
4.5. Covariance Analysis
4.6. Chromosomes Location, Gene Duplication, and Synteny Analysis
4.7. Enrichment Analysis of Stress-, Hormone-, and Light-Responsive Cis-Elements in the Promoter Region
4.8. Material and Stress Treatments
4.9. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis of PbUGT Genes
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Keegstra, K.; Raikhel, N. Plant glycosyltransferases. Curr. Opin. Plant Biol. 2001, 4, 219–224. [Google Scholar] [CrossRef]
- Hughes, J.; Hughes, M.A. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 1994, 5, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, S.; Kamikawa, S.; Ichihi, A.; Tanaka, Y.; Shimizu, K.; Koeda, S. Isolation of UDP:flavonoid 3-O-glycosyltransferase (UFGT)-like Genes and Expression Analysis of Genes Associated with Anthocyanin Accumulation in Mango ‘Irwin’ skin. Hortic. J. 2019, 88, 435–443. [Google Scholar] [CrossRef]
- Wang, B.; Jin, S.H.; Hu, H.Q.; Sun, Y.G.; Wang, Y.W.; Han, P.; Hou, B.K. UGT87A2, an Arabidopsis glycosyltransferase, regulates flowering time via FLOWERING LOCUS C. New Phytol. 2012, 194, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Paquette, S.; Møller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef]
- Xiao, X.; Lu, Q.; Liu, R.; Gong, J.; Gong, W.; Liu, A.; Ge, Q.; Li, J.; Shang, H.; Li, P.; et al. Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. 3 Biotech 2019, 9, 453. [Google Scholar] [CrossRef]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-Wide Identification, Expression Patterns, and Functional Analysis of UDP Glycosyltransferase Family in Peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. Cell Mol. Biol. 2012, 69, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Bartley, L.E.; Jung, K.; Ronald, P.C. Construction of a Rice Glycosyltransferase Phylogenomic Database and Identification of Rice-Diverged Glycosyltransferases. Mol. Plant 2008, 1, 858–877. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Liu, X.; Xu, K.; Zhang, B. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 2020, 20, 459. [Google Scholar] [CrossRef] [PubMed]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Li, J.; Huang, P.; Wei, F.; Cui, H.; van der Werff, H. Lauraceae. In Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2008; Volume 7. [Google Scholar]
- Wu, D.; Zhu, Z. Preliminary study on structure and spatial distribution pattern of Phoebe bournei in Luo Boyan nature reserve in Fujian province. Sci. Silvae Sin. 2003, 39, 23–30. [Google Scholar]
- Chen, S.P.; Sun, W.H.; Xiong, Y.F.; Jiang, Y.T.; Liu, X.D.; Liao, X.Y.; Zhang, D.Y.; Jiang, S.Z.; Li, Y.; Liu, B.; et al. The Phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 2020, 7, 146. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.; Han, S.; Chong, S.L.; Meng, G.; Song, M.; Wang, Y.; Zhou, S.; Liu, C.; Lou, L.; et al. The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 2022, 3, 100410. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, M.J.; Tong, Z.K.; Han, X.; Zhang, J.H.; Cheng, L.J.; Zhou, S.C. Identification of the PbWRKY gene family and its expression analysis under deficiency of phosphorus in Phoebe bournei. Sci. Silvae Sin. 2022, 58, 133–147. [Google Scholar]
- Han, S.; Han, X.; Li, Y.B.; Zhang, Y.T.; Zhang, J.H.; Tong, Z.K. Identification of NF-Y gene family and expression analysis in response to drought stress in Phoebe bournei. J. Agric. Biotechnol. 2022, 30, 1112–1127. [Google Scholar]
- Wang, Q.Q.; Zhang, Y.T.; Zhang, J.H.; Liu, H.; Tong, Z.K. Identification of PLR gene family and expression of responsive hormones in Phoebe bournei. J. Zhejiang AF Univ. 2022, 39, 1173–1182. [Google Scholar]
- Yin, Z.; Liao, W.; Li, J.; Pan, J.; Yang, S.; Chen, S.; Cao, S. Genome-Wide Identification of GATA Family Genes in Phoebe bournei and Their Transcriptional Analysis under Abiotic Stresses. Int. J. Mol. Sci. 2023, 24, 10342. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yin, K.; Liu, Y.; Li, Y.; Zhang, J.; Han, X.; Tong, Z. Co-expression network analysis reveals PbTGA4 and PbAPRR2 as core transcription factors of drought response in an important timber species Phoebe bournei. Front. Plant Sci. 2024, 14, 1297235. [Google Scholar] [CrossRef] [PubMed]
- Bressan, R.; Bohnert, H.; Zhu, J. Abiotic Stress Tolerance: From Gene Discovery in Model Organisms to Crop Improvement. Mol. Plant 2009, 2, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Li, P.; Wang, T.; Zhang, F.J.; Huang, X.X.; Hou, B.K. The maize secondary metabolism glycosyltransferase UFGT2 modifies flavonols and contributes to plant acclimation to abiotic stresses. Ann. Bot. 2018, 122, 1203–1217. [Google Scholar] [CrossRef]
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Involvement of Auxin in Growth and Stress Response of Cucumber. Veg. Res. 2022, 2, 13. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, N.; Gao, T.; Jin, J.; Jing, T.; Wang, J.; Wu, Y.; Wan, X.; Schwab, W.; Song, C. Sesquiterpene glucosylation mediated by glucosyltransferase UGT91Q2 is involved in the modulation of cold stress tolerance in tea plants. New Phytol. 2020, 226, 362–372. [Google Scholar] [CrossRef]
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Hormonal Interactions Underlying Parthenocarpic Fruit Formation in Horticultural Crops. Hortic. Res. 2022, 9, uhab024. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Ivica, L.; Tobias, D.; Peer, B. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Belanger, A.; Fournel-Gigleux, S.; Green, M.; Hum, D.W.; Iyanagi, T.; et al. The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 1997, 7, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Li, Y.; Lim, E.K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, reviews3004. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. Cell Mol. Biol. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A. The Genome of Black Cottonwood, Populus trichocarpa (Torr. &Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Sun, W.; Xiang, S.; Zhang, Q.; Xiao, L.; Zhang, D.; Zhang, P.; Chen, D.; Hao, Y.; Liu, D.; Ding, L. The camphor tree genome enhances the understanding of magnoliid evolution. J. Genet. Genom. 2022, 49, 249–253. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, Y.; Pan, Y.; Huang, H.; Li, C.; Li, G.; Tong, Z. Transcriptomic profiling and identification of candidate genes in two Phoebe bournei ecotypes with contrasting cold stress responses. Trees 2018, 32, 1315–1333. [Google Scholar] [CrossRef]


| Name | Average Molecular Weight | Theoretical pI | Asp + Glu | Arg + Lys | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|
| Distribution range | 14,948.04–113,398.82 | 4.62–9.8 | 17–127 | 11–110 | 32.1–72.59 | 71.5–102.3 | −0.467–0.273 |
| Average value | 50,994.75 | 5.87 | 54 | 44 | 44.31 | 88.18 | −0.121 |
| Minimum value member | PbUGT66 | PbUGT140 | PbUGT154 | PbUGT154 | PbUGT144 | PbUGT136 | PbUGT66 |
| Maximum value member | PbUGT89 | PbUGT1 | PbUGT89 | PbUGT9 | PbUGT122 | PbUGT75 | PbUGT154 |
| Primer | Sequence (5′~3′) |
|---|---|
| PbUGT14(OF21457-F) | TGCATGGGGCCTCAAAGG |
| PbUGT14(OF21457-R) | GGGCGAGCACTTCCAACT |
| PbUGT15(OF04928-F) | GTTTCCGCAGTGGGGTGA |
| PbUGT15(OF04928-R) | CGGTCCCCCAACAACCTC |
| PbUGT56(OF23900-F) | GTTCGTGGGGTGGAGTGG |
| PbUGT56(OF23900-R) | GGGTGGGCCAGAACTTCC |
| PbUGT60(OF22991-F) | TCGCCGCATCGAATCCTC |
| PbUGT60(OF22991-R) | AGGCGAGGGATTCAGGGT |
| PbUGT154(OF17153-F) | GCGGGCAGAGGTTCTTGT |
| PbUGT154(OF17153-R) | TTGCGACGCCCATGAGTT |
| Internal reference gene (EF1α-F) | CATTCAAGTATGCGTGGGT |
| Internal reference gene (EF1α-R) | ACGGTGACCAGGAGCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, H.; Zhang, Y.; Li, J.; Zhu, Z.; Chang, J.; Bakari, A.; Chen, S.; Zheng, K.; Cao, S. Analysis of the UDP-Glucosyltransferase (UGT) Gene Family and Its Functional Involvement in Drought and Salt Stress Tolerance in Phoebe bournei. Plants 2024, 13, 722. https://doi.org/10.3390/plants13050722
Guan H, Zhang Y, Li J, Zhu Z, Chang J, Bakari A, Chen S, Zheng K, Cao S. Analysis of the UDP-Glucosyltransferase (UGT) Gene Family and Its Functional Involvement in Drought and Salt Stress Tolerance in Phoebe bournei. Plants. 2024; 13(5):722. https://doi.org/10.3390/plants13050722
Chicago/Turabian StyleGuan, Hengfeng, Yanzi Zhang, Jingshu Li, Zhening Zhu, Jiarui Chang, Almas Bakari, Shipin Chen, Kehui Zheng, and Shijiang Cao. 2024. "Analysis of the UDP-Glucosyltransferase (UGT) Gene Family and Its Functional Involvement in Drought and Salt Stress Tolerance in Phoebe bournei" Plants 13, no. 5: 722. https://doi.org/10.3390/plants13050722
APA StyleGuan, H., Zhang, Y., Li, J., Zhu, Z., Chang, J., Bakari, A., Chen, S., Zheng, K., & Cao, S. (2024). Analysis of the UDP-Glucosyltransferase (UGT) Gene Family and Its Functional Involvement in Drought and Salt Stress Tolerance in Phoebe bournei. Plants, 13(5), 722. https://doi.org/10.3390/plants13050722







