Nuclear Magnetic Resonance Fingerprints and Mini DNA Markers for the Authentication of Cinnamon Species Ingredients Used in Food and Natural Health Products
Abstract
:1. Introduction
- Development of mini DNA markers—we utilized genome skimming to retrieve plastid regions through the shallow sequencing of two commercial cinnamon species: C. burmannii and C. cassia. Then, we assembled full-length chloroplast genomes for C. burmannii and C. cassia from high-throughput sequence data using the chloroplast genome of C. verum as a reference. The goal was to provide broader genome sampling that could be used in a phylogenetic approach to differentiate cinnamon species and further the development of mini DNA markers. These markers could then be properly validated [71] and used in on-site qPCR platforms for quick, affordable, quality assurance tools.
- Development of NMR Fingerprints—commercial samples, with their species identity verified using DNA methods, were used to develop NMR spectral fingerprints on a 400 MHz Bruker AVANCE III. The goal was to develop quick screening methods and more detailed analysis of cinnamon spectra for use in quality control systems that could verify species identity from raw to processed samples throughout the supply chain. We utilized NMR spectra for each species to quantify molecules of interest to provide further assessment of quality assurance and possible efficacy for health claims.
2. Results
2.1. Chloroplast Genome
2.1.1. Chloroplast Genome Assembly
2.1.2. Chloroplast Genome Characteristics
2.1.3. Chloroplast Genome Phylogeny
2.1.4. Development and Testing of New Mini DNA Markers
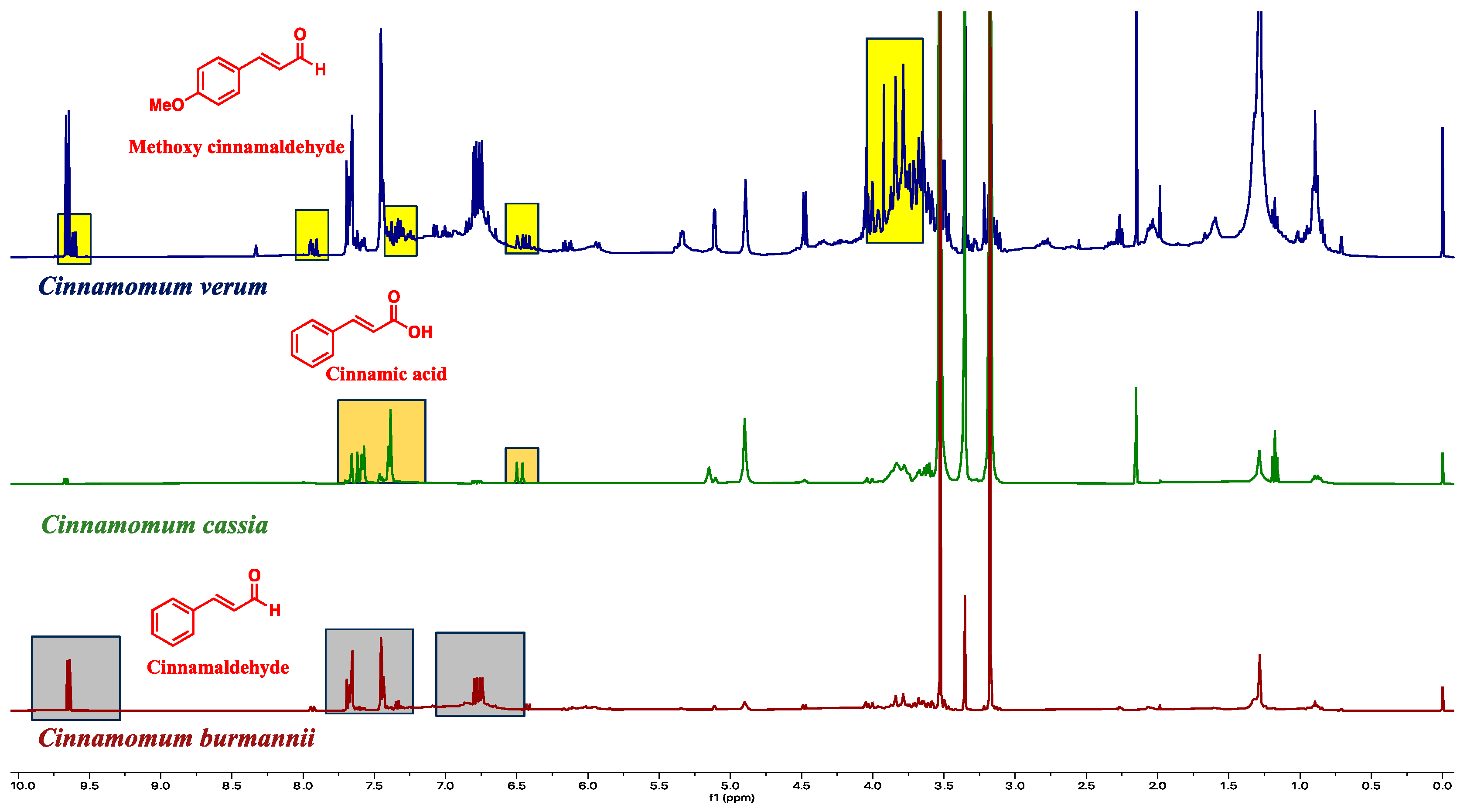
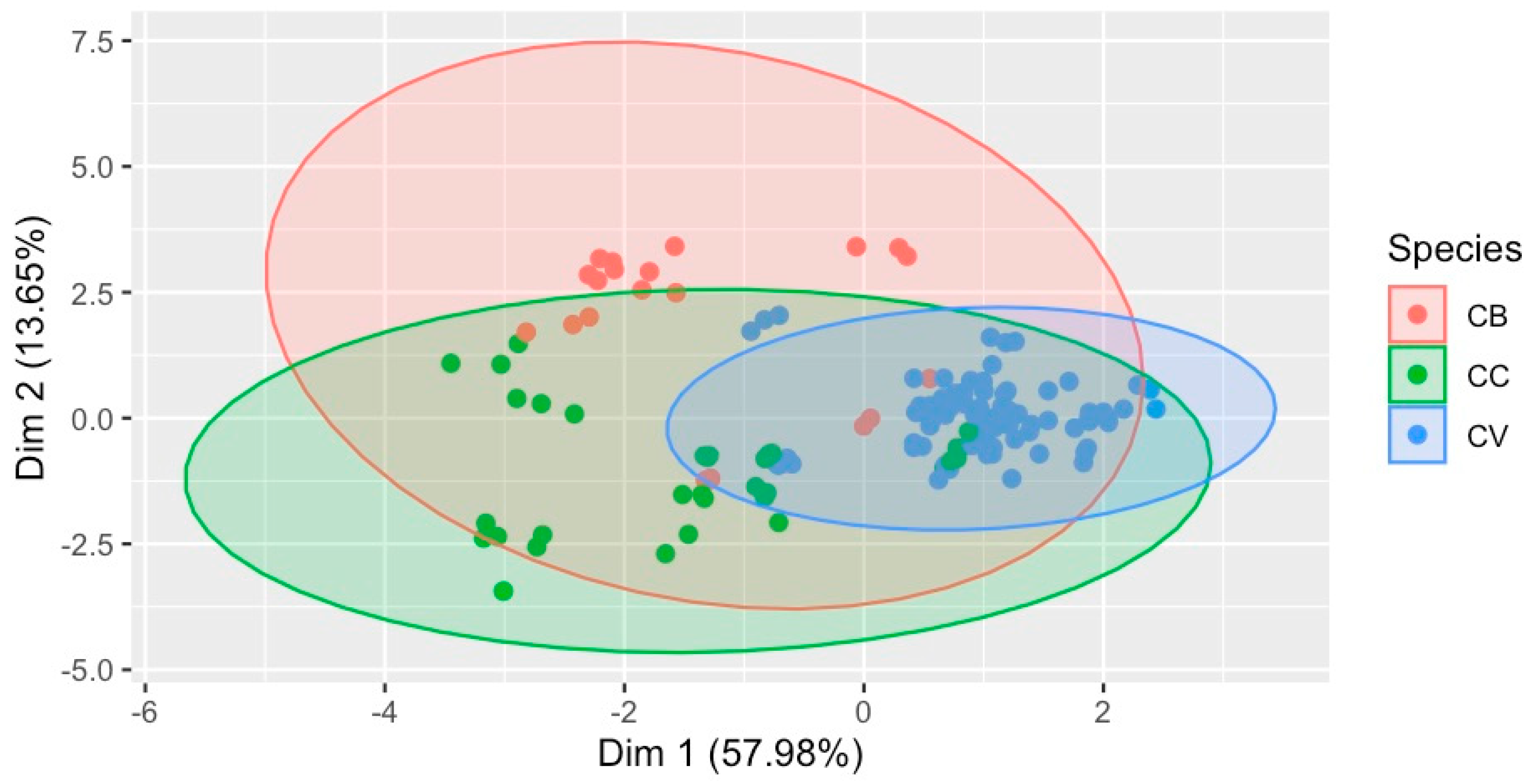
2.2. NMR Chemical Fingerprinting
2.3. Quantification of Metabolites
3. Discussion
3.1. Complete Chloroplast Genomes Reveal Important Phylogenetic Relationships in Cinnamon
3.2. Genome Skimming for Mini DNA Marker Development as an Authenticity Screening Tool
3.3. The Current Need to Develop Mini DNA Markers to Test Processed Ingredients and Products
3.4. NMR Fingerprints Provide an Efficient Multipurpose Quality Assurance Tool
4. Materials and Methods
4.1. DNA Samples Extraction and Sequencing
4.2. Assembly and Annotation of Chloroplast Genomes
4.3. Nuclear Magnetic Resonance (NMR) Sample Preparation Methods
4.4. Nuclear Magnetic Resonance (NMR) Spectroscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bandara, T.; Uluwaduge, I.; Jansz, E.R. Bioactivity of Cinnamon with Special Emphasis on Diabetes Mellitus: A Review. Int. J. Food Sci. Nutr. 2012, 63, 380–386. [Google Scholar] [CrossRef]
- Rafehi, H.; Ververis, K.; Karagiannis, T.C. Controversies Surrounding the Clinical Potential of Cinnamon for the Management of Diabetes. Diabetes Obes. Metab. 2012, 14, 493–499. [Google Scholar] [CrossRef]
- Ravindran, P.N.; Nirmal Babu, K. Introduction. In Cinnamon and Cassia: The Genus Cinnamomum; Ravindran, P.N., Nirmal Babu, K., Shylaja, M., Eds.; CRC Press: Boca Raton, FL, USA, 2004; pp. 1–13. [Google Scholar]
- Avula, B.; Smillie, T.J.; Wang, Y.-H.; Zweigenbaum, J.; Khan, I.A. Authentication of True Cinnamon (Cinnamon verum) Utilising Direct Analysis in Real Time (DART)-QToF-MS. Food Addit. Contam. A 2015, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yasmin, J.; Ahmed, M.R.; Lohumi, S.; Wakholi, C.; Lee, H.; Mo, C.; Cho, B.-K. Rapid Authentication Measurement of Cinnamon Powder Using FT-NIR and FT-IR Spectroscopic Techniques. Qual. Assur. Saf. Crops Foods 2019, 11, 257–267. [Google Scholar] [CrossRef]
- Ka, H.; Park, H.-J.; Jung, H.-J.; Choi, J.-W.; Cho, K.-S.; Ha, J.; Lee, K.-T. Cinnamaldehyde Induces Apoptosis by ROS-Mediated Mitochondrial Permeability Transition in Human Promyelocytic Leukemia HL-60 Cells. Cancer Lett. 2003, 196, 143–152. [Google Scholar] [CrossRef]
- Chou, S.-T.; Chang, W.-L.; Chang, C.-T.; Hsu, S.-L.; Lin, Y.-C.; Shih, Y. Cinnamomum cassia Essential Oil Inhibits α-MSH-Induced Melanin Production and Oxidative Stress in Murine B16 Melanoma Cells. Int. J. Mol. Sci. 2013, 14, 19186–19201. [Google Scholar] [CrossRef]
- Venugopala, K.N.; Rashmi, V.; Odhav, B. Review on Natural Coumarin Lead Compounds for Their Pharmacological Activity. BioMed Res. Int. 2013, 2013, 963248. [Google Scholar] [CrossRef]
- Mirmosayyeb, O.; Tanhaei, A.; Sohrabi, H.; Martins, R.; Tanhaei, M.; Najafi, M.; Safaei, A.; Meamar, R. Possible Role of Common Spices as a Preventive and Therapeutic Agent for Alzheimer’s Disease. Int. J. Prev. Med. 2017, 8, 5. [Google Scholar] [CrossRef]
- Shinjyo, N.; Waddell, G.; Green, J. A Tale of Two Cinnamons: A Comparative Review of the Clinical Evidence of Cinnamomum verum and C. cassia as Diabetes Interventions. J. Herb. Med. 2020, 21, 100342. [Google Scholar] [CrossRef]
- Doyle, A.A.; Stephens, J.C. A Review of Cinnamaldehyde and Its Derivatives as Antibacterial Agents. Fitoterapia 2019, 139, 104405. [Google Scholar] [CrossRef]
- Zhu, R.; Liu, H.; Liu, C.; Wang, L.; Ma, R.; Chen, B.; Li, L.; Niu, J.; Fu, M.; Zhang, D.; et al. Cinnamaldehyde in Diabetes: A Review of Pharmacology, Pharmacokinetics and Safety. Pharmacol. Res. 2017, 122, 78–89. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Dey, A.; Koirala, N.; Shaheen, S.; El Omari, N.; Salehi, B.; Goloshvili, T.; Cirone Silva, N.C.; Bouyahya, A.; Vitalini, S.; et al. Cinnamomum Species: Bridging Phytochemistry Knowledge, Pharmacological Properties and Toxicological Safety for Health Benefits. Front. Pharmacol. 2021, 12, 600139. [Google Scholar] [CrossRef]
- Matos, M.J.; Santana, L.; Uriarte, E.; Abreu, O.A.; Molina, E.; Yordi, E.G. Coumarins—An Important Class of Phytochemicals. In Phytochemicals—Isolation, Characterisation and Role in Human Health; Rao, A.V., Rao, L.G., Eds.; InTech: London, UK, 2015; pp. 113–140. [Google Scholar]
- Cohen, A.J. Critical Review of the Toxicology of Coumarin with Special Reference to Interspecies Differences in Metabolism and Hepatotoxic Response and Their Significance to Man. Food Cosmet. Toxicol. 1979, 17, 277–289. [Google Scholar] [CrossRef]
- Egan, D.; O’Kennedy, R.; Moran, E.; Cox, D.; Prosser, E.; Thornes, R.D. The Pharmacology, Metabolism, Analysis, and Applications of Coumarin and Coumarin-Related Compounds. Drug Metab. Rev. 1990, 22, 503–529. [Google Scholar] [CrossRef]
- Choi, J.; Lee, K.-T.; Ka, H.; Jung, W.-T.; Jung, H.-J.; Park, H.-J. Constituents of the Essential Oil of the Cinnamomum cassia Stem Bark and the Biological Properties. Arch. Pharm. Res. 2001, 24, 418–423. [Google Scholar] [CrossRef]
- Ranasinghe, P.; Pigera, S.; Premakumara, G.S.; Galappaththy, P.; Constantine, G.R.; Katulanda, P. Medicinal Properties of ‘True’ Cinnamon (Cinnamomum zeylanicum): A Systematic Review. BMC Complement. Altern. Med. 2013, 13, 275. [Google Scholar] [CrossRef]
- Farag, M.A.; Khaled, S.E.; El Gingeehy, Z.; Shamma, S.N.; Zayed, A. Comparative Metabolite Profiling and Fingerprinting of Medicinal Cinnamon Bark and Its Commercial Preparations via a Multiplex Approach of GC–MS, UV, and NMR Techniques. Metabolites 2022, 12, 614. [Google Scholar] [CrossRef]
- Suzuki, R.; Kasuya, Y.; Sano, A.; Tomita, J.; Maruyama, T.; Kitamura, M. Comparison of Various Commercially Available Cinnamon Barks Using NMR Metabolomics and the Quantification of Coumarin by Quantitative NMR Methods. J. Nat. Med. 2022, 76, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Negi, A.; Pare, A.; Meenatchi, R. Emerging Techniques for Adulterant Authentication in Spices and Spice Products. Food Control 2021, 127, 108113. [Google Scholar] [CrossRef]
- European Commission. Regulation (EC) No 1334/2008 of the European Parliament and of the Council of 16 December 2008 on Flavourings and Certain Food Ingredients with Flavouring Properties for Use in and on Foods and Amending Council Regulation (EEC) No 1601/91, Regulations (EC) No 2232/96 and (EC) No 110/2008 and Directive 2000/13/EC. The Official Journal of the European Union, 31 December 2008.
- Feltes, G.; Ballen, S.C.; Steffens, J.; Paroul, N.; Steffens, C. Differentiating True and False Cinnamon: Exploring Multiple Approaches for Discrimination. Micromachines 2023, 14, 1819. [Google Scholar] [CrossRef] [PubMed]
- Dugo, P.; Mondello, L.; Dugo, G. Thin-Layer (Planar) Chromatography. Essent. Oils 2000, 2755–2760. [Google Scholar] [CrossRef]
- Biringanine, G.; Chiarelli, M.; Faes, M.; Duez, P. A Validation Protocol for the HPTLC Standardization of Herbal Products: Application to the Determination of Acteoside in Leaves of Plantago palmata Hook. f.s. Talanta 2006, 69, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Leach, D.; Fryganas, C.; Mueller-Harvey, I.; Wohlmuth, H. Phytoequivalence of Therapeutic Cinnamon Barks and Extracts with Low Coumarin Levels. Planta Medica Int. Open 2017, 4, S1–S202. [Google Scholar] [CrossRef]
- Meena, A.K.; Narasimhaji, C.V.; Rekha, P.; Velvizhi, D.; Ilavarasan, R. Comparative Preliminary Phytochemical and HPTLC Fingerprint Profile Studies of Two Cinnamon Species Commonly Used in ASU Formulations. Asian J. Res. Chem. 2018, 11, 344–350. [Google Scholar] [CrossRef]
- Senanayake, U.M.; Wijesekera, R.O.B. Chemistry of Cinnamon and Cassia. In Cinnamon and Cassia: The Genus Cinnamomum; Ravindran, P.N., Nirmal Babu, K., Shylaja, M., Eds.; CRC Press: Boca Raton, FL, USA, 2004; pp. 80–120. [Google Scholar]
- Chen, P.; Sun, J.; Ford, P. Differentiation of the Four Major Species of Cinnamons (C. burmannii, C. verum, C. cassia, and C. loureiroi) Using a Flow Injection Mass Spectrometric (FIMS) Fingerprinting Method. J. Agric. Food Chem. 2014, 62, 2516–2521. [Google Scholar] [CrossRef]
- Cantarelli, M.Á.; Moldes, C.A.; Marchevsky, E.J.; Azcarate, S.M.; Camiña, J.M. Low-Cost Analytic Method for the Identification of Cinnamon Adulteration. Microchem. J. 2020, 159, 105513. [Google Scholar] [CrossRef]
- Lixourgioti, P.; Goggin, K.A.; Zhao, X.; Murphy, D.J.; Van Ruth, S.; Koidis, A. Authentication of Cinnamon Spice Samples Using FT-IR Spectroscopy and Chemometric Classification. LWT-Food Sci. Technol. 2022, 154, 112760. [Google Scholar] [CrossRef]
- Cruz-Tirado, J.P.; Lima Brasil, Y.; Freitas Lima, A.; Alva Pretel, H.; Teixeira Godoy, H.; Barbin, D.; Siche, R. Rapid and Non-Destructive Cinnamon Authentication by NIR-Hyperspectral Imaging and Classification Chemometrics Tools. Spectrochim. Acta A 2023, 289, 122226. [Google Scholar] [CrossRef] [PubMed]
- Ghidotti, M.; Papoci, S.; Pietretti, D.; Ždiniaková, T.; De La Calle Guntiñas, M.B. Use of Elemental Profiles Determined by Energy-Dispersive X-ray Fluorescence and Multivariate Analyses to Detect Adulteration in Ceylon Cinnamon. Anal. Bioanal. Chem. 2023, 415, 5437–5449. [Google Scholar] [CrossRef]
- Holzgrabe, U.; Wawer, I.; Diehl, B. NMR Spectroscopy in Pharmaceutical Analysis, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 1–528. [Google Scholar]
- Ferri, E.; Galimberti, A.; Casiraghi, M.; Airoldi, C.; Ciaramelli, C.; Palmioli, A.; Mezzasalma, V.; Bruni, I.; Labra, M. Towards a Universal Approach Based on Omics Technologies for the Quality Control of Food. BioMed Res. Int. 2015, 2015, 9160375. [Google Scholar] [CrossRef]
- Sobolev, A.; Mannina, L.; Proietti, N.; Carradori, S.; Daglia, M.; Giusti, A.; Antiochia, R.; Capitani, D. Untargeted NMR-Based Methodology in the Study of Fruit Metabolites. Molecules 2015, 20, 4088–4108. [Google Scholar] [CrossRef]
- Hatzakis, E. Nuclear Magnetic Resonance (NMR) Spectroscopy in Food Science: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef] [PubMed]
- Burton, I.W.; Martinez Farina, C.F.; Ragupathy, S.; Arunachalam, T.; Newmaster, S.; Berrué, F. Quantitative NMR Methodology for the Authentication of Roasted Coffee and Prediction of Blends. J. Agric. Food Chem. 2020, 68, 14643–14651. [Google Scholar] [CrossRef] [PubMed]
- Simmler, C.; Napolitano, J.G.; McAlpine, J.B.; Chen, S.-N.; Pauli, G.F. Universal Quantitative NMR Analysis of Complex Natural Samples. Curr. Opin. Biotech. 2014, 25, 51–59. [Google Scholar] [CrossRef]
- Martinez-Farina, C.F.; Driscoll, S.; Wicks, C.; Burton, I.; Wentzell, P.D.; Berrué, F. Chemical Barcoding: A Nuclear-Magnetic-Resonance-Based Approach To Ensure the Quality and Safety of Natural Ingredients. J. Agric. Food Chem. 2019, 67, 7765–7774. [Google Scholar] [CrossRef] [PubMed]
- Remaud, G.; Debon, A.A.; Martin, Y.; Martin, G.G.; Martin, G.J. Authentication of Bitter Almond Oil and Cinnamon Oil: Application of the SNIF-NMR Method to Benzaldehyde. J. Agric. Food Chem. 1997, 45, 4042–4048. [Google Scholar] [CrossRef]
- Wu, N.; Balayssac, S.; Assemat, G.; Danoun, S.; Déjean, S.; Malet-Martino, M.; Gilard, V. Evaluation of Low-Field versus High-Field Proton NMR Spectroscopy for Quality Control of Cinnamon Samples. J. Food Compos. Anal. 2021, 96, 103706. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Rudaz, S.; Choi, Y.H.; Kim, H.K. Plant Metabolomics: From Holistic Data to Relevant Biomarkers. Curr. Med. Chem. 2013, 20, 1056–1090. [Google Scholar] [CrossRef]
- Ding, Y.; Wu, E.Q.; Liang, C.; Chen, J.; Tran, M.N.; Hong, C.H.; Jang, Y.; Park, K.L.; Bae, K.; Kim, Y.H.; et al. Discrimination of Cinnamon Bark and Cinnamon Twig Samples Sourced from Various Countries Using HPLC-Based Fingerprint Analysis. Food Chem. 2011, 127, 755–760. [Google Scholar] [CrossRef]
- Farag, M.A.; Labib, R.M.; Noleto, C.; Porzel, A.; Wessjohann, L.A. NMR Approach for the Authentication of 10 Cinnamon Spice Accessions Analyzed via Chemometric Tools. LWT-Food Sci. Technol. 2018, 90, 491–498. [Google Scholar] [CrossRef]
- Kojoma, M.; Kurihara, K.; Yamada, K.; Sekita, S.; Satake, M.; Iida, O. Genetic Identification of Cinnamon (Cinnamomum spp.) Based on the trnL-trnF Chloroplast DNA. Planta Med. 2002, 68, 94–96. [Google Scholar] [CrossRef]
- Chiou, S.-J.; Yen, J.-H.; Fang, C.-L.; Chen, H.-L.; Lin, T.-Y. Authentication of Medicinal Herbs Using PCR-Amplified ITS2 with Specific Primers. Planta Med. 2007, 73, 1421–1426. [Google Scholar] [CrossRef]
- Lee, S.-C.; Lee, C.-H.; Lin, M.-Y.; Ho, K.-Y. Genetic Identification of Cinnamomum Species Based on Partial Internal Transcribed Spacer 2 of Ribosomal DNA. J. Food Drug Anal. 2010, 18, 225–231. [Google Scholar] [CrossRef]
- Lee, S.-C.; Chiou, S.-J.; Yen, J.-H.; Lin, T.-Y.; Hsieh, K.-T.; Yang, J.-C. DNA Barcoding Cinnamomum osmophloeum Kaneh. Based on the Partial Non-Coding ITS2 Region of Ribosomal Genes. J. Food Drug Anal. 2010, 18, 128–135. [Google Scholar] [CrossRef]
- Swetha, V.P.; Parvathy, V.A.; Sheeja, T.E.; Sasikumar, B. DNA Barcoding for Discriminating the Economically Important Cinnamomum verum from Its Adulterants. Food Biotechnol. 2014, 28, 183–194. [Google Scholar] [CrossRef]
- Yang, P.; Hong, Z.; Shuang-jiao, M.; Wei, S.; Yong-hua, L.; Hui, Y. Authentication of Raw Material for Edible and Medicinal Cinnamon Based on Plastid Intergenic Region psbA-trnH. Chin. Pharm. J. 2015, 50, 1496–1499. [Google Scholar] [CrossRef]
- Soulange, J.G.; Ranghoo-Sanmukhiya, V.M.; Seeburrun, S.D. Tissue Culture and RAPD Analysis of Cinnamomum camphora and Cinnamomum verum. Biotechnology 2007, 6, 239–244. [Google Scholar] [CrossRef]
- Abeysinghe, P.D.; Wijesinghe, K.G.G.; Tachida, H.; Yoshda, T. Molecular Characterization of Cinnamon (Cinnamomum verum Presl) Accessions and Evaluation of Genetic Relatedness of Cinnamon Species in Sri Lanka Based on TrnL Intron Region, Intergenic Spacers Between trnT-trnL, trnL-trnF, trnH-psbA and nuclear ITS. Res. J. Agric. Biol. Sci. 2009, 5, 1079–1088. [Google Scholar]
- Bhau, B.S.; Gogoi, G.; Baruah, D.; Ahmed, R.; Hazarika, G.; Borah, B.; Gogoi, B.; Sarmah, D.K.; Nath, S.C.; Wann, S.B. Development of an Effective and Efficient DNA Isolation Method for Cinnamomum Species. Food Chem. 2015, 188, 264–270. [Google Scholar] [CrossRef]
- Doh, E.J.; Kim, J.-H.; Oh, S.E.; Lee, G. Identification and Monitoring of Korean Medicines Derived from Cinnamomum spp. by Using ITS and DNA Marker. Genes Genom. 2017, 39, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Shi, Y.; Sun, W.; Wu, L.; Xiong, C.; Zhu, Z.; Zhao, H.; Zhang, B.; Wang, C.; Liu, X. An Efficient DNA Barcoding Based Method for the Authentication and Adulteration Detection of the Powdered Natural Spices. Food Control 2019, 106, 106745. [Google Scholar] [CrossRef]
- Yang, B.-C.; Lee, M.-S.; Sun, F.-C.; Chao, H.-H.; Chang, W.-T.; Lin, M.-K.; Chen, H.-J.; Lee, M.-S. Rapid Identification of the Indigenous Medicinal Crop Cinnamomum osmophloeum from Various Adulterant Cinnamomum Species by DNA Polymorphism Analysis. Pharmacogn. Mag. 2020, 16, S64–S69. [Google Scholar] [CrossRef]
- Wang, Y.; Harrington, P.D.B.; Chen, P. Metabolomic Profiling and Comparison of Major Cinnamon Species Using UHPLC–HRMS. Anal. Bioanal. Chem. 2020, 412, 7669–7681. [Google Scholar] [CrossRef]
- Ragupathy, S.; Faller, A.C.; Shanmughanandhan, D.; Kesanakurti, P.; Shaanker, R.U.; Ravikanth, G.; Sathishkumar, R.; Mathivanan, N.; Song, J.; Han, J.; et al. Exploring DNA Quantity and Quality from Raw Materials to Botanical Extracts. Heliyon 2019, 5, e01935. [Google Scholar] [CrossRef]
- Faller, A.C.; Ragupathy, S.; Shanmughanandhan, D.; Zhang, Y.; Lu, Z.; Chang, P.; Swanson, G.; Newmaster, S.G. DNA Quality and Quantity Analysis of Camellia sinensis Through Processing from Fresh Leaves to a Green Tea Extract. J. AOAC Int. 2019, 102, 1798–1807. [Google Scholar] [CrossRef]
- Dodsworth, S. Genome Skimming for Next-Generation Biodiversity Analysis. Trends Plant Sci. 2015, 20, 525–527. [Google Scholar] [CrossRef]
- Bandaranayake, P.C.G.; Naranpanawa, N.; Chandrasekara, C.H.W.M.R.B.; Samarakoon, H.; Lokuge, S.; Jayasundara, S.; Bandaranayake, A.U.; Pushpakumara, D.K.N.G.; Wijesundara, D.S.A. Chloroplast Genome, Nuclear ITS Regions, Mitogenome Regions, and Skmer Analysis Resolved the Genetic Relationship among Cinnamomum Species in Sri Lanka. PLoS ONE 2023, 18, e0291763. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zheng, Y.; Liu, S.; Zhong, Y.; Wu, Y.; Li, J.; Xu, L.-A.; Xu, M. The Complete Chloroplast Genome of Cinnamomum camphora and Its Comparison with Related Lauraceae Species. PeerJ 2017, 5, e3820. [Google Scholar] [CrossRef]
- Li, P.; Jia, G.; Xin, G.; Cai, X. The Complete Chloroplast Genome of Cinnamomum camphora (L.) Presl., a Unique Economic Plant to China. Mitochondrial DNA B 2019, 4, 2511–2512. [Google Scholar] [CrossRef]
- Song, K.; He, M.; Yu, J.; Guan, Y.; Bai, Y.; Xin, S.; Cao, T. Characterization of the Chloroplast Genome of the Family Lauraceae Plant Species, Cinnamomum cassia. Mitochondrial DNA B 2019, 4, 3906–3907. [Google Scholar] [CrossRef]
- Yang, Y.; Song, Y.; Xin, P. The Chloroplast Genome of Aromatic Plants Cinnamomum burmanni (Lauraceae). Mitochondrial DNA B 2019, 4, 3616–3617. [Google Scholar] [CrossRef]
- Xie, P.; Lin, S.; Lai, Q.; Lian, H.; Chen, J.; Zhang, Q.; He, B. The Complete Plastid Genome of Chinese Cinnamon, Cinnamomum aromaticum Nees (Lauraceae). Mitochondrial DNA B 2019, 4, 3831–3833. [Google Scholar] [CrossRef]
- Zheng, Y.; Luo, Y.; Li, Y.; Wang, Y. The Complete Chloroplast Genome Sequence of Cinnamomum longipetiolatum. Mitochondrial DNA B 2020, 5, 198–199. [Google Scholar] [CrossRef]
- Jiang, S.; Liu, Y.; Cui, S.; Gui, L.; Huang, J. The Complete Chloroplast Genome Analysis Of Cinnamomum cassia Presl. Bangladesh J. Bot. 2022, 51, 51–55. [Google Scholar] [CrossRef]
- Li, F.; Huang, S.; Mei, Y.; Wu, B.; Hou, Z.; Zhan, P.; Hou, Z.; Huang, W.; Zhao, J.; Wang, J. Genome Assembly Provided New Insights into the Cinnamomum burmannii Evolution and D-Borneol Biosynthesis Differences between Chemotypes. Ind. Crops Prod. 2022, 186, 115181. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Shanmughanandhan, D.; Kesanakurti, P.; Shehata, H.; Faller, A.; Noce, I.D.; Lee, J.Y.; Rudzinski, P.; Lu, Z.; Zhang, Y.; et al. Recommendations for Validation of Real-Time PCR Methods for Molecular Diagnostic Identification of Botanicals. J. AOAC Int. 2019, 102, 1767–1773. [Google Scholar] [CrossRef]
- Pitaro, M.; Croce, N.; Gallo, V.; Arienzo, A.; Salvatore, G.; Antonini, G. Coumarin-Induced Hepatotoxicity: A Narrative Review. Molecules 2022, 27, 9063. [Google Scholar] [CrossRef] [PubMed]
- Journal, T.E. Opinion of the Scientific Panel on Food Additives, Flavourings, Processing Aids and Materials in Contact with Food (AFC) Related to Coumarin. EFSA J. 2004, 2, 104. [Google Scholar] [CrossRef]
- Eugenol (Clove Oil). LiverTox: Clinical and Research Information on Drug-Induced Liver Injury; [Internet], [Updated 28 October 2019]; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2012; Available online: https://www.ncbi.nlm.ni (accessed on 28 November 2023).
- Coissac, E.; Hollingsworth, P.M.; Lavergne, S.; Taberlet, P. From Barcodes to Genomes: Extending the Concept of DNA Barcoding. Mol. Ecol. 2016, 25, 1423–1428. [Google Scholar] [CrossRef]
- Zeng, C.-X.; Hollingsworth, P.M.; Yang, J.; He, Z.-S.; Zhang, Z.-R.; Li, D.-Z.; Yang, J.-B. Genome Skimming Herbarium Specimens for DNA Barcoding and Phylogenomics. Plant Methods 2018, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- Alsos, I.G.; Lavergne, S.; Merkel, M.K.F.; Boleda, M.; Lammers, Y.; Alberti, A.; Pouchon, C.; Denoeud, F.; Pitelkova, I.; Pușcaș, M.; et al. The Treasure Vault Can Be Opened: Large-Scale Genome Skimming Works Well Using Herbarium and Silica Gel Dried Material. Plants 2020, 9, 432. [Google Scholar] [CrossRef]
- Nevill, P.G.; Zhong, X.; Tonti-Filippini, J.; Byrne, M.; Hislop, M.; Thiele, K.; Van Leeuwen, S.; Boykin, L.M.; Small, I. Large Scale Genome Skimming from Herbarium Material for Accurate Plant Identification and Phylogenomics. Plant Methods 2020, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Kesanakurti, P.; Thirugnanasambandam, A.; Ragupathy, S.; Newmaster, S.G. Genome Skimming and NMR Chemical Fingerprinting Provide Quality Assurance Biotechnology to Validate Sarsaparilla Identity and Purity. Sci. Rep. 2020, 10, 19192. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Du, Y.; Shen, C.; Li, R.; Lee, J.; Li, P. The Complete Chloroplast Genome of Papaver setigerum and Comparative Analyses in Papaveraceae. Genet. Mol. Biol. 2020, 43, e20190272. [Google Scholar] [CrossRef] [PubMed]
- Twyford, A.D.; Ness, R.W. Strategies for Complete Plastid Genome Sequencing. Mol. Ecol. Resour. 2017, 17, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Z.; Chen, S.-Y. Genome Skimming Reveals the Complete Chloroplast Genome of Ampelocalamus naibunensis (Poaceae: Bambusoideae: Arundinarieae) with Phylogenomic Implication. Mitochondrial DNA B 2016, 1, 635–637. [Google Scholar] [CrossRef]
- Grazina, L.; Amaral, J.S.; Mafra, I. Botanical Origin Authentication of Dietary Supplements by DNA-based Approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1080–1109. [Google Scholar] [CrossRef]
- Costa, J.; Amaral, J.S.; Fernandes, T.J.R.; Batista, A.; Oliveira, M.B.P.P.; Mafra, I. DNA Extraction from Plant Food Supplements: Influence of Different Pharmaceutical Excipients. Mol. Cell. Probes 2015, 29, 473–478. [Google Scholar] [CrossRef]
- Parveen, I.; Gafner, S.; Techen, N.; Murch, S.; Khan, I. DNA Barcoding for the Identification of Botanicals in Herbal Medicine and Dietary Supplements: Strengths and Limitations. Planta Med. 2016, 82, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Gangwar, H.; Gahlaut, V.; Chauhan, R.; Singh, S.; Jaiswal, V. Development of Species-Specific ISSR-Derived SCAR Marker for Early Discrimination between Cinnamomum verum and Cinnamomum cassia. Mol. Biol. Rep. 2023, 50, 6311–6321. [Google Scholar] [CrossRef] [PubMed]
- Paran, I.; Michelmore, R.W. Development of Reliable PCR-Based Markers Linked to Downy Mildew Resistance Genes in Lettuce. Theor. Appl. Genet. 1993, 85, 985–993. [Google Scholar] [CrossRef]
- Kiran, U.; Khan, S.; Mirza, K.J.; Ram, M.; Abdin, M.Z. SCAR Markers: A Potential Tool for Authentication of Herbal Drugs. Fitoterapia 2010, 81, 969–976. [Google Scholar] [CrossRef]
- Ganie, S.H.; Upadhyay, P.; Das, S.; Prasad Sharma, M. Authentication of Medicinal Plants by DNA Markers. Plant Gene 2015, 4, 83–99. [Google Scholar] [CrossRef]
- Ün, İ.; Ok, S. Analysis of Olive Oil for Authentication and Shelf Life Determination. J. Food Sci. Technol. 2018, 55, 2476–2487. [Google Scholar] [CrossRef]
- Hoppenreijs, L.J.G.; Berton-Carabin, C.C.; Dubbelboer, A.; Hennebelle, M. Evaluation of Oxygen Partial Pressure, Temperature and Stripping of Antioxidants for Accelerated Shelf-Life Testing of Oil Blends Using 1H NMR. Food Res. Int. 2021, 147, 110555. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.C.; De Oliveira, V.M.; Martins, F.T.; Lião, L.M.; Ferri, P.H.; Queiroz Júnior, L.H.K. Predicting Chemical Shelf Life of Mozzarella Cheese Submitted to Irregular Refrigeration Practices by Nuclear Magnetic Resonance Spectroscopy and Statistical Analysis. J. Food Compos. Anal. 2022, 105, 104229. [Google Scholar] [CrossRef]
- Wei, F.; Furihata, K.; Koda, M.; Hu, F.; Kato, R.; Miyakawa, T.; Tanokura, M. 13 C NMR-Based Metabolomics for the Classification of Green Coffee Beans According to Variety and Origin. J. Agric. Food Chem. 2012, 60, 10118–10125. [Google Scholar] [CrossRef] [PubMed]
- Consonni, R.; Cagliani, L.R.; Cogliati, C. NMR Based Geographical Characterization of Roasted Coffee. Talanta 2012, 88, 420–426. [Google Scholar] [CrossRef]
- Caligiani, A.; Palla, L.; Acquotti, D.; Marseglia, A.; Palla, G. Application of 1H NMR for the Characterisation of Cocoa Beans of Different Geographical Origins and Fermentation Levels. Food Chem. 2014, 157, 94–99. [Google Scholar] [CrossRef]
- Marseglia, A.; Acquotti, D.; Consonni, R.; Cagliani, L.R.; Palla, G.; Caligiani, A. HR MAS 1H NMR and Chemometrics as Useful Tool to Assess the Geographical Origin of Cocoa Beans—Comparison with HR 1H NMR. Food Res. Int. 2016, 85, 273–281. [Google Scholar] [CrossRef]
- Longobardi, F.; Ventrella, A.; Napoli, C.; Humpfer, E.; Schütz, B.; Schäfer, H.; Kontominas, M.G.; Sacco, A. Classification of Olive Oils According to Geographical Origin by Using 1H NMR Fingerprinting Combined with Multivariate Analysis. Food Chem. 2012, 130, 177–183. [Google Scholar] [CrossRef]
- Calò, F.; Girelli, C.R.; Wang, S.C.; Fanizzi, F.P. Geographical Origin Assessment of Extra Virgin Olive Oil via NMR and MS Combined with Chemometrics as Analytical Approaches. Foods 2022, 11, 113. [Google Scholar] [CrossRef]
- Viskić, M.; Bandić, L.M.; Korenika, A.-M.J.; Jeromel, A. NMR in the Service of Wine Differentiation. Foods 2021, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Masetti, O.; Sorbo, A.; Nisini, L. NMR Tracing of Food Geographical Origin: The Impact of Seasonality, Cultivar and Production Year on Data Analysis. Separations 2021, 8, 230. [Google Scholar] [CrossRef]
- Castro, R.C.; Ribeiro, D.S.M.; Santos, J.L.M.; Páscoa, R.N.M.J. Authentication/Discrimination, Identification and Quantification of Cinnamon Adulterants Using NIR Spectroscopy and Different Chemometric Tools: A Tutorial to Deal with Counterfeit Samples. Food Control 2023, 147, 109619. [Google Scholar] [CrossRef]
- Abreu, A.C.; Fernández, I. NMR Metabolomics Applied on the Discrimination of Variables Influencing Tomato (Solanum lycopersicum). Molecules 2020, 25, 3738. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R. Quality Assessment of Traditional Food by NMR Analysis. Food Control 2022, 142, 109226. [Google Scholar] [CrossRef]
- Ji, H.; Pu, D.; Yan, W.; Zhang, Q.; Zuo, M.; Zhang, Y. Recent Advances and Application of Machine Learning in Food Flavor Prediction and Regulation. Trends Food Sci. Tech. 2023, 138, 738–751. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Kuzmina, M.L.; Newmaster, S.G.; Hollingsworth, P.M. DNA Barcoding Methods for Land Plants. In DNA Barcodes: Methods and Protocols; Kress, W.J., Erickson, D.L., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 223–252. [Google Scholar]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Walker, J.F.; Jansen, R.K.; Zanis, M.J.; Emery, N.C. Sources of Inversion Variation in the Small Single Copy (SSC) Region of Chloroplast Genomes. Am. J. Bot. 2015, 102, 1751–1752. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-Based Metabolomic Analysis of Plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef]
- Manual, B. ERETIC2 User’s Guide-Preliminary; Bruker: Billerica, MA, USA, 2012. [Google Scholar]
- Elyashberg, M. Identification and Structure Elucidation by NMR Spectroscopy. TrAC-Trends Anal. Chem. 2015, 69, 88–97. [Google Scholar] [CrossRef]
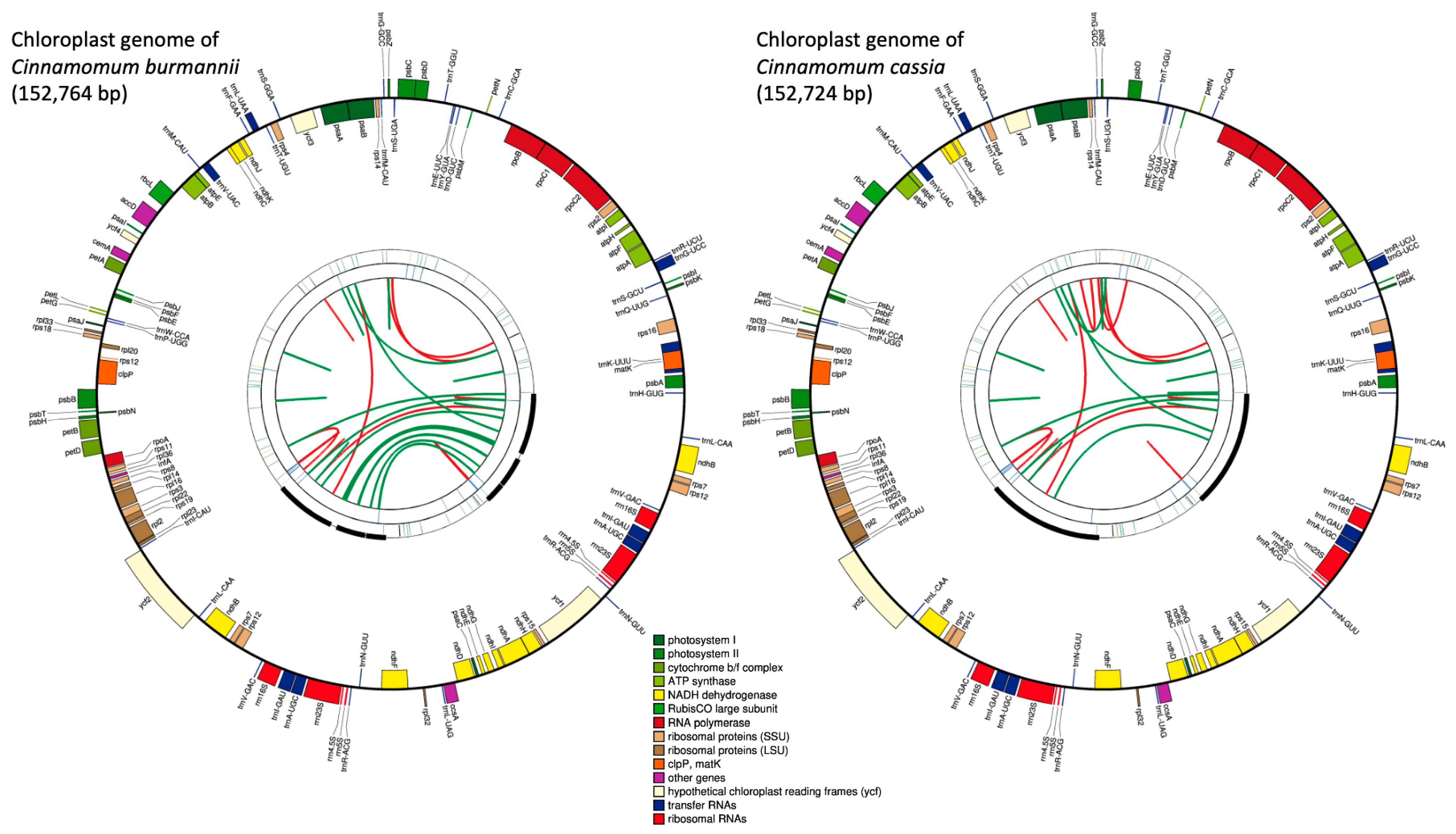


| Cinnamomum burmanii Exon/Intron Locations | ||||||||
| Gene | Strand | Start | End | ExonI | IntronI | ExonII | IntronII | ExonIII |
| trnK-UUU | − | 1786 | 4370 | 37 | 2513 | 35 | ||
| rps16 | − | 5193 | 6306 | 40 | 844 | 230 | ||
| trnG-UCC | + | 10,328 | 11,149 | 23 | 751 | 48 | ||
| atpF | − | 13,053 | 14,333 | 145 | 726 | 410 | ||
| rpoC1 | − | 21,858 | 24,650 | 453 | 720 | 1620 | ||
| ycf3 | − | 44,714 | 46,685 | 124 | 734 | 230 | 731 | 153 |
| trnL-UAA | + | 49,428 | 49,991 | 35 | 479 | 50 | ||
| trnV-UAC | − | 54,470 | 55,132 | 39 | 589 | 35 | ||
| clpP | − | 73,555 | 75,592 | 71 | 773 | 294 | 656 | 244 |
| petB | + | 78,470 | 79,905 | 6 | 788 | 642 | ||
| petD | + | 80,103 | 81,301 | 8 | 716 | 475 | ||
| rpl16 | − | 84,735 | 86,110 | 9 | 971 | 396 | ||
| rpl2 | − | 87,837 | 89,328 | 392 | 670 | 430 | ||
| ndhB | − | 97,853 | 100,033 | 721 | 702 | 758 | ||
| trnI-GAU | + | 105,605 | 106,620 | 37 | 944 | 35 | ||
| trnA-UGC | + | 106,685 | 107,555 | 38 | 798 | 35 | ||
| ndhA | − | 124,323 | 126,541 | 553 | 1127 | 539 | ||
| trnA-UGC | − | 138,914 | 139,784 | 38 | 798 | 35 | ||
| trnI-GAU | − | 139,849 | 140,864 | 37 | 944 | 35 | ||
| ndhB | + | 146,436 | 148,616 | 721 | 702 | 758 | ||
| Cinnamomum cassia Exon/Intron Locations | ||||||||
| Gene | Strand | Start | End | ExonI | IntronI | ExonII | IntronII | ExonIII |
| trnK-UUU | − | 1789 | 4373 | 37 | 2513 | 35 | ||
| rps16 | − | 5189 | 6302 | 40 | 844 | 230 | ||
| trnG-UCC | + | 10,317 | 11,138 | 23 | 751 | 48 | ||
| atpF | − | 13,042 | 14,324 | 145 | 728 | 410 | ||
| rpoC1 | − | 21,849 | 24,642 | 453 | 721 | 1620 | ||
| ycf3 | − | 44,706 | 46,677 | 124 | 735 | 230 | 730 | 153 |
| trnL-UAA | + | 49,419 | 49,982 | 35 | 479 | 50 | ||
| trnV-UAC | − | 54,461 | 55,124 | 39 | 590 | 35 | ||
| clpP | − | 73,541 | 75,576 | 71 | 772 | 294 | 655 | 244 |
| petB | + | 78,454 | 79,888 | 6 | 787 | 642 | ||
| petD | + | 80,086 | 81,284 | 8 | 716 | 475 | ||
| rpl16 | − | 84,720 | 86,094 | 9 | 970 | 396 | ||
| rpl2 | − | 87,818 | 89,309 | 392 | 670 | 430 | ||
| ndhB | − | 97,833 | 100,013 | 721 | 702 | 758 | ||
| trnI-GAU | + | 105,577 | 106,592 | 37 | 944 | 35 | ||
| trnA-UGC | + | 106,657 | 107,527 | 38 | 798 | 35 | ||
| ndhA | − | 124,295 | 126,516 | 553 | 1130 | 539 | ||
| trnA-UGC | − | 138,882 | 139,752 | 38 | 798 | 35 | ||
| trnI-GAU | − | 139,817 | 140,832 | 37 | 944 | 35 | ||
| ndhB | + | 146,396 | 148,576 | 721 | 702 | 758 | ||
| Sample_ID | Cinnamaldehyde | Coumarin | Methoxy Cinnamaldehyde | Cinnamic Acid | Benzoic Acid | Methyl Salicylate | Quinic Acid | Eugenol | α-Glucose | β-Glucose | Fructose | Formic Acid | Choline | Shikimic ACID | Succinic Acid | Alanine | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | |
| Cin_veru_01 | 9.57 | 0.05 | 0.8 | 0.04 | 10.43 | 0.07 | 3.95 | 0.03 | 0.09 | 0 | 0.7 | 0.01 | 9.16 | 0.04 | 6.44 | 0.04 | 3.07 | 0.11 | 4.74 | 0.02 | 3.94 | 0 | 0.06 | 0 | 0.55 | 0 | 1.95 | 0.02 | 3.4 | 0.17 | 1.6 | 0.02 |
| Cin_veru_02 | 0.13 | 0 | ND | ND | 0.32 | 0 | 0.11 | 0 | ND | ND | ND | ND | 0.18 | 0 | 0.16 | 0 | 0.08 | 0 | 0.11 | 0 | 0.08 | 0 | ND | ND | ND | ND | 0.12 | 0 | 0.13 | 0.07 | 0.18 | 0 |
| Cin_veru_03 | 1.14 | 0.01 | 0.17 | 0.01 | 0.8 | 0.05 | 0.3 | 0.02 | ND | ND | 0.12 | 0.01 | 0.58 | 0.03 | 0.7 | 0.04 | 0.21 | 0 | 0.35 | 0.02 | 0.24 | 0 | ND | ND | ND | ND | 0.25 | 0.01 | ND | ND | 0.22 | 0.01 |
| Cin_cass_04 | 2.88 | 0.05 | 0.79 | 0.02 | 2.03 | 0.04 | 0.72 | 0.02 | ND | ND | 0.13 | 0 | 2.51 | 0.06 | 2.02 | 0.04 | 0.79 | 0.02 | 1.29 | 0.03 | 1 | 0 | ND | ND | ND | ND | 0.34 | 0.01 | ND | ND | 0.21 | 0.01 |
| Cin_burm_05 | 1.61 | 0.05 | 0.58 | 0.01 | 1.79 | 0.03 | 0.62 | 0.01 | ND | ND | 0.12 | 0 | 2.03 | 0.03 | 1.54 | 0.02 | 0.75 | 0.02 | 1.1 | 0.02 | 0.83 | 0 | ND | ND | 0.06 | 0 | 0.35 | 0.01 | ND | ND | 0.27 | 0.01 |
| Cin_cass_06 | ND | ND | 0.09 | 0.02 | 0.98 | 0.22 | 0.16 | 0.04 | ND | ND | ND | ND | 0.76 | 0.16 | 0.42 | 0.1 | 0.39 | 0.05 | 0.51 | 0.1 | 0.3 | 0.1 | ND | ND | 0.07 | 0 | 0.11 | 0.03 | ND | ND | ND | ND |
| Cin_cass_07 | ND | ND | ND | ND | 1.79 | 0.07 | 0.05 | 0 | ND | ND | 0.16 | 0.01 | 0.34 | 0.02 | 0.09 | 0 | 1.93 | 0.21 | 0.15 | 0 | 0.14 | 0 | ND | ND | 0.14 | 0 | ND | ND | ND | ND | ND | ND |
| Cin_veru_08 | 0.15 | 0.01 | 0.07 | 0.01 | 0.36 | 0.03 | 0.12 | 0.01 | ND | ND | ND | ND | 0.2 | 0.02 | 0.18 | 0.02 | 0.09 | 0.01 | 0.13 | 0.01 | 0.09 | 0 | ND | ND | ND | ND | 0.13 | 0.01 | 0.11 | 0.01 | 0.2 | 0.02 |
| Cin_burm_09 | 0.21 | 0.02 | 0.15 | 0.01 | 0.48 | 0.05 | 0.25 | 0.02 | ND | ND | ND | ND | 0.61 | 0.07 | 0.51 | 0.06 | 0.23 | 0.01 | 0.33 | 0.03 | 0.23 | 0 | ND | ND | 0.31 | 0.1 | 0.1 | 0.02 | 0.48 | 0.06 | 0.06 | 0.01 |
| Cin_veru_10 | ND | ND | 0.11 | 0 | 0.74 | 0.01 | 0.19 | 0.01 | ND | ND | 0.06 | 0 | 0.66 | 0.01 | 0.25 | 0 | 0.25 | 0.01 | 0.33 | 0.01 | 0.25 | 0 | ND | ND | 0.4 | 0 | 0.16 | 0.01 | 0.65 | 0.47 | 0.11 | 0 |
| Cin_cass_11 | ND | ND | 1.13 | 0.02 | 32.64 | 0.38 | 2.59 | 0.04 | ND | ND | 1.81 | 0.03 | 27.46 | 0.36 | 3.59 | 0.05 | 11.46 | 0.07 | 14.89 | 0.16 | 10.67 | 0.1 | ND | ND | 1.84 | 0.1 | 1.46 | 0.02 | 0.09 | 0 | 0.54 | 0.01 |
| Cin_cass_12 | 24.63 | 0.14 | 6.94 | 0.04 | 15.76 | 0.01 | 6.37 | 0.02 | 0.08 | 0 | 1.06 | 0.01 | 17.89 | 0.06 | 14.97 | 0.06 | 5.84 | 0.25 | 9.23 | 0.05 | 7.1 | 0 | 0.07 | 0 | 0.35 | 0 | 2.82 | 0.02 | 0.12 | 0 | 1.79 | 0.01 |
| Cin_veru_13 | 1.92 | 0.23 | 0.3 | 0.02 | 16.52 | 0.09 | 4.33 | 0.05 | 0.88 | 0.01 | 1.43 | 0.03 | 6.95 | 0.04 | 3.53 | 0.04 | 2.34 | 0.09 | 3.81 | 0.02 | 3.05 | 0 | 0.09 | 0 | 0.44 | 0 | 4.7 | 0.05 | 0.35 | 0 | 6.09 | 0.05 |
| Cin_veru_14 | 1.16 | 0.21 | 0.65 | 0.05 | 6.44 | 0.11 | 0.79 | 0.05 | 2.62 | 0.21 | 0.06 | 0 | 0.51 | 0.03 | 20.93 | 8.3 | 2.98 | 0.29 | 0.71 | 0.05 | 0.34 | 0.1 | ND | ND | 0.32 | 0 | 0.39 | 0.02 | 0.3 | 0.01 | 5.98 | 0.47 |
| Cin_cass_15 | 23.86 | 0.64 | 2.6 | 0.09 | 10.86 | 0.29 | 3.87 | 0.11 | ND | ND | 4.55 | 0.11 | 10.23 | 0.35 | 6.7 | 0.19 | 4.33 | 0.38 | 4.86 | 0.19 | 3.55 | 0.1 | ND | ND | 0.39 | 0 | 2.76 | 0.09 | 1.03 | 0.11 | 4.06 | 0.12 |
| Cin_veru_16 | ND | ND | ND | ND | 5.56 | 0.04 | ND | ND | ND | ND | 0.19 | 0 | 0.36 | 0.01 | 0.06 | 0 | 1.03 | 0.04 | 0.8 | 0.01 | 0.16 | 0 | ND | ND | 0.38 | 0 | ND | ND | ND | ND | ND | ND |
| Cin_cass_17 | ND | ND | 0.15 | 0.01 | 4.73 | 0.36 | 0.35 | 0.03 | ND | ND | 0.25 | 0.02 | 3.82 | 0.29 | 0.49 | 0.04 | 1.62 | 0.15 | 2.12 | 0.14 | 1.54 | 0.1 | ND | ND | 0.16 | 0 | 0.18 | 0.02 | 0.14 | 0.06 | 0.07 | 0 |
| Cin_cass_18 | 0.27 | 0.01 | 0.16 | 0 | 0.58 | 0 | 0.23 | 0 | ND | ND | ND | ND | 0.64 | 0 | 0.39 | 0 | 0.18 | 0 | 0.32 | 0 | 0.26 | 0 | ND | ND | 0.06 | 0 | 0.09 | 0 | 0.06 | 0 | 0.06 | 0 |
| Cin_cass_19 | 0.3 | 0 | 0.16 | 0 | 0.56 | 0 | 0.23 | 0 | ND | ND | ND | ND | 0.6 | 0.01 | 0.45 | 0 | 0.17 | 0 | 0.3 | 0 | 0.24 | 0 | ND | ND | 0.06 | 0 | 0.1 | 0 | 0.07 | 0.01 | 0.07 | 0 |
| Cin_cass_20 | 0.35 | 0 | 9.2 | 0.09 | 28.97 | 0.3 | 13.19 | 0.13 | 0.4 | 0.01 | 2.38 | 0.02 | 18.95 | 0.17 | 10.89 | 0.09 | 5.19 | 0.03 | 9.46 | 0.08 | 8.44 | 0.1 | 0.15 | 0 | 0.53 | 0 | 8.42 | 0.08 | 0.68 | 0.01 | 11.31 | 0.12 |
| Cin_cass_21 | 1.97 | 0.03 | 3.72 | 0.01 | 12.04 | 0.03 | 3.76 | 0.02 | 0.13 | 0 | 0.89 | 0 | 6.6 | 0.02 | 2.68 | 0 | 2.38 | 0.04 | 3.59 | 0.02 | 2.82 | 0 | 0.13 | 0 | 0.52 | 0 | 2.14 | 0 | 0.17 | 0 | 2.59 | 0.01 |
| Cin_veru_22 | ND | ND | ND | ND | 4.95 | 0.02 | ND | ND | ND | ND | 0.16 | 0 | 0.33 | 0 | ND | ND | 0.85 | 0.01 | 0.75 | 0.01 | 0.15 | 0 | ND | ND | 0.33 | 0 | ND | ND | ND | ND | ND | ND |
| Cin_veru_23 | ND | ND | ND | ND | 5.04 | 0.05 | ND | ND | ND | ND | 0.16 | 0 | 0.33 | 0.01 | 0.06 | 0 | 0.91 | 0.04 | 0.77 | 0.01 | 0.15 | 0 | ND | ND | 0.33 | 0 | 0.05 | 0.01 | 0.09 | 0.01 | ND | ND |
| Cin_veru_24 | 1.28 | 0.05 | 0.51 | 0.02 | 2.66 | 0.12 | 0.92 | 0.03 | 0.16 | 0.01 | 0.3 | 0.01 | 1.48 | 0.06 | 1.28 | 0.05 | 0.72 | 0.03 | 0.96 | 0.03 | 0.63 | 0 | 0.13 | 0 | 0.37 | 0 | 0.94 | 0.03 | 1.15 | 0.07 | 1.48 | 0.05 |
| Cin_veru_25 | 5.23 | 0.12 | 0.5 | 0.11 | 20.81 | 0.88 | 4.96 | 0.15 | 0.14 | 0.01 | 2.63 | 0.07 | 19.13 | 0.78 | 6.79 | 0.16 | 6.78 | 0.18 | 8.91 | 0.36 | 7.5 | 0.3 | 0.3 | 0.01 | 0.95 | 0.1 | 4.4 | 0.12 | 1.44 | 0.05 | 3.23 | 0.1 |
| Cin_veru_26 | 14.98 | 0.2 | 0.66 | 0.03 | 27.15 | 0.37 | 7.52 | 0.05 | 0.26 | 0 | 4.03 | 0.02 | 19.16 | 0.17 | 6.07 | 0.01 | 7.48 | 0.03 | 9.36 | 0.11 | 7.51 | 0.1 | 0.25 | 0 | 0.67 | 0 | 6.48 | 0.03 | 1.16 | 0.36 | 6.55 | 0.07 |
| Cin_veru_27 | 2.99 | 0.12 | 0.97 | 0.07 | 13.58 | 0.42 | 3.72 | 0.12 | 0.77 | 0.02 | 1.54 | 0.05 | 6.1 | 0.19 | 3 | 0.09 | 1.88 | 0.06 | 3.28 | 0.11 | 2.7 | 0.1 | 0.07 | 0 | 0.43 | 0 | 4.06 | 0.11 | 0.31 | 0.01 | 5.35 | 0.12 |
| Cin_veru_28 | 8.75 | 0.04 | 0.25 | 0.01 | 6.28 | 0.02 | 2.17 | 0.02 | 0.29 | 0 | 0.92 | 0.01 | 5.01 | 0.03 | 5.29 | 0.05 | 1.88 | 0.05 | 2.91 | 0.03 | 2.01 | 0 | ND | ND | 0.45 | 0 | 1.95 | 0.03 | 0.08 | 0 | 1.46 | 0.02 |
| Cin_veru_29 | 0.35 | 0 | 0.58 | 0.05 | 61.79 | 1.21 | 42.99 | 1.15 | 0.4 | 0.01 | 6.39 | 0.17 | 18.65 | 0.4 | 2.59 | 0.06 | 12.1 | 0.4 | 7.75 | 0.14 | 9.34 | 0.2 | 0.28 | 0.01 | 0.78 | 0 | 5.57 | 0.15 | 0.55 | 0.27 | 1.48 | 0.05 |
| Cin_burm_30 | 18.81 | 0.11 | 4.75 | 0.05 | 8.55 | 0.07 | 3.72 | 0.04 | 0.07 | 0 | 1.19 | 0 | 8.38 | 0.05 | 6.3 | 0.06 | 3.43 | 0.08 | 4.29 | 0.02 | 3.29 | 0 | 0.06 | 0 | 0.43 | 0 | 2.29 | 0.03 | 0.36 | 0.33 | 2.55 | 0.02 |
| Cin_burm_31 | 21.99 | 0.2 | 5.63 | 0.07 | 11.42 | 0.15 | 4.67 | 0.06 | 0.08 | 0 | 0.83 | 0.01 | 12.3 | 0.15 | 4.7 | 0.12 | 4.42 | 0.08 | 6.47 | 0.06 | 5.01 | 0.1 | 0.07 | 0 | 0.41 | 0 | 2.12 | 0.03 | 0.43 | 0.14 | 1.73 | 0.02 |
| Cin_burm_32 | 21.77 | 0.08 | 5.73 | 0.04 | 12.64 | 0.05 | 5.19 | 0.04 | 0.08 | 0 | 0.87 | 0 | 14.14 | 0.05 | 1.33 | 0.09 | 5.08 | 0.12 | 7.39 | 0.02 | 5.68 | 0 | 0.08 | 0 | 0.39 | 0 | 2.41 | 0.02 | 1.01 | 0.2 | 1.86 | 0.02 |
| Cin_burm_33 | 22.57 | 0.15 | 5.87 | 0.03 | 12.34 | 0.18 | 4.89 | 0.03 | 0.08 | 0 | 0.86 | 0.01 | 13.34 | 0.18 | 1.03 | 0.06 | 4.41 | 0.08 | 6.99 | 0.06 | 5.43 | 0.1 | 0.08 | 0 | 0.41 | 0 | 2.19 | 0.02 | 0.48 | 0.27 | 1.76 | 0.01 |
| Cin_burm_34 | 18.98 | 0.25 | 4.6 | 0 | 7.95 | 0.07 | 3.4 | 0.05 | 0.06 | 0 | 1.14 | 0 | 7.64 | 0.07 | 5.5 | 0.01 | 3.29 | 0.07 | 3.88 | 0.05 | 2.96 | 0 | ND | ND | 0.48 | 0 | 3.58 | 0.01 | 0.97 | 0.39 | 2.98 | 0.01 |
| Cin_veru_35 | ND | ND | ND | ND | 3.17 | 0.09 | ND | ND | ND | ND | 0.12 | 0 | 0.3 | 0.01 | ND | ND | 0.67 | 0.01 | 0.53 | 0.02 | 0.13 | 0 | ND | ND | 0.36 | 0 | 0.06 | 0 | 1.56 | 1.32 | 0.06 | 0 |
| Cin_veru_36 | ND | ND | ND | ND | 57.62 | 0.88 | 40.41 | 0.63 | 0.36 | 0 | 5.82 | 0.12 | 17.22 | 0.25 | 2.38 | 0.04 | 11.5 | 0.02 | 7.44 | 0.08 | 8.58 | 0.1 | 0.25 | 0 | 0.77 | 0 | 5.17 | 0.06 | 0.34 | 0.13 | 1.43 | 0.02 |
| Cin_veru_37 | 13.47 | 1.23 | 1.12 | 0.09 | 3.2 | 0.24 | 1.92 | 0.14 | 0.17 | 0.01 | 0.72 | 0.07 | 3.86 | 0.3 | 6.68 | 0.51 | 1.46 | 0.06 | 2.34 | 0.18 | 1.44 | 0.1 | ND | ND | 0.29 | 0 | 2.11 | 0.13 | 0.32 | 0.19 | 1.63 | 0.09 |
| Cin_veru_38 | ND | ND | ND | ND | 2.1 | 0.11 | ND | ND | ND | ND | 0.08 | 0 | 0.22 | 0 | ND | ND | 0.4 | 0.02 | 0.39 | 0.01 | 0.1 | 0 | ND | ND | 0.38 | 0 | ND | ND | 0.19 | 0.09 | 0.07 | 0.02 |
| Cin_veru_39 | 0.31 | 0.01 | 0.49 | 0.17 | 65.4 | 0.51 | 36.83 | 0.34 | 0.35 | 0 | 6.21 | 0.05 | 19.86 | 0.14 | 4.69 | 0.06 | 13.86 | 0.24 | 9.34 | 0.06 | 9.63 | 0.1 | 0.24 | 0 | 0.8 | 0 | 5.49 | 0.06 | 0.78 | 0.06 | 1.41 | 0.01 |
| Cin_cass_40 | ND | ND | 0.09 | 0.03 | 4.49 | 0.53 | 1.65 | 0.82 | ND | ND | 0.17 | 0.02 | 1.21 | 0.17 | 0.32 | 0.03 | 1.18 | 0.17 | 1.14 | 0.15 | 0.49 | 0.1 | ND | ND | 0.48 | 0 | 0.11 | 0.02 | 0.36 | 0.12 | 0.1 | 0.02 |
| Cin_veru_41 | ND | ND | ND | ND | 2.01 | 0.04 | ND | ND | ND | ND | 0.08 | 0 | 0.21 | 0.01 | ND | ND | 0.43 | 0.01 | 0.35 | 0.01 | 0.09 | 0 | ND | ND | 0.32 | 0 | ND | ND | 0.69 | 0.03 | 0.06 | 0 |
| Cin_veru_42 | ND | ND | ND | ND | 1.91 | 0.03 | ND | ND | ND | ND | 0.07 | 0 | 0.19 | 0.01 | ND | ND | 0.36 | 0 | 0.35 | 0 | 0.09 | 0 | ND | ND | 0.37 | 0 | ND | ND | ND | ND | 0.06 | 0 |
| Cin_veru_43 | ND | ND | ND | ND | 1.91 | 0.02 | ND | ND | ND | ND | 0.07 | 0 | 0.19 | 0 | ND | ND | 0.36 | 0 | 0.34 | 0.01 | 0.09 | 0 | ND | ND | 0.35 | 0 | ND | ND | ND | ND | ND | ND |
| Cin_veru_44 | ND | ND | ND | ND | 2.2 | 0.02 | ND | ND | ND | ND | 0.08 | 0 | 0.22 | 0 | ND | ND | 0.42 | 0 | 0.4 | 0.01 | 0.09 | 0 | ND | ND | 0.24 | 0 | ND | ND | 0.53 | 0.1 | ND | ND |
| Cin_veru_45 | 9.61 | 0.14 | 0.21 | 0.02 | 6.04 | 0.12 | 2.19 | 0.04 | 0.23 | 0 | 0.9 | 0.02 | 5.7 | 0.08 | 5.37 | 0.07 | 2.05 | 0.05 | 3.14 | 0.04 | 2.22 | 0 | ND | ND | 0.41 | 0 | 2.07 | 0.03 | 0.4 | 0.02 | 1.57 | 0.02 |
| Cin_veru_46 | 24.95 | 0.37 | 0.21 | 0.06 | 9.15 | 0.2 | 5.22 | 0.11 | 0.09 | 0 | 2.1 | 0.04 | 7.08 | 0.16 | 4.06 | 0.08 | 3.4 | 0.07 | 3.76 | 0.07 | 2.76 | 0.1 | 0.06 | 0 | 0.4 | 0 | 2.68 | 0.06 | 0.4 | 0.01 | 3.02 | 0.07 |
| Cin_veru_47 | ND | ND | 0.38 | 0.09 | 68.01 | 0.6 | 38.28 | 0.57 | 0.36 | 0 | 6.57 | 0.06 | 21.08 | 0.18 | 5.12 | 0.04 | 13.94 | 0.19 | 9.82 | 0.05 | 10.19 | 0.1 | 0.25 | 0 | 0.83 | 0 | 5.71 | 0.06 | 0.51 | 0.21 | 1.47 | 0.02 |
| Cin_veru_48 | ND | ND | ND | ND | 4.71 | 0.07 | ND | ND | ND | ND | 0.16 | 0 | 0.31 | 0.01 | ND | ND | 0.84 | 0.02 | 0.68 | 0.01 | 0.14 | 0 | ND | ND | 0.36 | 0 | ND | ND | 0.52 | 0.18 | 0.09 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragupathy, S.; Thirugnanasambandam, A.; Vinayagam, V.; Newmaster, S.G. Nuclear Magnetic Resonance Fingerprints and Mini DNA Markers for the Authentication of Cinnamon Species Ingredients Used in Food and Natural Health Products. Plants 2024, 13, 841. https://doi.org/10.3390/plants13060841
Ragupathy S, Thirugnanasambandam A, Vinayagam V, Newmaster SG. Nuclear Magnetic Resonance Fingerprints and Mini DNA Markers for the Authentication of Cinnamon Species Ingredients Used in Food and Natural Health Products. Plants. 2024; 13(6):841. https://doi.org/10.3390/plants13060841
Chicago/Turabian StyleRagupathy, Subramanyam, Arunachalam Thirugnanasambandam, Varathan Vinayagam, and Steven G. Newmaster. 2024. "Nuclear Magnetic Resonance Fingerprints and Mini DNA Markers for the Authentication of Cinnamon Species Ingredients Used in Food and Natural Health Products" Plants 13, no. 6: 841. https://doi.org/10.3390/plants13060841
APA StyleRagupathy, S., Thirugnanasambandam, A., Vinayagam, V., & Newmaster, S. G. (2024). Nuclear Magnetic Resonance Fingerprints and Mini DNA Markers for the Authentication of Cinnamon Species Ingredients Used in Food and Natural Health Products. Plants, 13(6), 841. https://doi.org/10.3390/plants13060841







