Transcriptomic Analysis for Diurnal Temperature Differences Reveals Gene-Regulation-Network Response to Accumulation of Bioactive Ingredients of Protocorm-like Bodies in Dendrobium officinale
Abstract
1. Introduction
2. Results
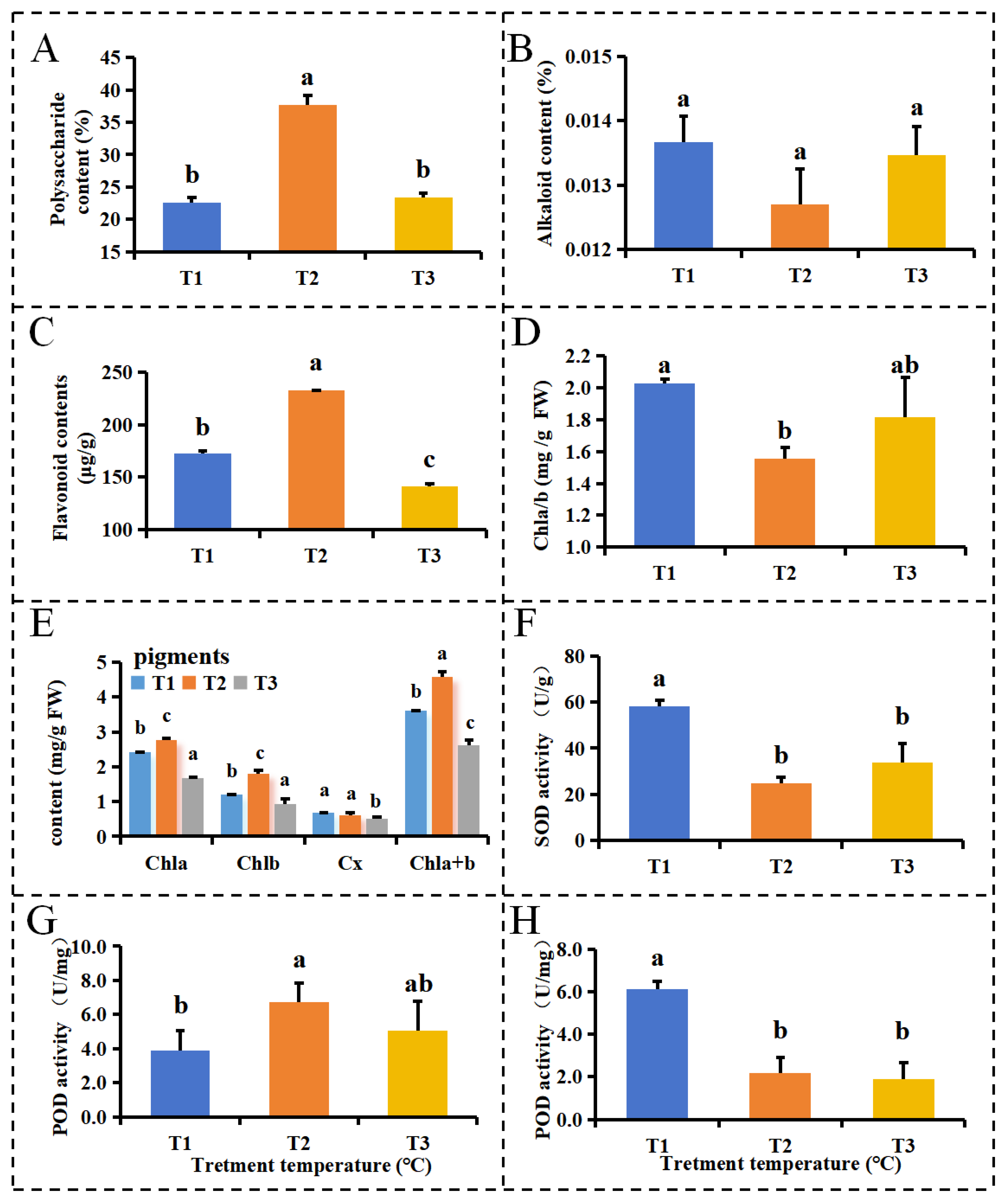
2.1. DIF’s Affects on Functional Metabolites in D. officinale PLBs
2.2. Analysis of the Effect of the DIF on the Physiological Indices of D. officinale PLBs
2.3. Data Filtering and Reference-Sequence Alignment in D. officinale PLBs
2.3.1. Analysis of Transcriptome-Sequencing Basic Data
2.3.2. Numbers of DEGs
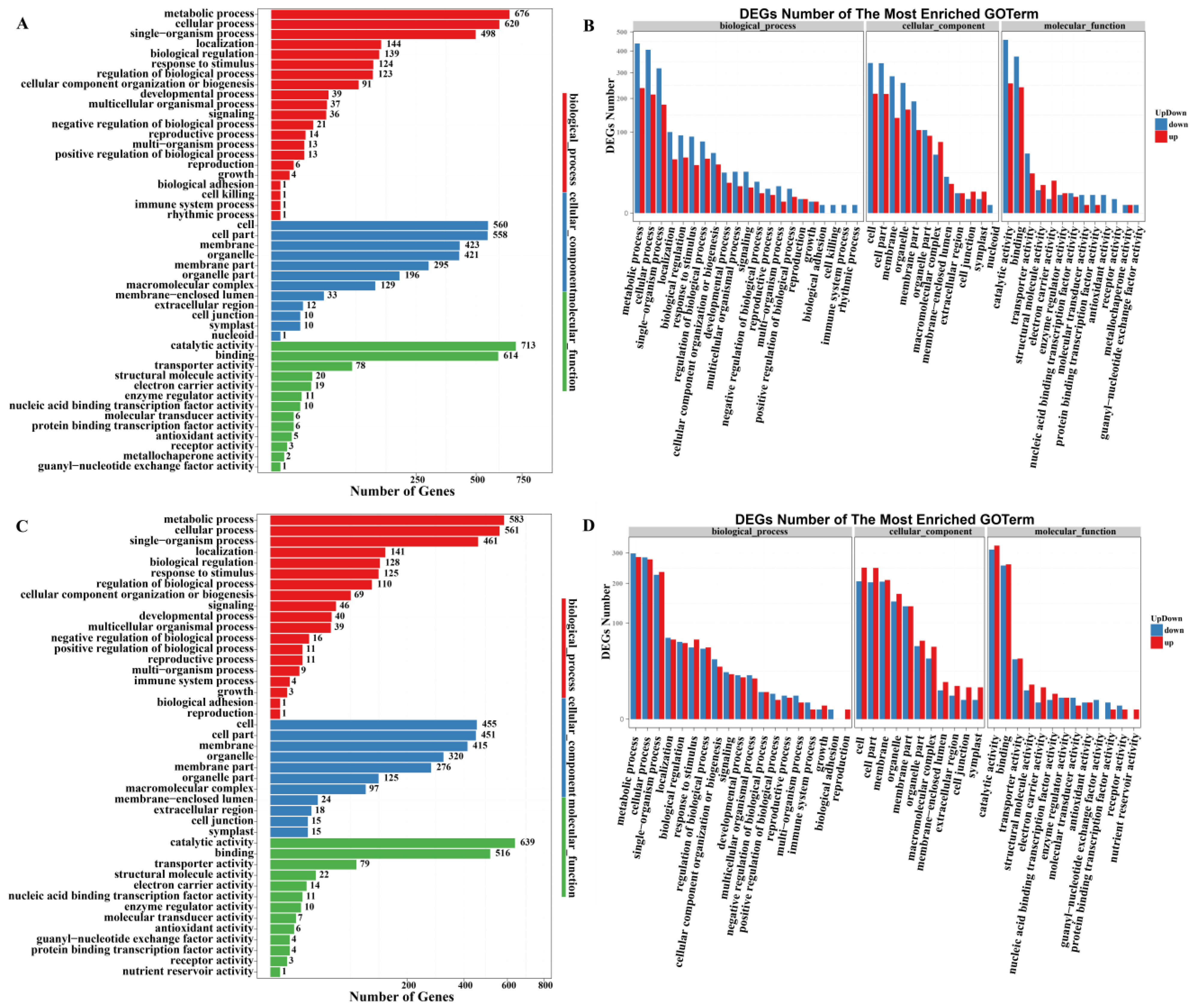
2.3.3. Gene Ontology (GO) Analysis of DEGs
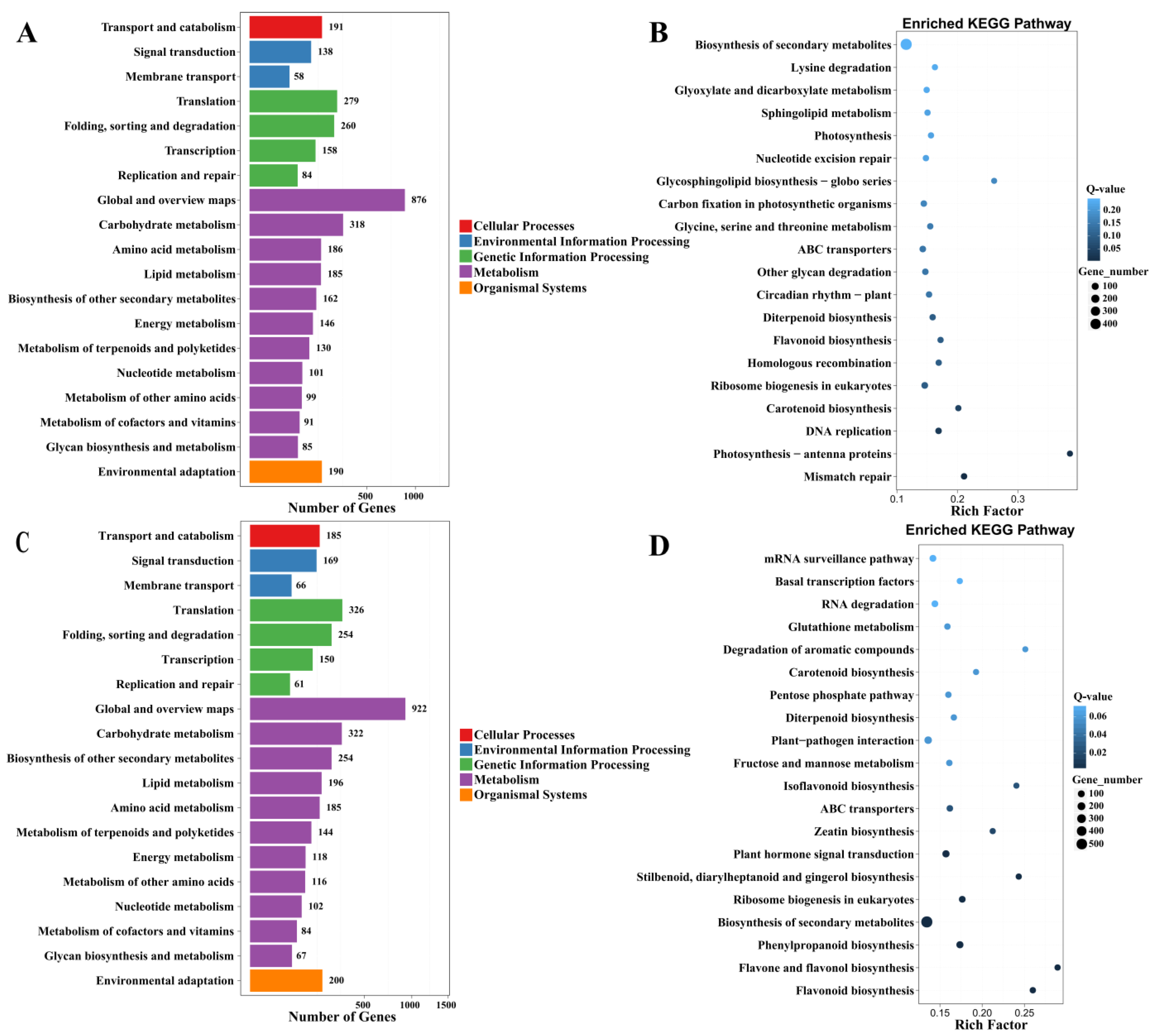
2.3.4. Kyoto Encyclopedia of Genes and Genomes Pathway (KEGG) Analysis
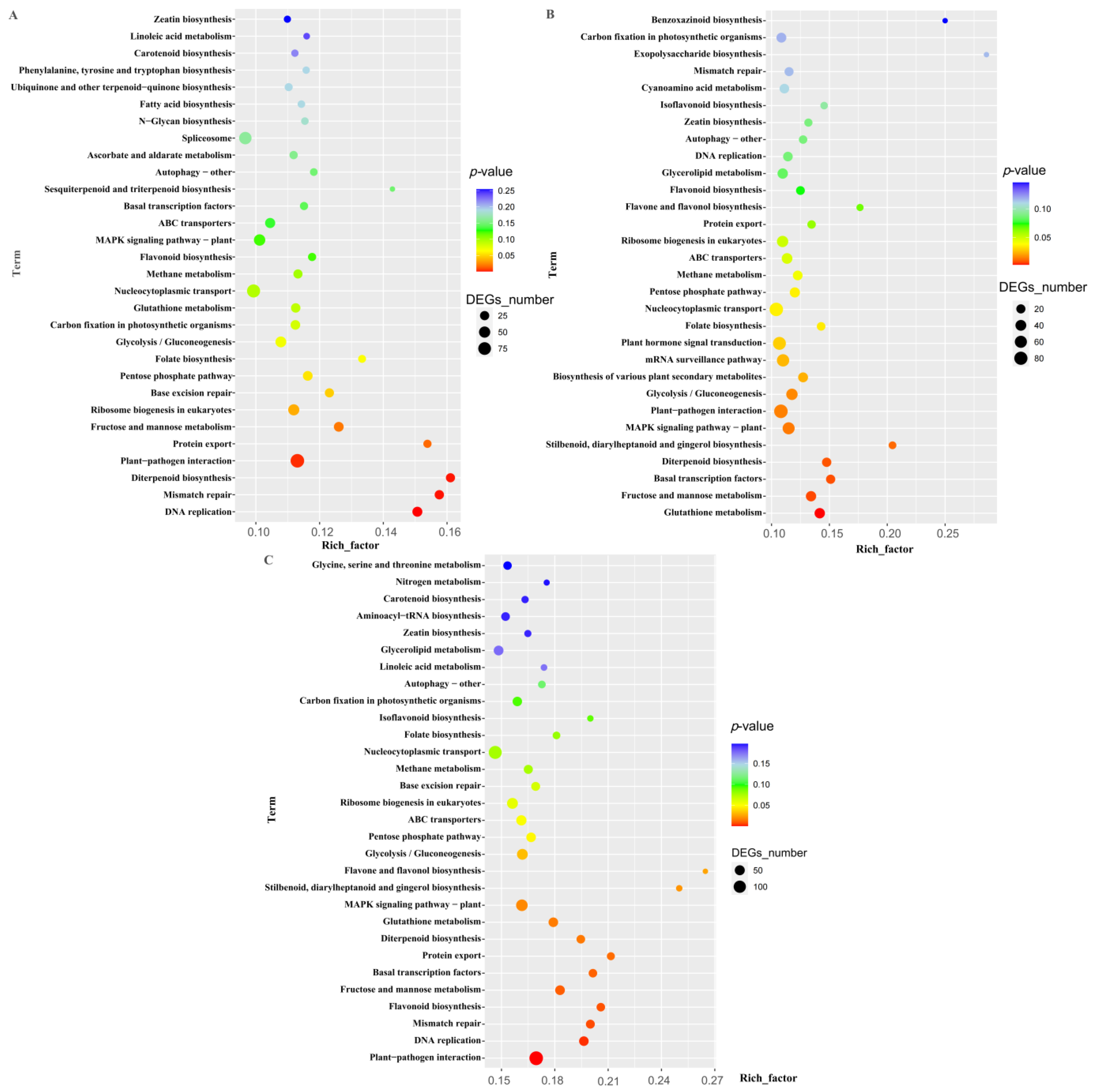
2.3.5. KEGG Analysis of DEGs in DIF-Specific Response
2.4. DEGs Annotated by MapMan in D. officinale PLBs
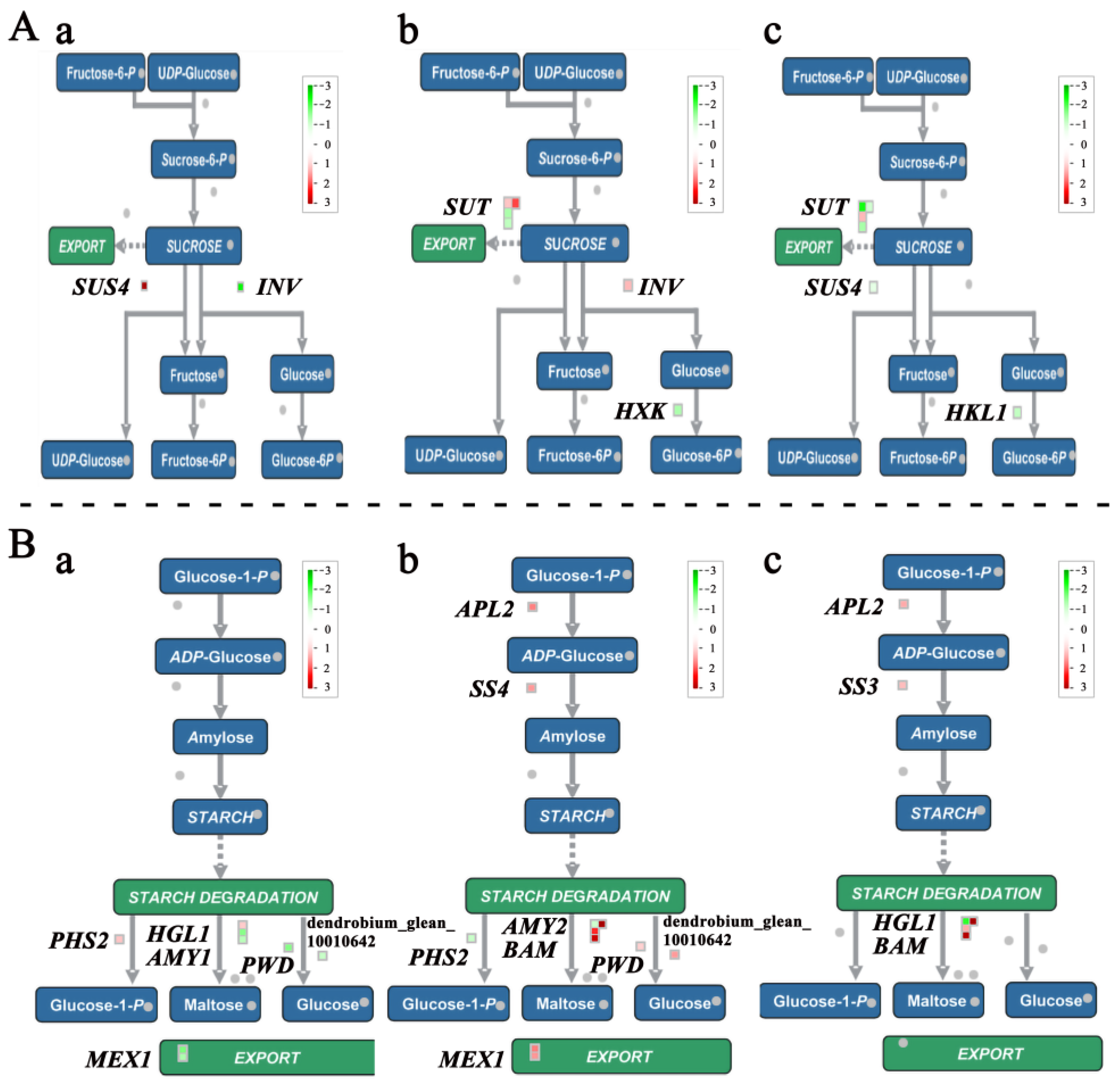
2.5. The DEGs Related to Polysaccharide Metabolism
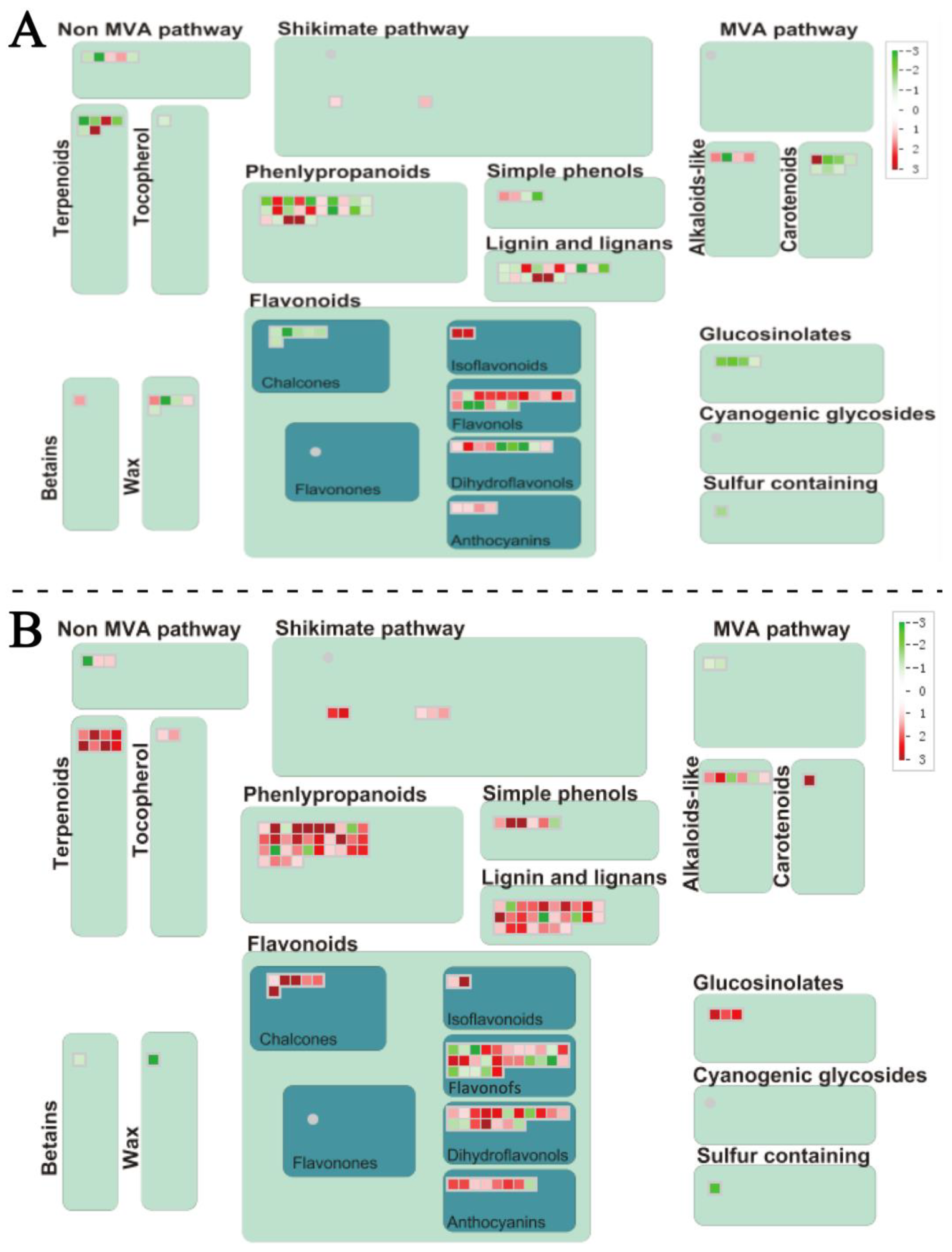
2.6. The DEGs Related to Secondary Metabolism
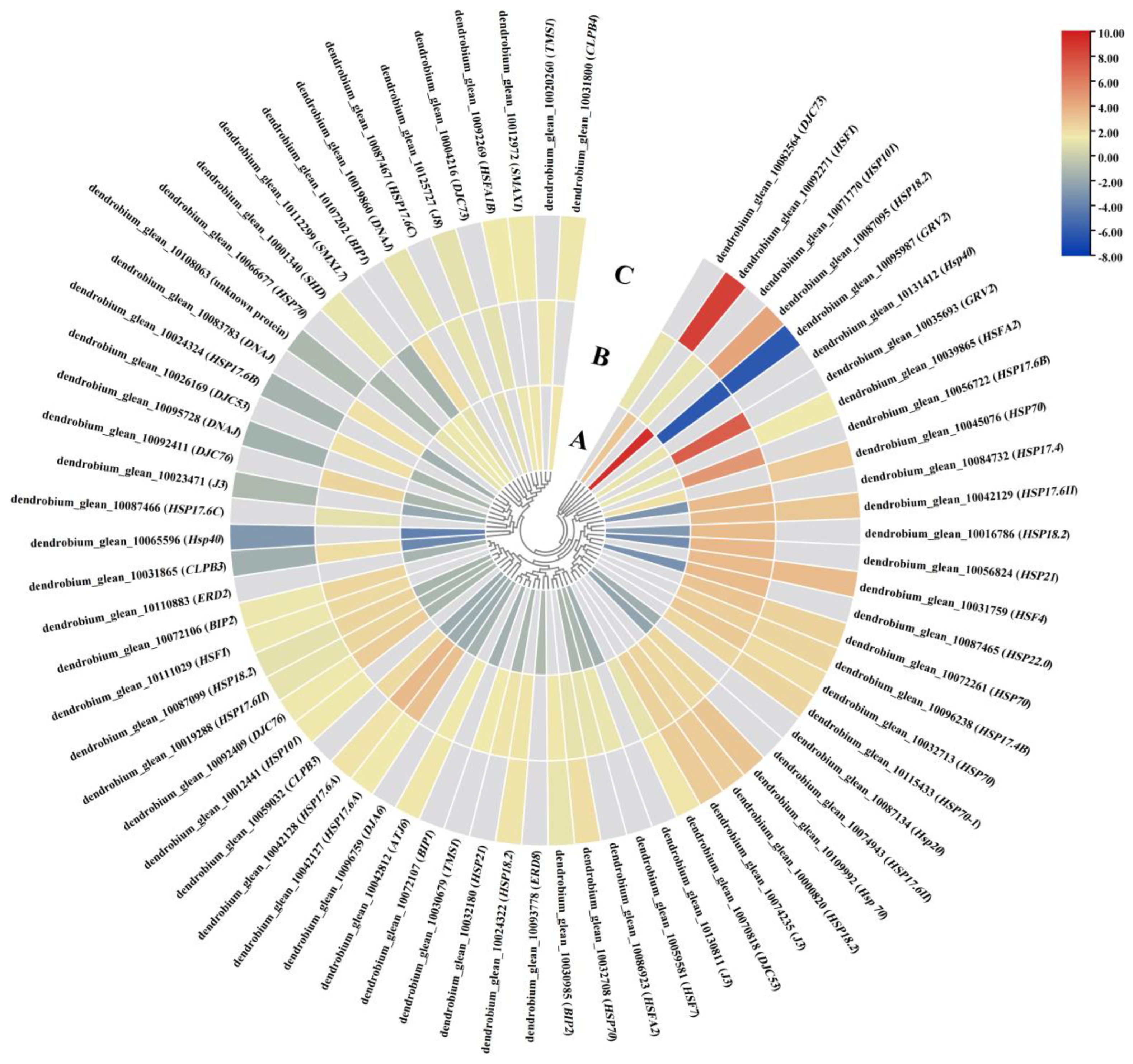
2.7. The Impact of the DIF on DEGs Related to Heat Stress
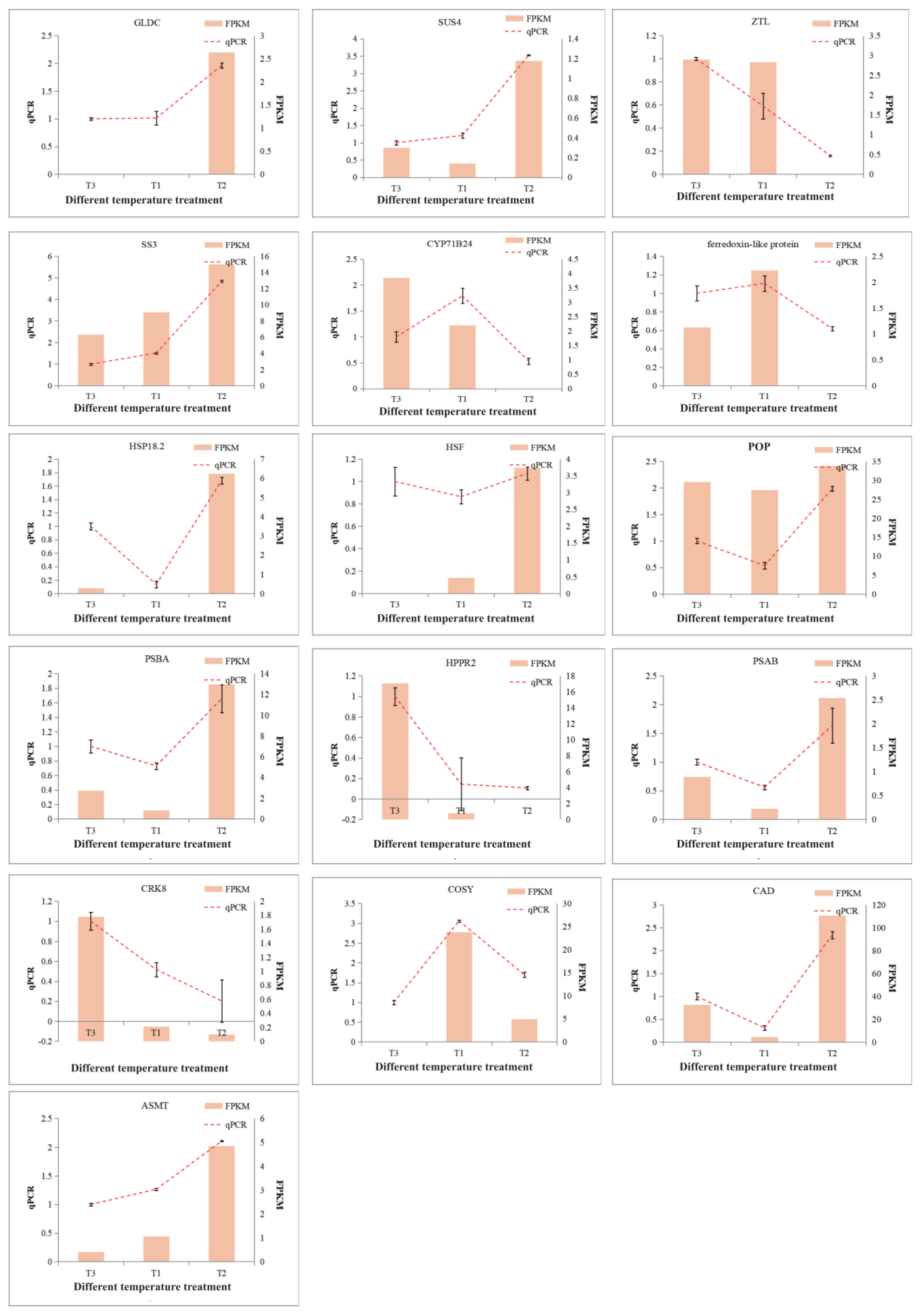
2.8. Verification and Expression Analysis of the Key DEGs in D. officinale PLBs
3. Discussion
3.1. The DIF Affects the Physiological State in D. officinale PLBs
3.2. The DIF Promotes the Accumulation of Major Functional Components in in D. officinale PLBs
3.3. The Polysaccharide Metabolism Process in Response to Temperature Change in D. officinale PLBs
3.4. Secondary Metabolic Processes in Response to DIF in D. officinale PLBs
3.5. Genes Involved in the Heat-Stress Pathway Play an Important Role in D. officinale PLBs’ Response to DIF
4. Materials and Methods
4.1. Materials
4.2. Determination of Polysaccharide and Flavonoid
4.3. Determination of Chlorophyll and Carotenoids
4.4. Measurement of Alkaloid
4.5. RNA Isolation, Library Preparation, and Sequencing
4.6. Transcriptome Mapping, Annotation, and Differential Expression Analysis
4.7. Synthesis Pathway Identification via RT-PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yuan, Y.; Zuo, J.; Zhang, H.; Zu, M.; Yu, M.; Liu, S. Transcriptome and metabolome profiling unveil the accumulation of flavonoids in Dendrobium officinale. Genomics 2022, 114, 110324. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.; Lyu, P.; Chen, L.; Shen, C.; Sun, C. Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale. J. Plant Res. 2019, 132, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Lin, Y.; Xie, H.; Farag, M.A.; Feng, S.; Li, J.; Shao, P. Dendrobium officinale leaf polysaccharides ameliorated hyperglycemia and promoted gut bacterial associated SCFAs to alleviate type 2 diabetes in adult mice. Food Chem. X 2022, 13, 100207. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Cai, H.; Li, W.; Cai, B. Ultrafiltration coupled with high-performance liquid chromatography and quadrupole-time-of-flight mass spectrometry for screening lipase binders from different extracts of Dendrobium officinale. Anal. Bioanal. Chem. 2015, 407, 6081–6093. [Google Scholar] [CrossRef]
- Xu, X.-F.; Dai, D.-C.; Yan, H.; Zhang, Y. Chemical constituents from the Dendrobium officinale and their chemotaxonomic significance. Biochem. Syst. Ecol. 2022, 102, 104420. [Google Scholar] [CrossRef]
- Zhu, N.; Han, S.; Yang, C.; Qu, J.; Sun, Z.; Liu, W.; Zhang, X. Element-tracing of mineral matters in Dendrobium officinale using ICP-MS and multivariate analysis. SpringerPlus 2016, 5, 979. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.; Nie, W.; Yu, G.; Li, Y.; Hu, Y.; Lu, J.; Jin, L. Antitumor and immunomodulatory activity of polysaccharides from Sargassum fusiforme. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2012, 50, 695–700. [Google Scholar] [CrossRef]
- Xie, H.; Fang, J.; Farag, M.A.; Li, Z.; Sun, P.; Shao, P. Dendrobium officinale leaf polysaccharides regulation of immune response and gut microbiota composition in cyclophosphamide-treated mice. Food Chem. X 2022, 13, 100235. [Google Scholar] [CrossRef]
- Chan, C.F.; Wu, C.T.; Huang, W.Y.; Lin, W.S.; Wu, H.W.; Huang, T.K.; Chang, M.Y.; Lin, Y.S. Antioxidation and Melanogenesis Inhibition of Various Dendrobium tosaense Extracts. Molecules 2018, 23, 1810. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Liu, J.; Liang, J.; Si, J.; Wu, S. Dendrobium officinale leaves as a new antioxidant source. J. Funct. Foods 2017, 37, 400–415. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, H.; Ding, X.; Liu, J.; Wang, X.; Hu, L.; Liu, M.; Zhang, C. Anti-inflammatory octahydroindolizine alkaloid enantiomers from Dendrobium crepidatum. Bioorg. Chem. 2020, 100, 103809. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Yang, C.Y.; Wei, S.H. Enhancement of the differentiation of protocorm-like bodies of Dendrobium officinale to shoots by ultrasound treatment. J. Plant Physiol. 2012, 169, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Lucidos, J.G.; Ryu, K.B.; Younis, A.; Kim, C.-K.; Hwang, Y.-J.; Son, B.-G.; Lim, K.-B. Different day and night temperature responses in Lilium hansonii in relation to growth and flower development. Hortic. Environ. Biotechnol. 2013, 54, 405–411. [Google Scholar] [CrossRef]
- Chen, X.; Qin, X.; Guo, L.; He, S. Effects of day-night Temperature variation on the growth of spathiphyllum plantlets in vitro. J. Northwest For. Univ. 2017, 32, 165–169. [Google Scholar] [CrossRef]
- He, S.-l. Effects of Day-night Temperature Difference on the Growth of Gerbera jamesonii Plantlets in vitro. J. Northwest For. Univ. 2012, 27, 88–92. [Google Scholar] [CrossRef]
- Chunli, Z. Cloning DoGAUT1 and DoPGSIP6 from Protocorms and the Expression Analysis by qPCR Under Day-and-night Temperature Differences in Dendrobium officinale. Chin. J. Trop. Crops 2015, 36, 456–465. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.J. Chlorophyll interactions and light conversion in photosynthesis. Naturwissenschaften 1973, 60, 32–39. [Google Scholar] [CrossRef]
- Nielsen, E. The Chlorophyll Content and the Light Utilization in Communities of Plankton Algae and Terrestrial Higher Plants. Physiol. Plant. 2006, 10, 1009–1021. [Google Scholar] [CrossRef]
- Cockshull, K.E.; Langton, F.A.; Cave, C.R.J. Differential effects of different DIF treatments on chrysanthemum and poinsettia. Acta Hortic. 1995, 378, 15–26. [Google Scholar] [CrossRef]
- Yuan, X.K. Effect of day/night temperature difference on chlorophyll content, photosynthesis and fluorescence parameters of tomato at fruit stage. Photosynthetica 2016, 54, 475–477. [Google Scholar] [CrossRef]
- Ma, X.; Song, L.; Yu, W.; Hu, Y.; Liu, Y.; Wu, J.; Ying, Y. Growth, physiological, and biochemical responses of Camptotheca acuminata seedlings to different light environments. Front. Plant Sci. 2015, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Raza, A.; Gao, A.; Jia, Z.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X. Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants 2021, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, M.; Tian, S.; Lu, L.; Shohag, M.J.I.; Yang, X. The activities of SOD, POD CAT and the content of H2O2 in the shoot and roots of wild-type and transgenic tobacco plants. PLoS ONE 2014, 9, e102750. [Google Scholar] [CrossRef]
- Wei, H.; Ai, H.S.; Fu, S.Y.; Yuan, F.Z.; Shan-Zhen, L.; Zhao, D.Z. Effects of sub-optimal temperatures and low light intensity on growth and anti-oxidant enzyme activities in watermelon (Citrullus lanatus) seedlings. J. Hortic. Sci. Biotechnol. 2015, 90, 92–98. [Google Scholar] [CrossRef]
- Xu, J.; Li, S.-L.; Yue, R.-Q.; Ko, C.-H.; Hu, J.-M.; Liu, J.; Ho, H.-M.; Yi, T.; Zhao, Z.-Z.; Zhou, J.; et al. A novel and rapid HPGPC-based strategy for quality control of saccharide-dominant herbal materials: Dendrobium officinale, a case study. Anal. Bioanal. Chem. 2014, 406, 6409–6417. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Peng, X.; Zhao, X.; Yuan, X.; Han, X. Effect of difference between day and night temperature on nutrients and dry mass partitioning of tomato in climate chamber. Trans. Chin. Soc. Agric. Eng. (Trans. CSAE) 2014, 30, 138–147. [Google Scholar] [CrossRef]
- Kang, Y.; Qiang, Y. The effect of endogenous hormones on plant morphology and fruit quality of tomato under difference between day and night temperature. Hortic. Sci. 2018, 45, 131–138. [Google Scholar] [CrossRef]
- Ouyang, W.H.; Xiao, Y.; Lu, Q.J.; Cheng, D.; Liu, L.P. Effects of Different Drying Processes on Effective Components and Antioxidant Activity of Dendrobium officinale Kimura et Migo. Chin. J. Pharm. Biotechnol. 2017, 24, 133–136. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, X.; Jeong, B.R. Manipulating the Difference between the Day and Night Temperatures Can Enhance the Quality of Astragalus membranaceus and Codonopsis lanceolata Plug Seedlings. Agronomy 2019, 9, 654. [Google Scholar] [CrossRef]
- Korzekwa, K. Ecophysiological Responses of Tall Fescue Genotypes to Fungal Endophyte Infection and Elevated Temperature and Precipitation. CSA News 2016, 61, 10–11. [Google Scholar] [CrossRef]
- Guo, X.-r.; Yang, L.; Yu, J.-h.; Tang, Z.-h.; Zu, Y.-g. Alkaloid variations in Catharanthus roseus seedlings treated by different temperatures in short term and long term. J. For. Res. 2007, 18, 313–315. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Decker, D.; Wilczynska, M. UDP-sugar pyrophosphorylase: A new old mechanism for sugar activation. Plant Physiol. 2011, 156, 3–10. [Google Scholar] [CrossRef]
- Hirose, T.; Scofield, G.N.; Terao, T. An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 2008, 174, 534–543. [Google Scholar] [CrossRef]
- Zhang, J.; Arro, J.; Chen, Y.; Ming, R. Haplotype analysis of sucrose synthase gene family in three Saccharumspecies. BMC Genom. 2013, 14, 314. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Tang, C.; Fang, Y.; Yang, M.; Zhou, B.; Qi, J.; Zhang, Y. Structure and expression profile of the sucrose synthase gene family in the rubber tree: Indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 2014, 281, 291–305. [Google Scholar] [CrossRef]
- Boris, K.V.; Ryzhova, N.N.; Kochieva, E.Z. Identification and characterization of intraspecific variability of the sucrose synthase gene Sus4 of potato (Solanum tuberosum). Russ. J. Genet. 2011, 47, 168–175. [Google Scholar] [CrossRef]
- Fan, J.; Wang, H.; Li, X.; Sui, X.; Zhang, Z. Down-Regulating Cucumber Sucrose Synthase 4 (CsSUS4) Suppresses the Growth and Development of Flowers and Fruits. Plant Cell Physiol. 2019, 60, 752–764. [Google Scholar] [CrossRef]
- Shi, Y.; Xu, H.; Shen, Q.; Lin, J.; Wang, Y.; Hua, X.; Yao, W.; Yu, Q.; Ming, R.; Zhang, J. Comparative Analysis of SUS Gene Family between Saccharum officinarum and Saccharum spontaneum. Trop. Plant Biol. 2019, 12, 174–185. [Google Scholar] [CrossRef]
- Sun, L.; Deng, R.; Liu, J.; Lai, M.; Wu, J.; Liu, X.; Shahid, M.Q. An overview of sucrose transporter (SUT) genes family in rice. Mol. Biol. Rep. 2022, 49, 5685–5695. [Google Scholar] [CrossRef] [PubMed]
- Shiratake, K. Genetics of Sucrose Transporter in Plants. Genes Genomes Genom. 2007, 1, 73–80. [Google Scholar]
- Li, Y.; Wang, R.; Liu, X.; Lu, J.X.; Yang, Q.S.; Li, Y.H. Preliminary Function Analysis of PsSUT2 Involved in Sucrose Transportation in the Peony. Russ. J. Plant Physiol. 2022, 69, 65. [Google Scholar] [CrossRef]
- Chung, P.; Hsiao, H.-H.; Chen, H.-J.; Chang, C.-W.; Wang, S.-J. Influence of temperature on the expression of the rice sucrose transporter 4 gene, OsSUT4, in germinating embryos and maturing pollen. Acta Physiol. Plant. 2014, 36, 217–229. [Google Scholar] [CrossRef]
- Gong, X.; Liu, M.; Zhang, L.; Ruan, Y.; Ding, R.; Ji, Y.; Zhang, N.; Zhang, S.; Farmer, J.; Wang, C. Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway. Physiol. Plant. 2014, 153, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Bilska-Kos, A.; Grzybowski, M.; Jończyk, M.; Sowiński, P. In situ localization and changes in the expression level of transcripts related to intercellular transport and phloem loading in leaves of maize (Zea mays L.) treated with low temperature. Acta Physiol. Plant. 2016, 38, 123. [Google Scholar] [CrossRef][Green Version]
- Barker, L.; Kühn, C.; Weise, A.; Schulz, A.; Gebhardt, C.; Hirner, B.; Hellmann, H.; Schulze, W.; Ward, J.M.; Frommer, W.B. SUT2, a Putative Sucrose Sensor in Sieve Elements. Plant Cell 2000, 12, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Terao, T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 2004, 220, 9–16. [Google Scholar] [CrossRef]
- Hannah, L.C.; James, M. The complexities of starch biosynthesis in cereal endosperms. Curr. Opin. Biotechnol. 2008, 19, 160–165. [Google Scholar] [CrossRef]
- Szydlowski, N.; Ragel, P.; Raynaud, S.; Lucas, M.M.; Roldán, I.; Montero, M.; Muñoz, F.J.; Ovecka, M.; Bahaji, A.; Planchot, V.R.; et al. Starch Granule Initiation in Arabidopsis Requires the Presence of Either Class IV or Class III Starch Synthases. Plant Cell 2009, 21, 2443–2457. [Google Scholar] [CrossRef]
- Yue, C.; Cao, H.L.; Wang, L.; Zhou, Y.H.; Huang, Y.T.; Hao, X.Y.; Wang, Y.C.; Wang, B.; Yang, Y.J.; Wang, X.C. Effects of cold acclimation on sugar metabolism and sugar-related gene expression in tea plant during the winter season. Plant Mol. Biol. 2015, 88, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.-Z.; Pan, G.; Wang, F.-B.; Wei, K.-S.; Li, Z.-W.; Shi, C.-H.; Wei, G.; Cheng, F.-M. Effect of high temperature on the expressions of genes encoding starch synthesis enzymes in developing rice endosperms. J. Integr. Agric. 2015, 14, 642–659. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Langlois-Meurinne, M.; Gachon, C.M.; Saindrenan, P. Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 2005, 139, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Messner, B.; Nakajima, J.; Schäffner, A.R.; Saito, K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 43910–43918. [Google Scholar] [CrossRef] [PubMed]
- Husar, S.; Berthiller, F.; Fujioka, S.; Rozhon, W.; Khan, M.; Kalaivanan, F.; Elias, L.; Higgins, G.S.; Li, Y.; Schuhmacher, R.; et al. Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Stone, J.M.; Heard, J.E.; Kovtun, Y.; Yorgey, P.; Sheen, J.; Ausubel, F.M. Fumonisin B1-induced cell death in arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 2000, 12, 1823–1836. [Google Scholar] [CrossRef]
- Yanping, L.; Shu, W. Studies on Glucosyltransferase CsUGT73D1 Gene in Camellia sinensis. J. Hainan Norm. Univ. (Nat. Sci.) 2019, 32, 268–280. [Google Scholar] [CrossRef]
- Ceylan, Y.; Altunoglu, Y.C.; Horuz, E. HSF and Hsp Gene Families in sunflower: A comprehensive genome-wide determination survey and expression patterns under abiotic stress conditions. Protoplasma 2023, 260, 1473–1491. [Google Scholar] [CrossRef]
- Agarwal, P.; Khurana, P. Functional characterization of HSFs from wheat in response to heat and other abiotic stress conditions. Funct. Integr. Genom. 2019, 19, 497–513. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Chen, J.; Guo, L.; Li, X.; Li, W.; Yu, Z.; Deng, J.; Zhang, P.; Zhang, K.; et al. Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol. Biochem. 2013, 64, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, Y.; Li, X.; Yu, P. Purification and structural characterization of Chinese yam polysaccharide and its activities. Carbohydr. Polym. 2015, 117, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, X.; You, X.; Li, Y.; Long, S.; Wen, S.; Liu, Q.; Liu, T.; Guo, H.; Xu, Y. Hydrogen sulfide improves tall fescue photosynthesis response to low-light stress by regulating chlorophyll and carotenoid metabolisms. Plant Physiol. Biochem. 2022, 170, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Si, J.P.; Zhang, Y.; Luo, Y.B.; Liu, J.J.; Liu, Z.J. Herbal textual research on relationship between Chinese medicine “Shihu” (Dendrobium spp.) and “Tiepi Shihu” (D. catenatum). Zhongguo Zhong Yao Za Zhi = Zhongguo Zhongyao Zazhi = China J. Chin. Mater. Medica 2017, 42, 2001–2005. [Google Scholar] [CrossRef]
- Zhang, G.-Q.; Xu, Q.; Bian, C.; Tsai, W.-C.; Yeh, C.-M.; Liu, K.-W.; Yoshida, K.; Zhang, L.-S.; Chang, S.-B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, M.M.; Zhang, D.; Guo, S.X. Reference gene selection for real-time quantitative PCR analysis of Dendrobium officinale. Chin. Pharm. J. 2013, 48, 1664–1668. [Google Scholar] [CrossRef]
- Ling, H.; Zeng, X.; Guo, S. Functional insights into the late embryogenesis abundant (LEA) protein family from Dendrobium officinale (Orchidaceae) using an Escherichia coli system. Sci. Rep. 2016, 6, 39693. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Song, Z.; Wei, L.; Li, L. Molecular characterization and expression analysis of WRKY family genes in Dendrobium officinale. Genes Genom. 2018, 40, 265–279. [Google Scholar] [CrossRef]
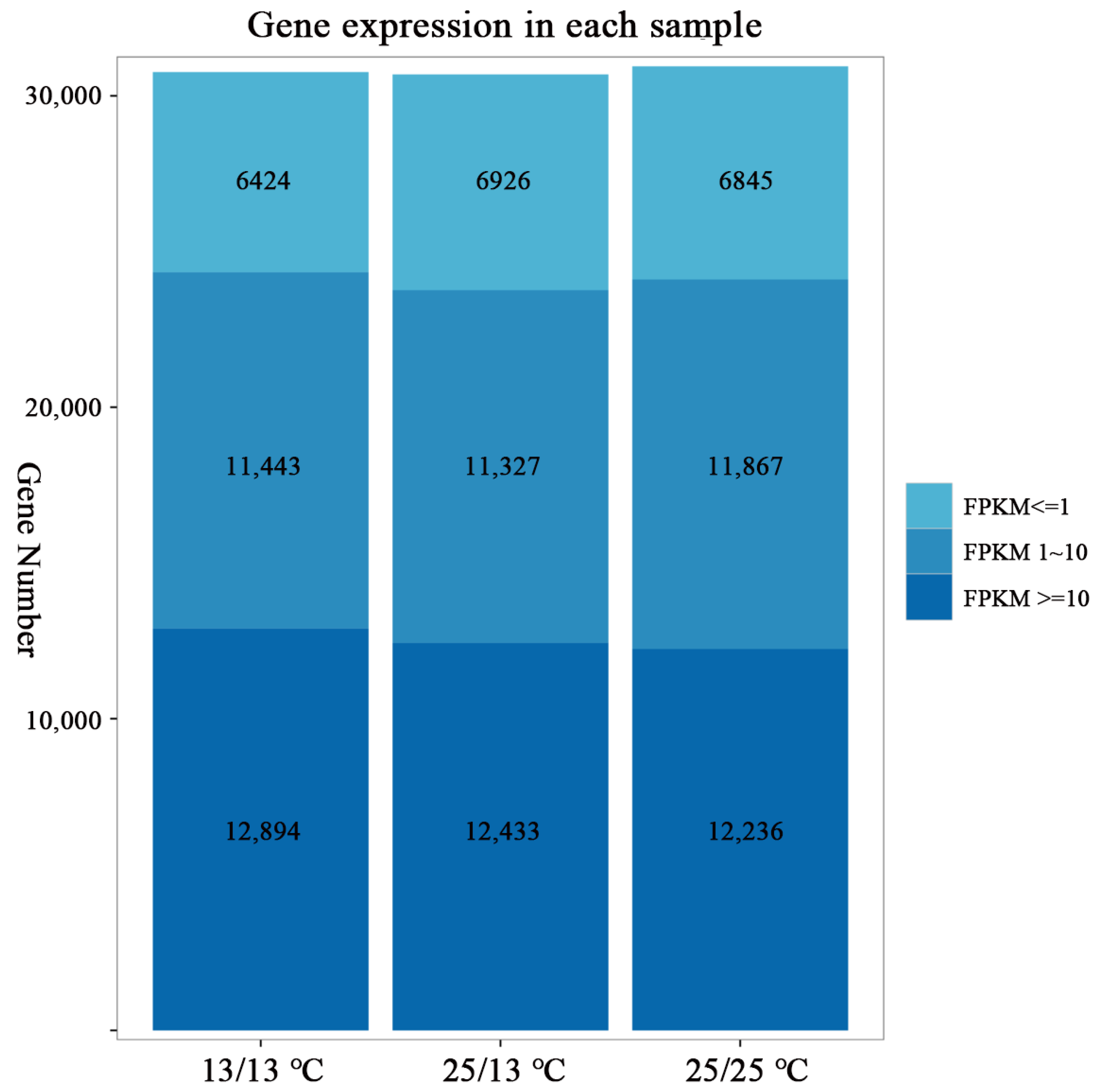
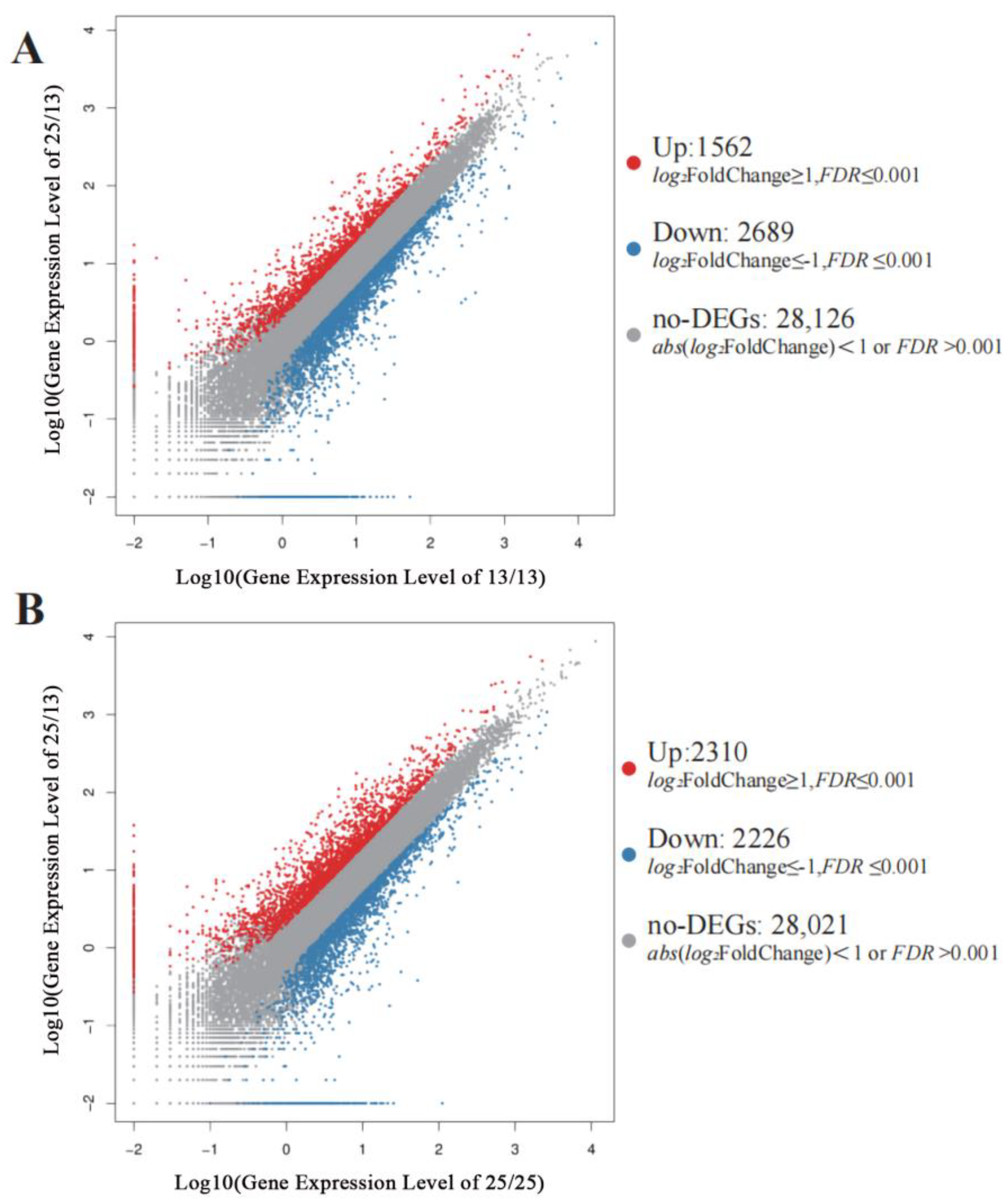
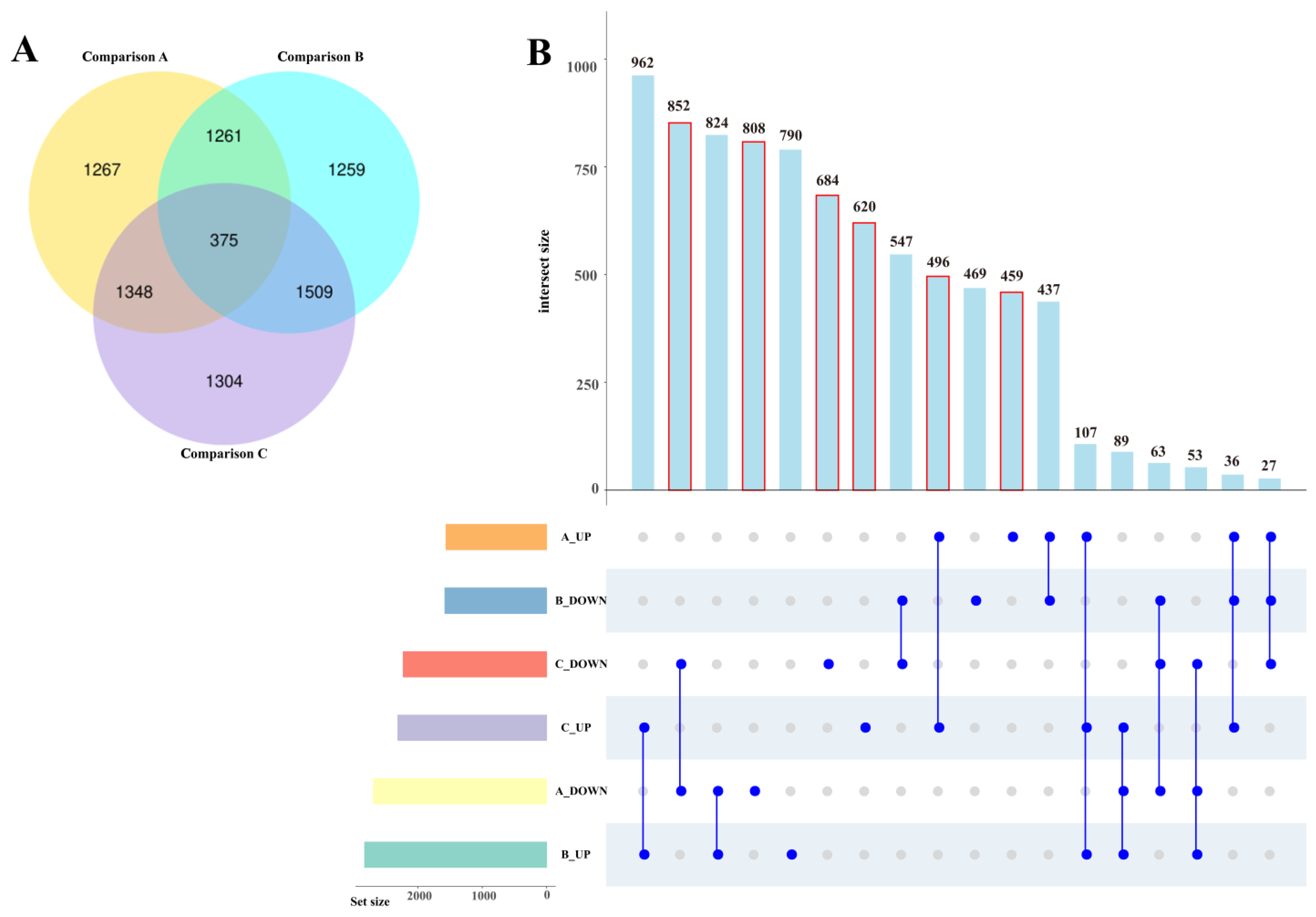

| Sample | T1 (13/13 °C) | T2 (25/13 °C) | T3 (25/25 °C) |
|---|---|---|---|
| Total Raw Reads (Mb) | 69.97 | 69.97 | 69.97 |
| Total Clean Reads (Mb) | 66.98 | 66.54 | 66.66 |
| Total Clean Bases (Gb) | 6.7 | 6.65 | 6.67 |
| Clean Reads Q20 (%) | 99.38 | 99.35 | 99.36 |
| Clean Reads Q30 (%) | 97.88 | 97.77 | 97.8 |
| Clean Reads Ratio (%) | 95.73 | 95.11 | 95.27 |
| Total Mapping Ratio (%) | 82.16 | 82.40 | 81.39 |
| Uniquely Mapping Ratio (%) | 54.89 | 54.24 | 53.10 |
| NO | Name | Primers | Sequences (5′–3′) |
|---|---|---|---|
| 1 | CRK8 | dendrobium_glean_10017232-F | TATCGCCTCATGCTTCCACTA |
| dendrobium_glean_10017232-R | GACACTCTCTCCGCTCTCTT | ||
| 2 | CAD6 | dendrobium_glean_10107234-F | CTATGGGGATGACTGCCACT |
| dendrobium_glean_10107234-R | CTACCGATTGCCTCCTTCAG | ||
| 3 | COSY | dendrobium_glean_10128767-F | ACTACGAAAAGGGCATCACG |
| dendrobium_glean_10128767-R | CTCACCTTCCGATTCAGCAT | ||
| 4 | CYP71B24 | dendrobium_glean_10021134-F | CCAGAGTTGCGATGGAAGAT |
| dendrobium_glean_10021134-R | TGGCTTATCCCAGTTTTTGG | ||
| 5 | ASMT | dendrobium_glean_10048936-F | GCGAGTGCTACATGACTGGA |
| dendrobium_glean_10048936-R | CTGTCCTCTCTTTCCCACCA | ||
| 6 | PSAB | dendrobium_glean_10008093-F | TACAATCGGATTACGCACCA |
| dendrobium_glean_10008093-R | AACGCTTGGTTTCCATTTTG | ||
| 7 | PSBA | dendrobium_glean_10135332-F | GGGTCGTGAGTGGGAACTTA |
| dendrobium_glean_10135332-R | TGTGCTCTGCCTGGAATACA | ||
| 8 | CHS | dendrobium_glean_10103920-F | TACTGAACGACCGCTTTTCC |
| dendrobium_glean_10103920-R | TGCCTCCACGAGACTCTTTT | ||
| 9 | HSF1 | dendrobium_glean_10092271-F | AAACGGCAAGGACAACAAAG |
| dendrobium_glean_10092271-R | GCAGGATGTGAACGAAACCT | ||
| 10 | HPPR2 | dendrobium_glean_10101025-F | TGTGTATTGATCGCCTCTGC |
| dendrobium_glean_10101025-R | AAGACCAATGATGCCTACCG | ||
| 11 | PFK3 | dendrobium_glean_10138387-F | GCAAGCAGAGATGTGGACTG |
| dendrobium_glean_10138387-R | CACCCTCAGCAACAACGATA | ||
| 12 | POP | dendrobium_glean_10121871-F | CCGCCAGAGATGTGTTGTAG |
| dendrobium_glean_10121871-R | CTGTAACCAGGCTGACTGAATC | ||
| 13 | HSP18.2 | dendrobium_glean_10087095-F | CGATTCCGACTTCCTGAGAA |
| dendrobium_glean_10087095-R | GGCTTCTTGACCTCCTCCTT | ||
| 14 | ZTL | dendrobium_glean_10106356-F | CCCAACAGAGGAGAAACCAA |
| dendrobium_glean_10106356-R | GCAATTCGCTTAGCATCCAT | ||
| 15 | SUS4 | dendrobium_glean_10105017-F | GGTGGTCCAGCAGAGATCAT |
| dendrobium_glean_10105017-R | CCAGCCAAGGTCATCAATCT | ||
| 16 | SS3 | dendrobium_glean_10016137-F | GGATTTCCATGCCGCTATAA |
| dendrobium_glean_10016137-R | CAAACGTGCTGCTTTCTCAG | ||
| 17 | GLDC | dendrobium_glean_10031276-F | CTCTGGGCTTCTTCAACAGC |
| dendrobium_glean_10031276-R | TGAGCATGGTCAGTGATTCC | ||
| 18 | ferredoxin-like protein | dendrobium_glean_10136528-F | TGGTGTCCTAGCCCAGACTC |
| dendrobium_glean_10136528-R | CCGACCTCTGTGTGGTTTTT |
| Treatment | DIF Treatment Phase (5 Days) | ||
|---|---|---|---|
| T1 | T2 | T3 (Control) | |
| 8:00~20:00 | 13 °C | 25 °C | 25 °C |
| 20:00~8:00 | 13 °C | 13 °C | 25 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Zhang, C.; Chen, Y.; Wang, C.; Lai, Z. Transcriptomic Analysis for Diurnal Temperature Differences Reveals Gene-Regulation-Network Response to Accumulation of Bioactive Ingredients of Protocorm-like Bodies in Dendrobium officinale. Plants 2024, 13, 874. https://doi.org/10.3390/plants13060874
Chen Q, Zhang C, Chen Y, Wang C, Lai Z. Transcriptomic Analysis for Diurnal Temperature Differences Reveals Gene-Regulation-Network Response to Accumulation of Bioactive Ingredients of Protocorm-like Bodies in Dendrobium officinale. Plants. 2024; 13(6):874. https://doi.org/10.3390/plants13060874
Chicago/Turabian StyleChen, Qingqing, Chunyu Zhang, Yukun Chen, Congqiao Wang, and Zhongxiong Lai. 2024. "Transcriptomic Analysis for Diurnal Temperature Differences Reveals Gene-Regulation-Network Response to Accumulation of Bioactive Ingredients of Protocorm-like Bodies in Dendrobium officinale" Plants 13, no. 6: 874. https://doi.org/10.3390/plants13060874
APA StyleChen, Q., Zhang, C., Chen, Y., Wang, C., & Lai, Z. (2024). Transcriptomic Analysis for Diurnal Temperature Differences Reveals Gene-Regulation-Network Response to Accumulation of Bioactive Ingredients of Protocorm-like Bodies in Dendrobium officinale. Plants, 13(6), 874. https://doi.org/10.3390/plants13060874







