The Combination of Low-Cost, Red–Green–Blue (RGB) Image Analysis and Machine Learning to Screen for Barley Plant Resistance to Net Blotch
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
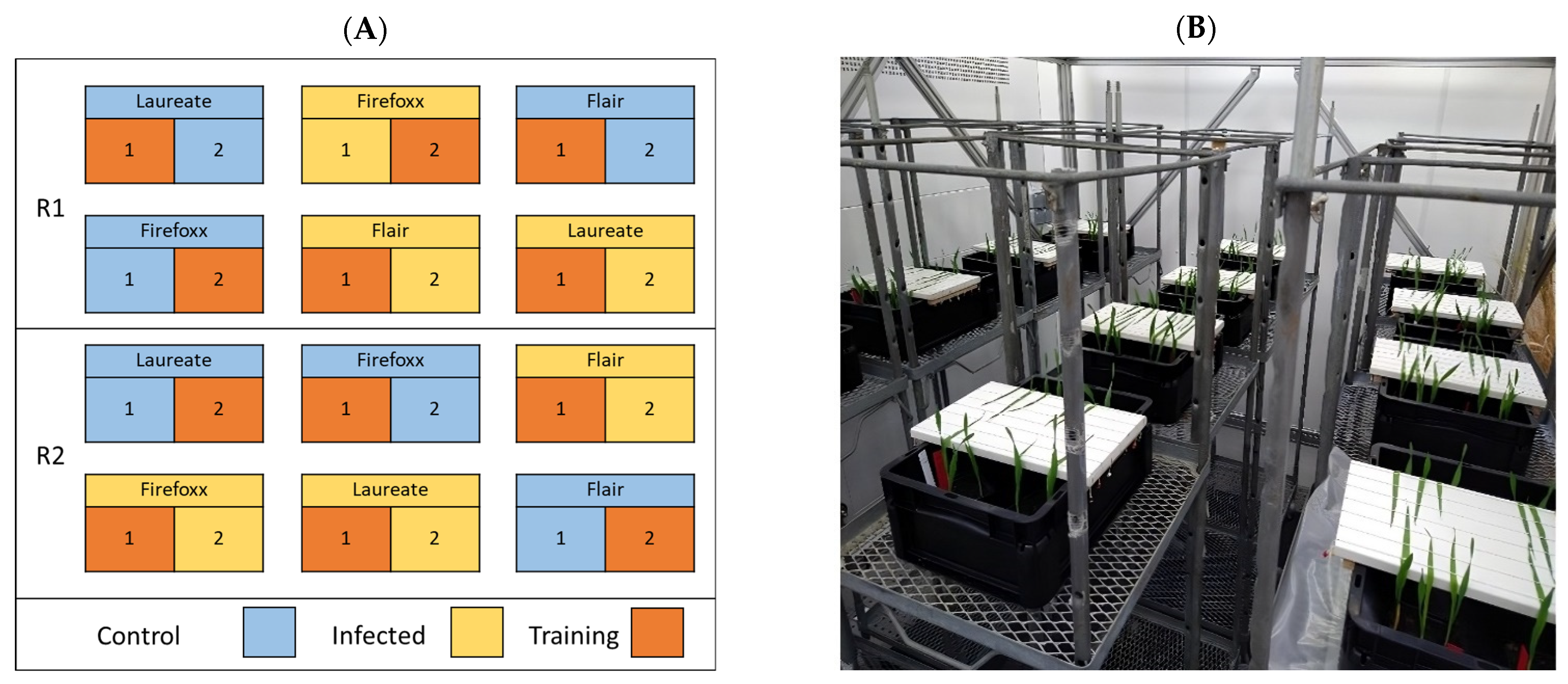
2.2. Inoculation Protocol
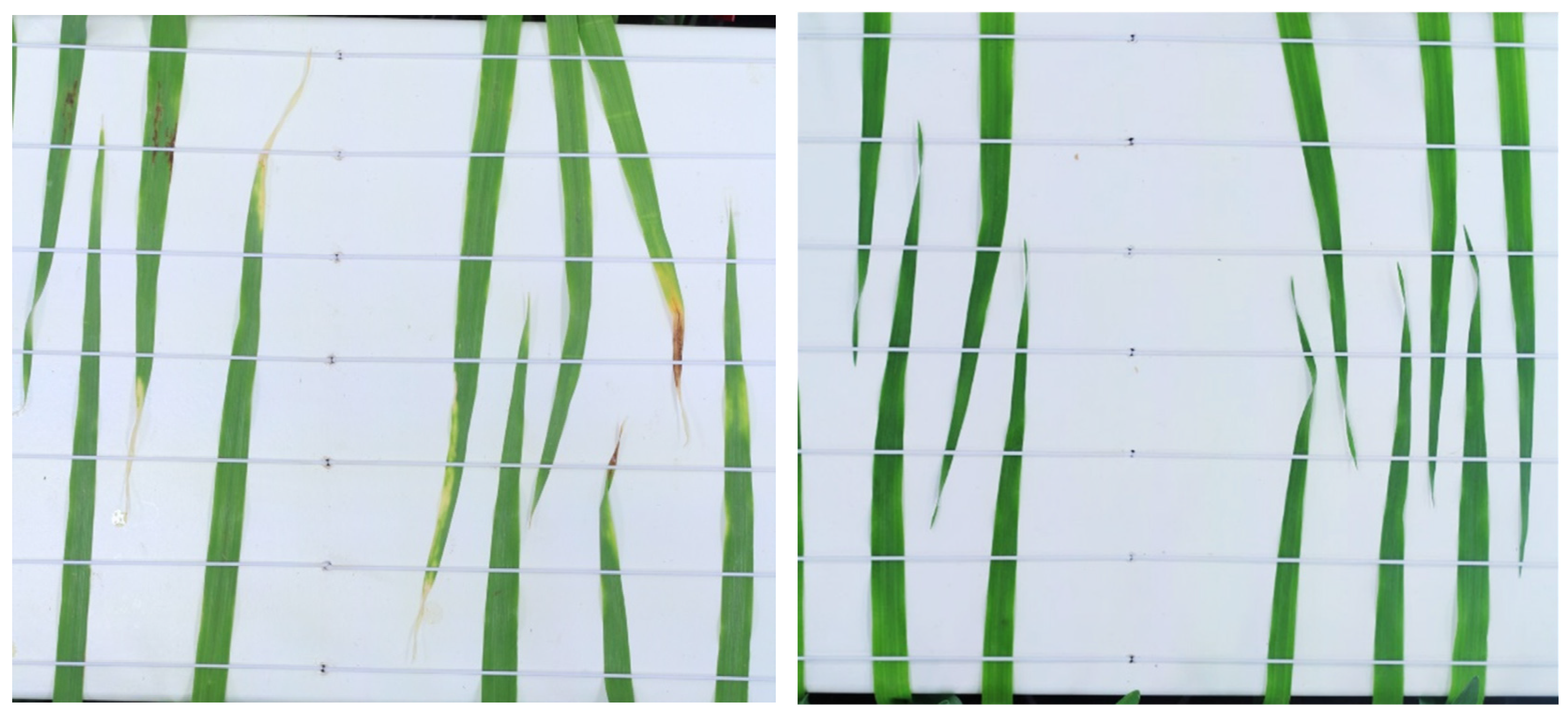
2.3. Image Acquisition
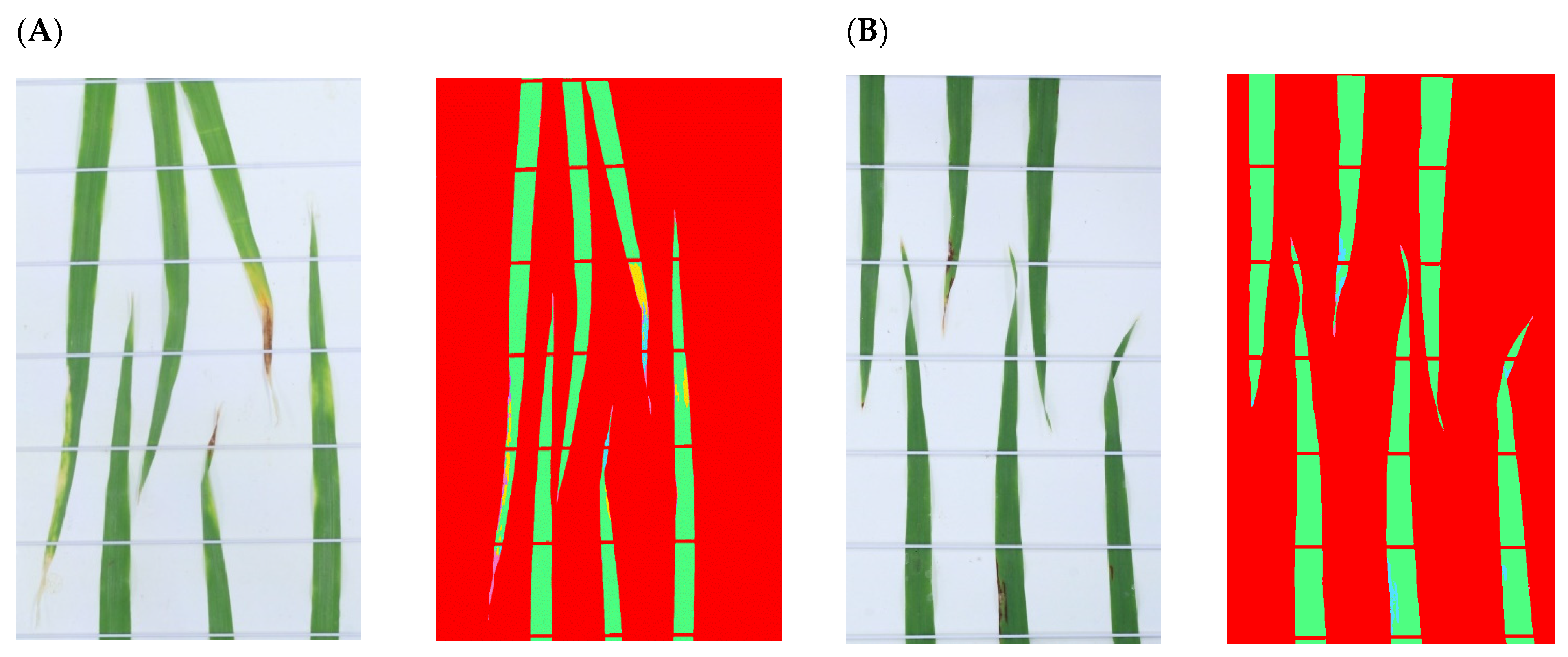
2.4. Image Processing
2.5. Model Performance
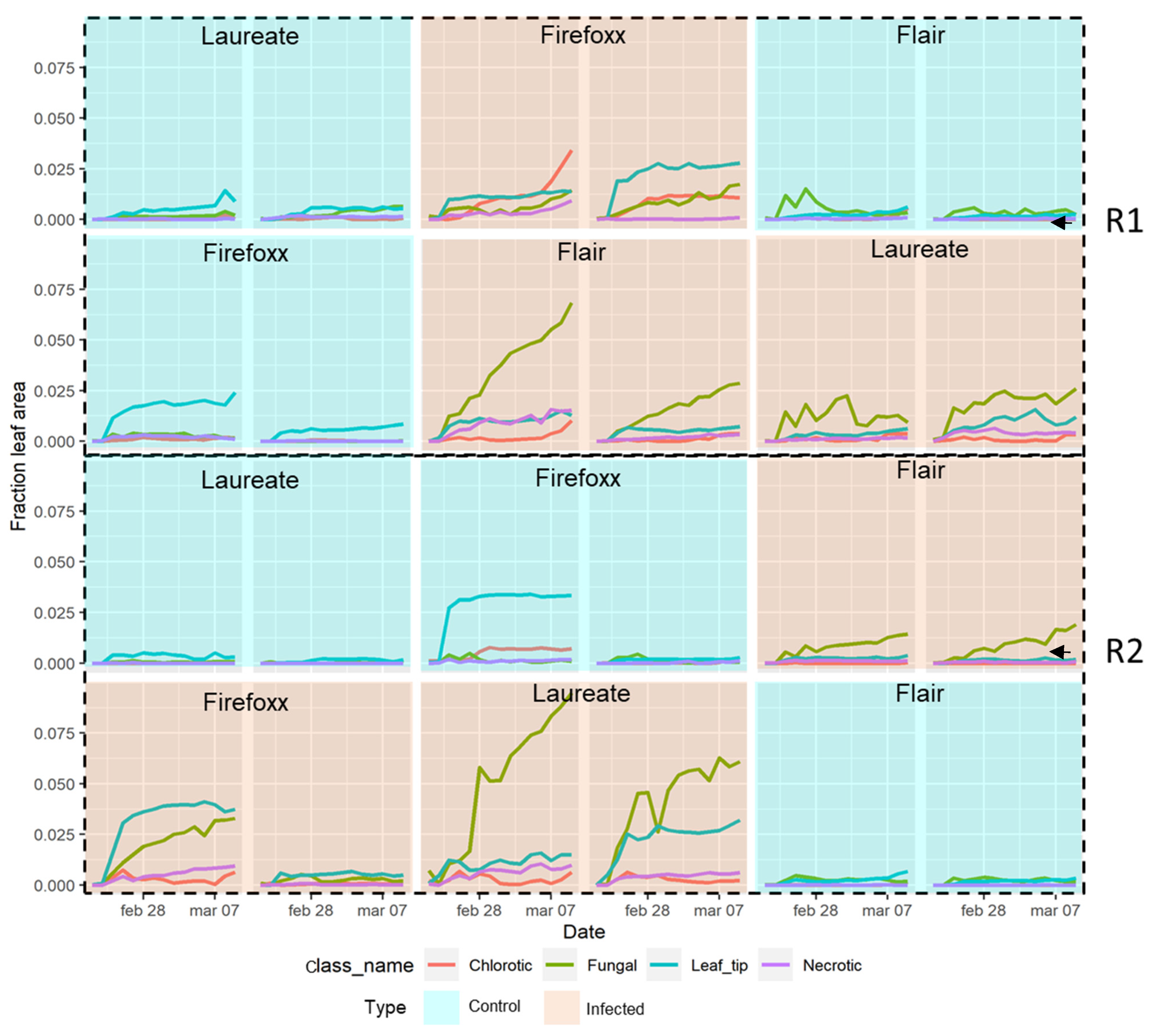
3. Results
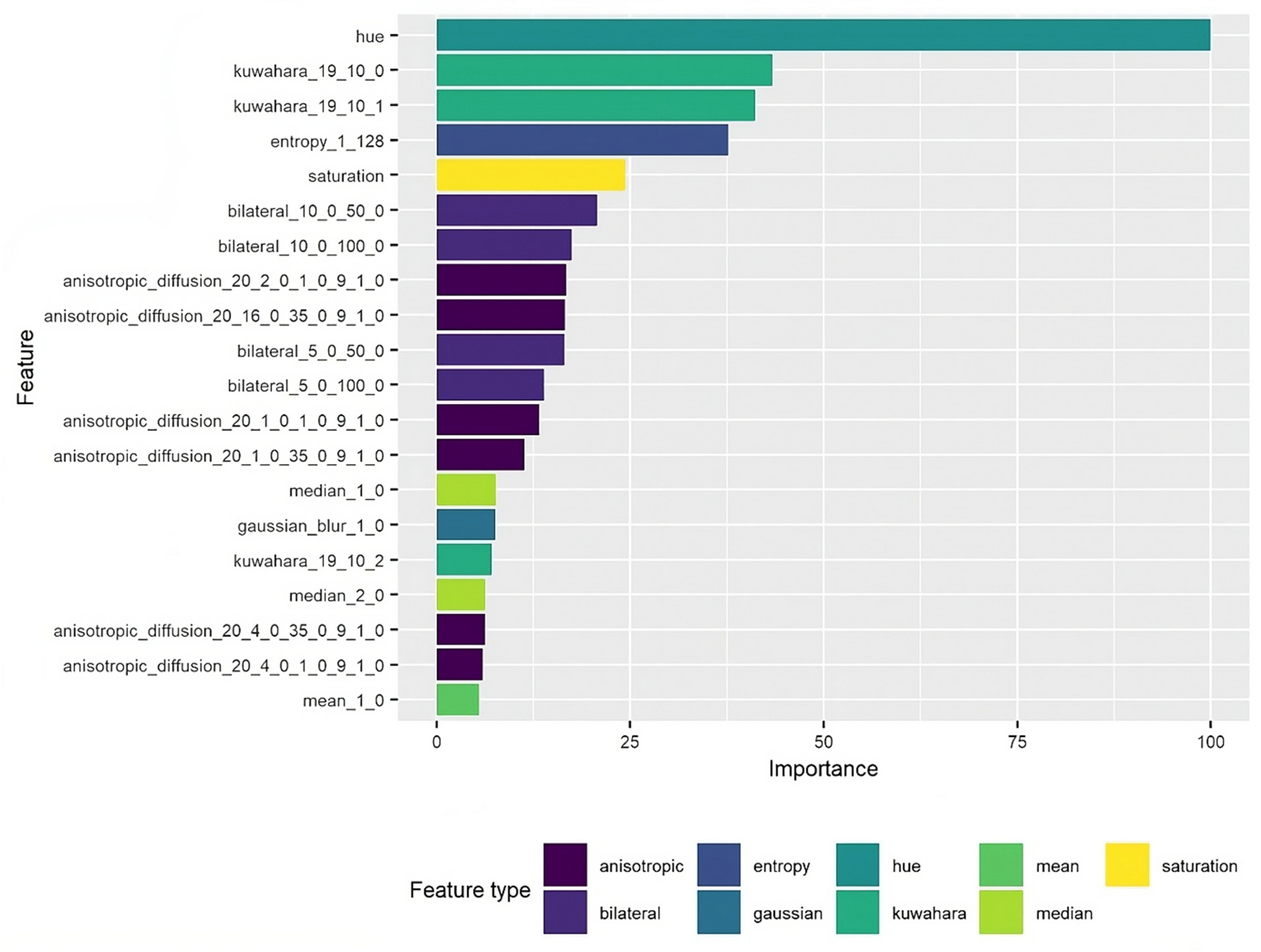
4. Feature Importance
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tscharntke, T.; Clough, Y.; Wanger, T.C.; Jackson, L.; Motzke, I.; Perfecto, I.; Vandermeer, J.; Whitbread, A. Global food security, biodiversity conservation and the future of agricultural intensification. Biol. Conserv. 2012, 151, 53–59. [Google Scholar] [CrossRef]
- Molotoks, A.; Smith, P.; Dawson, T.P. Impacts of land use, population, and climate change on global food security. Food Energy Secur. 2021, 10, e261. [Google Scholar] [CrossRef]
- Ben Mahmoud, K.; Abdelkefi, F.; Mezzapessa, G.N.; Perrino, E.V. Beta macrocarpa Guss. in Tunisia: Nutritional and functional properties of the underutilized wild beet in relation to soil characteristics. Euro-Mediterr. J. Environ. Integr. 2024. [Google Scholar]
- Clemente-Villalba, J.; Burló, F.; Hernández, F.; Carbonell-Barrachina, Á.A. Valorization of wild edible plants as food ingredients and their economic value. Foods 2023, 12, 1012. [Google Scholar] [CrossRef] [PubMed]
- Schulp, C.J.; Thuiller, W.; Verburg, P.H. Wild food in Europe: A synthesis of knowledge and data of terrestrial wild food as an ecosystem service. Ecol. Econ. 2014, 105, 292–305. [Google Scholar] [CrossRef]
- Owino, V.; Kumwenda, C.; Ekesa, B.; Parker, M.E.; Ewoldt, L.; Roos, N.; Lee, W.T.; Tome, D. The impact of climate change on food systems, diet quality, nutrition, and health outcomes: A narrative review. Front. Clim. 2022, 4, 941842. [Google Scholar] [CrossRef]
- Thuraga, V.; Martinsson, U.D.; Vetukuri, R.R.; Chawade, A. Delineation of genotype: Environment interaction for grain yield in spring barley under untreated and fungicide-treated environments. Plants 2023, 12, 715. [Google Scholar] [CrossRef]
- Abenavoli, L.; Milanovic, M.; Procopio, A.C.; Spampinato, G.; Maruca, G.; Perrino, E.V.; Mannino, G.C.; Fagoonee, S.; Luzza, F.; Musarella, C.M. Ancient wheats: Beneficial effects on insulin resistance. Minerva Med. 2020, 112, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Ruusunen, O.; Jalli, M.; Jauhiainen, L.; Ruusunen, M.; Leiviskä, K. Advanced data analysis as a tool for net blotch density estimation in spring barley. Agriculture 2020, 10, 179. [Google Scholar] [CrossRef]
- Tricase, C.; Amicarelli, V.; Lamonaca, E.; Rana, R.L. Economic analysis of the barley market and related uses. In Grasses as Food and Feed; Tadele, Z., Ed.; InTechOpen: London, UK, 2018; pp. 25–46. [Google Scholar]
- Nowosad, K.; Tratwal, A.; Bocianowski, J. Genotype by environment interaction for grain yield in spring barley using additive main effects and multiplicative interaction model. Cereal Res. Commun. 2018, 46, 729–738. [Google Scholar] [CrossRef]
- McDonald, W. Heterothallism in Pyrenophora teres. Phytopathology 1963, 53, 771–780. [Google Scholar]
- Tini, F.; Covarelli, L.; Ricci, G.; Balducci, E.; Orfei, M.; Beccari, G. Management of Pyrenophora teres f. teres, the causal agent of net form net blotch of barley, in a two-year field experiment in central Italy. Pathogens 2022, 11, 291. [Google Scholar] [CrossRef]
- Backes, A.; Guerriero, G.; Barka, E.A.; Jacquard, C. Pyrenophora teres: Taxonomy, morphology, interaction with barley, and mode of control. Front. Plant Sci. 2021, 12, 614951. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, H.J.L.; Lübeck, P.S.; Thordal-Christensen, H.; de Neergaard, E.; Smedegaard-Petersen, V. Mechanisms of induced resistance in barley against Drechslera teres. Phytopathology 1998, 88, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.R.; Avrova, A.; Bingham, I.J.; Burnett, F.J.; Fountaine, J.; Havis, N.D.; Hoad, S.P.; Hughes, G.; Looseley, M.; Oxley, S.J.P.; et al. Control of foliar diseases in barley: Towards an integrated approach. Eur. J. Plant Pathol. 2012, 133, 33–73. [Google Scholar] [CrossRef]
- Shipton, W. Effect of net blotch infection of barley on grain yield and quality. Aust. J. Exp. Agric. 1966, 6, 437–440. [Google Scholar] [CrossRef]
- Abebe, W. Barley net blotch disease management: A review. Int. J. Environ. Agric. Res. 2021, 7, 69–81. [Google Scholar]
- Geschele, E. The response of barley to parasitic fungi Helminthosporium teres Sacc. Bull. Appl. Bot. Genet. Plant Breed. 1928, 19, 371–384. [Google Scholar]
- Douiyssi, A.; Rasmusson, D.; Roelfs, A. Responses of barley cultivars and lines to isolates of Pyrenophora teres. Plant Dis. 1998, 82, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, A.; Turkington, T.K.; Askarian, H.; Tekauz, A.; Xi, K.; Tucker, J.R.; Kutcher, H.R.; Strelkov, S.E. Virulence of Pyrenophora teres populations in western Canada. Can. J. Plant Pathol. 2016, 38, 183–196. [Google Scholar] [CrossRef]
- Tekauz, A. A numerical scale to classify reactions of barley to Pyrenophora teres. Can. J. Plant Pathol. 1985, 7, 181–183. [Google Scholar] [CrossRef]
- Pavicic, M.; Overmyer, K.; Rehman, A.U.; Jones, P.; Jacobson, D.; Himanen, K. Image-based methods to score fungal pathogen symptom progression and severity in excised Arabidopsis leaves. Plants 2021, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef]
- Ahmed, H.G.M.-D.; Zeng, Y.; Fiaz, S.; Rashid, A.R. Applications of high-throughput phenotypic phenomics. In Sustainable Agriculture in the Era of the OMICs Revolution; Springer: Berlin/Heidelberg, Germany, 2023; pp. 119–134. [Google Scholar]
- Basavaraj, P.; Rane, J.; Prathibha, M.; Boraiah, K.; Kumar, M. Advances in High-Throughput Phenotyping of Maize (Zea Mays L.) for Climate Resilience. In Maize Improvement: Current Advances in Yield, Quality, and Stress Tolerance under Changing Climatic Scenarios; Springer: Berlin/Heidelberg, Germany, 2023; pp. 259–288. [Google Scholar]
- Gehan, M.A.; Fahlgren, N.; Abbasi, A.; Berry, J.C.; Callen, S.T.; Chavez, L.; Doust, A.N.; Feldman, M.J.; Gilbert, K.B.; Hodge, J.G.; et al. PlantCV v2: Image analysis software for high-throughput plant phenotyping. PeerJ 2017, 5, e4088. [Google Scholar] [CrossRef]
- Kim, J.; Chung, Y.S. A short review of RGB sensor applications for accessible high-throughput phenotyping. J. Crop Sci. Biotechnol. 2021, 24, 1–5. [Google Scholar] [CrossRef]
- Laflamme, B.; Middleton, M.; Lo, T.; Desveaux, D.; Guttman, D.S. Image-based quantification of plant immunity and disease. Mol. Plant Microbe Interact. 2016, 29, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Pavicic, M.; Mouhu, K.; Hautsalo, J.; Jacobson, D.; Jalli, M.; Himanen, K. Image-based time series analysis to establish differential disease progression for two Fusarium head blight pathogens in oat spikelets with variable resistance. Front. Plant Sci. 2023, 14, 1126717. [Google Scholar] [CrossRef] [PubMed]
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-throughput field-phenotyping tools for plant breeding and precision agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Lee, U.; Silva, R.R.; Kim, C.; Kim, H.; Heo, S.; Park, I.S.; Kim, W.; Jansky, S.; Chung, Y.S. Image analysis for measuring disease symptom to bacterial soft rot in potato. Am. J. Potato Res. 2019, 96, 303–313. [Google Scholar] [CrossRef]
- Leiva, F.; Abdelghafour, F.; Alsheikh, M.; Nagy, N.E.; Davik, J.; Chawade, A. ScabyNet, a user-friendly application for detecting common scab in potato tubers using deep learning and morphological traits. Sci. Rep. 2024, 14, 1277. [Google Scholar] [CrossRef]
- Leiva, F.; Vallenback, P.; Ekblad, T.; Johansson, E.; Chawade, A. Phenocave: An Automated, Standalone, and Affordable Phenotyping System for Controlled Growth Conditions. Plants 2021, 10, 1817. [Google Scholar] [CrossRef]
- Wright, H.C.; Lawrence, F.A.; Ryan, A.J.; Cameron, D.D. Free and open-source software for object detection, size, and colour determination for use in plant phenotyping. Plant Methods 2023, 19, 126. [Google Scholar] [CrossRef] [PubMed]
- Cgrain_AB. Cgrain Value TM, The New Standard for Analysis Grain Quality. Available online: https://www.cgrain.se (accessed on 5 June 2023).
- Alexandersson, E.; Keinänen, M.; Chawade, A.; Himanen, K. Nordic Research Infrastructures for Plant Phenotyping. Agric. Food Sci. 2018, 27, 7–16. [Google Scholar] [CrossRef][Green Version]
- Rathore, R.; Ahamad, S.; Soni, A.; Dewangan, A.K.; Jain, A. Agriculture Field Area Calculation Using Drone Camera: Method and Framework Design. SN Comput. Sci. 2024, 5, 267. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Garcia, S.M.; Ryckewaert, M.; Abdelghafour, F.; Metz, M.; Moura, D.; Feilhes, C.; Prezman, F.; Bendoula, R. Combination of multivariate curve resolution with factorial discriminant analysis for the detection of grapevine diseases using hyperspectral imaging. A case study: Flavescence dorée. Analyst 2021, 146, 7730–7739. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Seung, H.S. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef]
- Técher, D. Using superpixel-or pixel-based segmentation for efficient green roof digital image classification and rapid estimation of plant species cover. Urban For. Urban Green. 2022, 76, 127722. [Google Scholar] [CrossRef]
- Sekulska-Nalewajko, J.; Gocławski, J.; Chojak-Koźniewska, J.; Kuźniak, E. Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 2016, 109, 114–122. [Google Scholar] [CrossRef]
- Salum, P.; Güven, O.; Aydemir, L.Y.; Erbay, Z. Microscopy-assisted digital image analysis with Trainable Weka Segmentation (TWS) for emulsion droplet size determination. Coatings 2022, 12, 364. [Google Scholar] [CrossRef]
- Varga, T.; McKinney, A.L.; Bingham, E.; Handakumbura, P.P.; Jansson, C. Rapid phenotyping of root systems of Brachypodium plants using X-ray Computed Tomography: A comparative study of soil types and segmentation tools. In Proceedings of the AGU Fall Meeting Abstracts. (American Geophysical Union, 2017), New Orleans, LA, USA, 15 November 2017. [Google Scholar]
- Kendler, S.; Aharoni, R.; Young, S.; Sela, H.; Kis-Papo, T.; Fahima, T.; Fishbain, B. Detection of crop diseases using enhanced variability imagery data and convolutional neural networks. Comput. Electron. Agric. 2022, 193, 106732. [Google Scholar] [CrossRef]
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. 2014. Available online: https://cran.r-project.org/web/packages/agricolae/agricolae.pdf (accessed on 3 April 2023).
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; Team, R.C. Package ‘caret’. R J. 2020, 223, 7. [Google Scholar]
- Cherif, M.; Rezgui, S.; Devaux, P.; Harrabi, M. Genotype × environment interactions and heritability of quantitative resistance to net blotch in Tunisian barley. J. Plant Breed. Crop Sci. 2010, 2, 110–116. [Google Scholar]
| Cultivar | No. Inoculated Leaves | Percentage Infected |
|---|---|---|
| Firefoxx | 23 | 43.5% |
| Flair | 23 | 69.6% |
| Laureate | 19 | 52.6% |
| Reference | |||||||
| Background | Healthy | Necrotic | Chlorotic | Fungal | Leaf tip necrosis | ||
| Prediction | Background | 3,797,616 | 125 | 19 | 0 | 21 | 159 |
| Healthy | 468 | 1,988,363 | 37 | 59 | 318 | 55 | |
| Necrotic | 27 | 870 | 91,082 | 73 | 958 | 470 | |
| Chlorotic | 36 | 225 | 36 | 66,011 | 40 | 162 | |
| Fungal | 251 | 2319 | 217 | 45 | 300,801 | 147 | |
| Leaf tip necrosis | 1971 | 400 | 16 | 29 | 184 | 223,780 | |
| Overall accuracy | 0.999 | 0.998 | 0.996 | 0.997 | 0.995 | 0.996 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leiva, F.; Dhakal, R.; Himanen, K.; Ortiz, R.; Chawade, A. The Combination of Low-Cost, Red–Green–Blue (RGB) Image Analysis and Machine Learning to Screen for Barley Plant Resistance to Net Blotch. Plants 2024, 13, 1039. https://doi.org/10.3390/plants13071039
Leiva F, Dhakal R, Himanen K, Ortiz R, Chawade A. The Combination of Low-Cost, Red–Green–Blue (RGB) Image Analysis and Machine Learning to Screen for Barley Plant Resistance to Net Blotch. Plants. 2024; 13(7):1039. https://doi.org/10.3390/plants13071039
Chicago/Turabian StyleLeiva, Fernanda, Rishap Dhakal, Kristiina Himanen, Rodomiro Ortiz, and Aakash Chawade. 2024. "The Combination of Low-Cost, Red–Green–Blue (RGB) Image Analysis and Machine Learning to Screen for Barley Plant Resistance to Net Blotch" Plants 13, no. 7: 1039. https://doi.org/10.3390/plants13071039
APA StyleLeiva, F., Dhakal, R., Himanen, K., Ortiz, R., & Chawade, A. (2024). The Combination of Low-Cost, Red–Green–Blue (RGB) Image Analysis and Machine Learning to Screen for Barley Plant Resistance to Net Blotch. Plants, 13(7), 1039. https://doi.org/10.3390/plants13071039









