Dissecting the Genetic Basis of Yield Traits and Validation of a Novel Quantitative Trait Locus for Grain Width and Weight in Rice
Abstract
:1. Introduction
2. Results
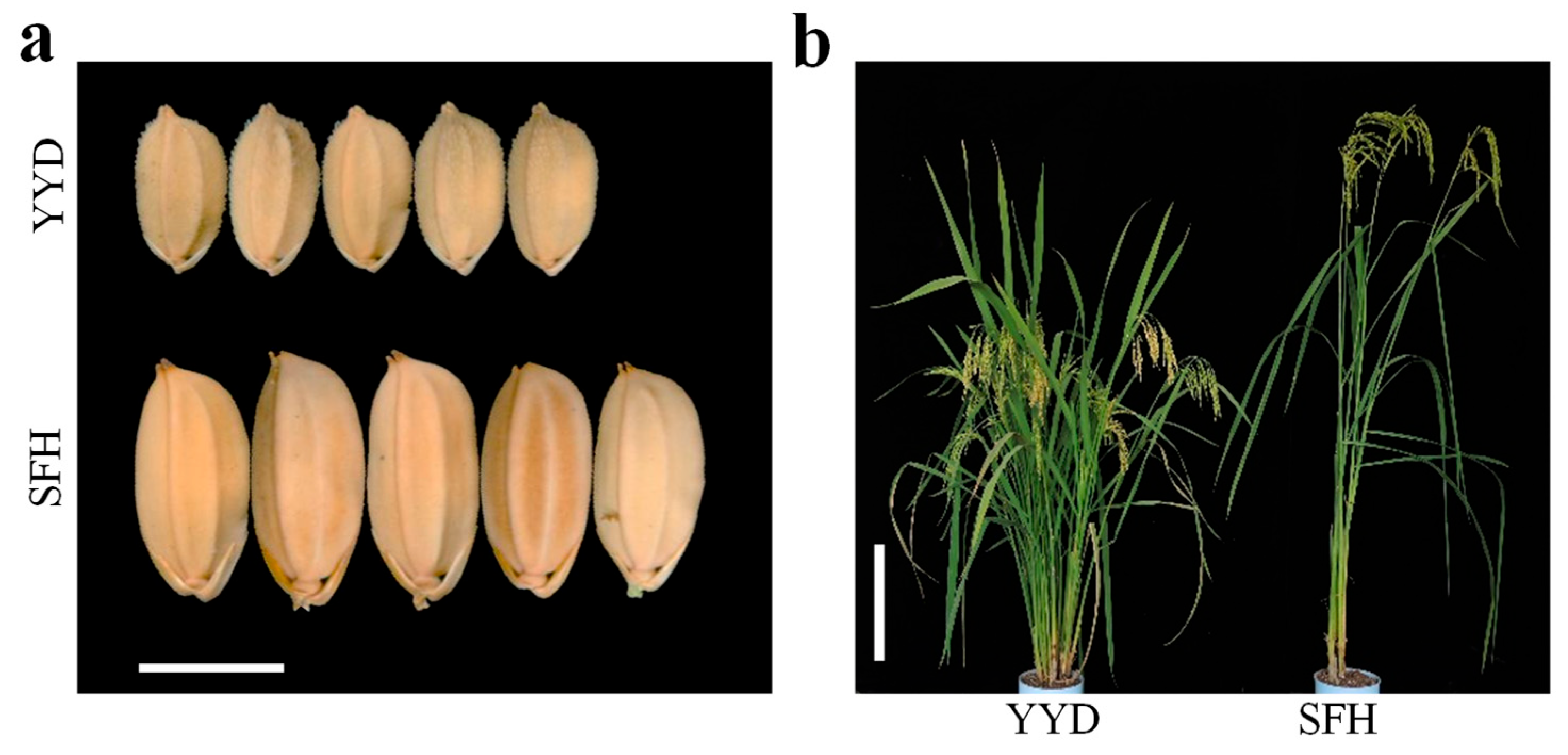
2.1. Phenotypic Variation
2.2. Correlations between Yield Traits
2.3. Creation of a Linkage Map
2.4. QTLs for Rice Yield Traits
2.4.1. QTLs for GYPP and NPPP
2.4.2. QTLs for Grain Number Traits
2.4.3. QTLs for Grain Size and Weight
2.5. Validation of qGW2-1
3. Discussion
3.1. QTLs Dissect the Genetic Basis of Rice Yield Traits
3.2. Construction of Genetic Linkage Maps Is Important to Identify QTLs
3.3. Complex Correlations among Rice Yield and Yield Traits
3.4. qGW2-1 Is Confirmed to Be a Novel QTL for Grain Width and Weight
4. Materials and Methods
4.1. Plant Materials and Field Trials
4.2. Phenotypic Measurement
4.3. DNA Extraction and DNA Marker Analysis
4.4. Construction of the Linkage Map
4.5. QTL Analysis and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Verneau, F.; Amato, M.; La Barbera, F. Edible Insects and Global Food Security. Insects 2021, 12, 472. [Google Scholar] [CrossRef]
- IFPRI. 2019 Global Food Policy Report; IFPRI: Washington, DC, USA, 2019. [Google Scholar]
- Sade, N.; Peleg, Z. Future challenges for global food security under climate change. Plant Sci. 2020, 295, 110467. [Google Scholar] [CrossRef]
- Kuroha, T.; Ashikari, M. Molecular mechanisms and future improvement of submergence tolerance in rice. Mol. Breed. 2020, 40, 41. [Google Scholar] [CrossRef]
- Fukagawa, N.K.; Ziska, L.H. Rice: Importance for Global Nutrition. J. Nutr. Sci. Vitaminol. 2019, 65, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Kobayashi, K.; Loladze, I.; Zhu, J.; Jiang, Q.; Xu, X.; Liu, G.; Seneweera, S.; Ebi, K.L.; Drewnowski, A.; et al. Carbon dioxide (CO(2)) levels this century will alter the protein, micronutrients, and vitamin content of rice grains with potential health consequences for the poorest rice-dependent countries. Sci. Adv. 2018, 4, eaaq1012. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 17031. [Google Scholar] [CrossRef]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhu, H.; Liu, S.; Zeng, R.; Zhang, Z.; Li, W.; Ding, X.; Zhao, F.; Zhang, G. Unconditional and conditional QTL mapping for the developmental behavior of tiller number in rice (Oryza sativa L.). Genetica 2010, 138, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Takai, T. Potential of rice tillering for sustainable food production. J. Exp. Bot. 2024, 75, 708–720. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J. Branching in rice. Curr. Opin. Plant Biol. 2011, 14, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Koumoto, T.; Shimada, H.; Kusano, H.; She, K.-C.; Iwamoto, M.; Takano, M. Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1,6-bisphosphatase. Plant Biotechnol. 2013, 30, 47–56. [Google Scholar] [CrossRef]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wang, D.; Yan, S.; Liu, S.; Liu, B.; Kang, H.; Wang, G.L. Dissection of the Genetic Architecture of Rice Tillering using a Genome-wide Association Study. Rice 2019, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Yan, C.; Zheng, J.; Nan, W.; Qin, X.; Zhang, H. Locating QTL associated with spike traits of Dongxiang wild rice (Oryza rufipogon Griff.). Euphytica 2019, 215, 26. [Google Scholar] [CrossRef]
- Cui, K.H.; Peng, S.B.; Xing, Y.Z.; Yu, S.B.; Xu, C.G. Genetic analysis of the panicle traits related to yield sink size of rice. Yi Chuan Xue Bao Acta. Genet. Sin. 2002, 29, 144–152. [Google Scholar]
- Zhang, X.; Jia, H.; Li, T.; Wu, J.; Nagarajan, R.; Lei, L.; Powers, C.; Kan, C.C.; Hua, W.; Liu, Z.; et al. TaCol-B5 modifies spike architecture and enhances grain yield in wheat. Science 2022, 376, 180–183. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Li, J.; Xu, P.; Wu, Z.; Deng, X.; Pu, Q.; Lv, Y.; Elgamal, W.; Maniruzzaman, S.; et al. Three QTL from Oryza meridionalis Could Improve Panicle Architecture in Asian Cultivated Rice. Rice 2023, 16, 22. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Kadam, N.N.; Struik, P.C.; Rebolledo, M.C.; Yin, X.; Jagadish, S.V.K. Genome-wide association reveals novel genomic loci controlling rice grain yield and its component traits under water-deficit stress during the reproductive stage. J. Exp. Bot. 2018, 69, 4017–4032. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Mi, X.F.; Shan, J.X.; Li, X.M.; Xu, J.L.; Lin, H.X. The QTL GNP1 Encodes GA20ox1, Which Increases Grain Number and Yield by Increasing Cytokinin Activity in Rice Panicle Meristems. PLoS Genet. 2016, 12, e1006386. [Google Scholar] [CrossRef]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef]
- Li, R.; Li, M.; Ashraf, U.; Liu, S.; Zhang, J. Exploring the Relationships Between Yield and Yield-Related Traits for Rice Varieties Released in China From 1978 to 2017. Front. Plant Sci. 2019, 10, 543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Yu, J.; Xiong, H.; Zhu, X.; Zhang, H.; Li, H.; Miao, J.; Wang, W.; Tang, Z.; Zhang, Z.; Yao, G.; et al. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 2017, 15, 28. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S.; et al. A Rare Allele of GS2 Enhances Grain Size and Grain Yield in Rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Li, Q.; Lu, L.; Liu, H.; Bai, X.; Zhou, X.; Wu, B.; Yuan, M.; Yang, L.; Xing, Y. A minor QTL, SG3, encoding an R2R3-MYB protein, negatively controls grain length in rice. Theor. Appl. Genet. 2020, 133, 2387–2399. [Google Scholar] [CrossRef]
- Liu, Q.; Han, R.; Wu, K.; Zhang, J.; Ye, Y.; Wang, S.; Chen, J.; Pan, Y.; Li, Q.; Xu, X.; et al. G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018, 9, 852. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Lin, Y.S.; Song, X.J.; Shen, J.B.; Huang, W.; Shan, J.X.; Zhu, M.Z.; Jiang, L.; Gao, J.P.; Lin, H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.Z.; Ma, M.; Bai, C.; Huang, X.H.; Liu, J.L.; Fan, Y.Y.; Song, X.J. TGW3, a Major QTL that Negatively Modulates Grain Length and Weight in Rice. Mol. Plant 2018, 11, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liu, X.; Wang, M.; Meyer, R.S.; Luo, X.; Ndjiondjop, M.N.; Tan, L.; Zhang, J.; Wu, J.; Cai, H.; et al. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 2017, 3, 17064. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Song, X.J.; Kuroha, T.; Ayano, M.; Furuta, T.; Nagai, K.; Komeda, N.; Segami, S.; Miura, K.; Ogawa, D.; Kamura, T.; et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Hou, Q.; Si, L.; Huang, X.; Luo, J.; Lu, D.; Zhu, J.; Shangguan, Y.; Miao, J.; Xie, Y.; et al. The PLATZ Transcription Factor GL6 Affects Grain Length and Number in Rice. Plant Physiol. 2019, 180, 2077–2090. [Google Scholar] [CrossRef]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.; Shang, L.; Zhang, B.; Hu, J.; Wang, Y.; Lin, H.; Zhang, A.; Liu, C.; Peng, Y.; Zhu, L.; et al. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 2020, 227, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.L.; Dong, N.Q.; Guo, T.; Ye, W.W.; Shan, J.X.; Lin, H.X. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Zhao, X.; Qin, Y.; Irfan, M.; Kim, T.H.; Wang, B.; Wang, S.; Keun Sohn, J. Quantitative trait loci mapping of panicle traits in rice. Mol. Biol. Res. Commun. 2019, 8, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Ahmadizadeh, M.; Babaeian-Jelodar, N.; Mohammadi-Nejad, G.; Bagheri, N.; Kumar Singh, R. High-density linkage mapping for agronomic and physiological traits of rice (Oryza sativa L.) under reproductive-stage salt stress. J. Genet. 2021, 100, 51. [Google Scholar] [CrossRef]
- Li, G.; Cheng, Y.; Yin, M.; Yang, J.; Ying, J.; Zhu, C. Detection of QTLs for panicle-related traits using an indica x japonica recombinant inbred line population in rice. PeerJ 2021, 9, e12504. [Google Scholar] [CrossRef]
- Huang, M.; Zou, Y.-B.; Jiang, P.; Xia, B.; Md, I.; Ao, H.-J. Relationship between Grain Yield and Yield Components in Super Hybrid Rice. Agric. Sci. China 2011, 10, 1537–1544. [Google Scholar] [CrossRef]
- Marathi, B.; Guleria, S.; Mohapatra, T.; Parsad, R.; Mariappan, N.; Kurungara, V.K.; Atwal, S.S.; Prabhu, K.V.; Singh, N.K.; Singh, A.K. QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 137. [Google Scholar] [CrossRef]
- Ying, J.Z.; Shan, J.X.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. Identification of quantitative trait Loci for lipid metabolism in rice seeds. Mol. Plant 2012, 5, 865–875. [Google Scholar] [CrossRef]
- Su, J.; Xu, K.; Li, Z.; Hu, Y.; Hu, Z.; Zheng, X.; Song, S.; Tang, Z.; Li, L. Genome-wide association study and Mendelian randomization analysis provide insights for improving rice yield potential. Sci. Rep. 2021, 11, 6894. [Google Scholar] [CrossRef]
- Ying, J.Z.; Gao, J.P.; Shan, J.X.; Zhu, M.Z.; Shi, M.; Lin, H.X. Dissecting the genetic basis of extremely large grain shape in rice cultivar ‘JZ1560’. J. Genet. Genom. 2012, 39, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.W.; Fan, Y.Y.; Zhu, Y.J.; Chen, J.Y.; Yu, S.B.; Zhuang, J.Y. Dissection of the qTGW1.1 region into two tightly-linked minor QTLs having stable effects for grain weight in rice. BMC Genet. 2016, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Huang, N.; Bennett, J.; Khush, G.S. PCR-based marker-assisted selection in rice breeding. In IRRI Discussion Paper Series No. 12; International Rice Research Institute: Los Banos, Philippines, 1995. [Google Scholar]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newberg, L.A. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2006. [Google Scholar]






| Traits | QTL | Chr. | Marker Interval | Position | LOD | A | D | R2 (%) |
|---|---|---|---|---|---|---|---|---|
| GYPP | qGYPP3 | 3 | JD3011-YS3041 | 22.34 | 5.86 | −5.37 | 1.12 | 12.35 |
| qGYPP6 | 6 | RM19472-RM19572 | 30.43 | 4.43 | −4.29 | −1.11 | 8.95 | |
| qGYPP7 | 7 | RM22039-JD7015 | 111.44 | 4.34 | −3.83 | −3.11 | 10.04 | |
| NPPP | qNPPP3 | 3 | RM14849-RM15029 | 89.92 | 7.61 | 5.5 | −4.1 | 17.85 |
| qNPPP4 | 4 | RM17096-RM17303 | 176.06 | 5.83 | −3.0 | 0.3 | 10.82 | |
| qNPPP10 | 10 | RM25367-RM25717 | 59.62 | 5.35 | −3.3 | −0.3 | 10.76 | |
| qNPPP12 | 12 | JD12010-RM27507 | 11.64 | 3.76 | 1.6 | 2.0 | 7.58 | |
| TNSP | qTNSP1 | 1 | JD1016-JD1007 | 24.55 | 3.37 | −243.3 | 27.6 | 6.19 |
| qTNSP3 | 3 | RM15029-RM15217 | 114.19 | 3.61 | 492.0 | −462.3 | 11.30 | |
| qTNSP7 | 7 | RM21719-RM21993 | 80.99 | 4.18 | −300.3 | 17.1 | 8.56 | |
| qTNSP10 | 10 | RM25367-RM25717 | 59.70 | 3.75 | −330.9 | −170.3 | 13.15 | |
| qTNSP12 | 12 | JD12010-RM27507 | 11.64 | 4.05 | 341.9 | −5.8 | 8.30 | |
| NFGPP | qNFGPP3 | 3 | JD3011-YS3041 | 22.34 | 5.44 | −200.6 | 46.1 | 11.36 |
| qNFGPP6 | 6 | RM19472-RM19572 | 30.43 | 3.86 | −146.1 | −53.7 | 7.71 | |
| qNFGPP7 | 7 | RM22039-JD7015 | 111.44 | 6.19 | −179.1 | −148.1 | 16.54 | |
| qNFGPP12 | 12 | YS12002-JD12019 | 50.69 | 3.20 | 7.9 | 218.0 | 8.16 | |
| NSPP | qNSPP1 | 1 | RM11570-JD1004 | 133.83 | 5.03 | −18.8 | −15.4 | 10.40 |
| qNSPP3 | 3 | RM15029-RM15217 | 114.15 | 4.47 | −26.7 | 16.3 | 7.19 | |
| qNSPP4 | 4 | RM17096-RM17303 | 176.00 | 3.39 | 16.9 | −5.4 | 6.76 | |
| qNSPP7 | 7 | RM22039-JD7015 | 111.44 | 11.92 | −31.0 | −11.4 | 25.11 | |
| SSR | qSSR1 | 1 | RM11811-RM3403 | 174.36 | 4.55 | −8.82 | −5.22 | 8.70 |
| qSSR3 | 3 | JD3011-YS3041 | 22.32 | 3.86 | −10.07 | −1.27 | 9.16 | |
| qSSR6 | 6 | RM19472-RM19572 | 30.53 | 5.00 | −5.18 | −12.31 | 11.88 | |
| qSSR10 | 10 | RM25367-RM25717 | 59.66 | 4.00 | 13.88 | −5.66 | 10.64 | |
| qSSR12 | 12 | JD12014-JD12010 | 0.04 | 3.35 | −0.84 | −13.08 | 8.08 | |
| GL | qGL3 | 3 | RM3513-RM15981 | 148.73 | 9.40 | 0.50 | −0.41 | 36.01 |
| qGL7 | 7 | RM22039-JD7015 | 111.46 | 3.75 | 0.20 | 0.18 | 9.71 | |
| qGL11 | 11 | RM3133-RM26567 | 38.72 | 3.49 | 0.28 | 0.05 | 12.06 | |
| GW | qGW1 | 1 | JD1007-RM10609 | 43.43 | 3.47 | −0.01 | 0.10 | 8.26 |
| qGW2-1 | 2 | JD2001-JD2029 | 70.21 | 9.02 | 0.12 | 0.02 | 23.58 | |
| qGW2-2 | 2 | JD2030-RM1367 | 99.12 | 9.85 | 0.13 | −0.01 | 25.89 | |
| qGW3 | 3 | RM16-RM15576 | 140.87 | 4.34 | 0.11 | −0.04 | 10.81 | |
| qGW10 | 10 | RM25367-RM25717 | 59.69 | 3.89 | 0.06 | 0.10 | 17.81 | |
| TGW | qTGW2 | 2 | JD2030-RM1367 | 99.14 | 4.40 | 1.62 | −0.43 | 11.51 |
| qTGW3 | 3 | RM15029-RM15353 | 114.32 | 8.01 | 2.68 | −0.73 | 20.67 | |
| qTGW7 | 7 | RM22039-JD7015 | 111.32 | 3.94 | 0.97 | 1.35 | 10.07 |
| Population | Trait | Interval | Sample | LOD | A | D | R2 (%) |
|---|---|---|---|---|---|---|---|
| YM1 | GL | W236-YS2006 | 128 | ns | ns | ns | ns |
| GW | W236-YS2006 | 128 | 8.14 | 0.09 | −0.01 | 31.54 | |
| TGW | W236-YS2006 | 128 | 4.10 | 1.21 | −0.55 | 19.07 | |
| YM2 | GL | YS2024-JD2029 | 140 | ns | ns | ns | ns |
| GW | YS2024-JD2029 | 140 | 13.07 | 0.05 | −0.01 | 45.79 | |
| TGW | YS2024-JD2029 | 140 | 3.56 | 0.57 | −0.04 | 14.70 | |
| YM3 | GL | W236-YS2010 | 179 | ns | ns | ns | ns |
| GW | W236-YS2010 | 179 | 3.33 | 0.06 | 0.01 | 11.17 | |
| TGW | W236-YS2010 | 179 | 4.24 | 0.97 | −0.18 | 17.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, M.; Tong, X.; Yang, J.; Cheng, Y.; Zhou, P.; Li, G.; Wang, Y.; Ying, J. Dissecting the Genetic Basis of Yield Traits and Validation of a Novel Quantitative Trait Locus for Grain Width and Weight in Rice. Plants 2024, 13, 770. https://doi.org/10.3390/plants13060770
Yin M, Tong X, Yang J, Cheng Y, Zhou P, Li G, Wang Y, Ying J. Dissecting the Genetic Basis of Yield Traits and Validation of a Novel Quantitative Trait Locus for Grain Width and Weight in Rice. Plants. 2024; 13(6):770. https://doi.org/10.3390/plants13060770
Chicago/Turabian StyleYin, Man, Xiaohong Tong, Jinyu Yang, Yichen Cheng, Panpan Zhou, Guan Li, Yifeng Wang, and Jiezheng Ying. 2024. "Dissecting the Genetic Basis of Yield Traits and Validation of a Novel Quantitative Trait Locus for Grain Width and Weight in Rice" Plants 13, no. 6: 770. https://doi.org/10.3390/plants13060770
APA StyleYin, M., Tong, X., Yang, J., Cheng, Y., Zhou, P., Li, G., Wang, Y., & Ying, J. (2024). Dissecting the Genetic Basis of Yield Traits and Validation of a Novel Quantitative Trait Locus for Grain Width and Weight in Rice. Plants, 13(6), 770. https://doi.org/10.3390/plants13060770








